Abstract
Prostate cancer is one of the major health care problems, but the molecular pathogenesis has been relatively insufficiently elucidated. Recently, whole exome sequencing of prostate cancer identified recurrent mutations involving MED12 in Caucasian patients, which finding was not reproduced in one subsequent study by Sanger sequencing. Thus, we investigated mutation status of MED12 in exons 2 and 26 by Sanger sequencing in 102 radical prostatectomy cases from Korean patients. The analysis found the mutation in none of the cases. Therefore, MED12 mutation does not appear to represent a significant molecular alteration in this cohort of patients according to the analysis by the traditional “gold standard.”
Keywords: mediator complex subunit 12, mutation analysis, prostate cancer, prostate neoplasms
INTRODUCTION
Prostate cancer (PCa) represents the second most frequently diagnosed new cancer in men worldwide1,2 and recently has been rapidly increasing in incidence in the Far East Asian countries including Korea.3,4,5 Molecular pathogenesis of PCa has been relatively, insufficiently elucidated compared to other major cancers and it has been suggested that substantial difference in molecular alteration is present between different ethnic groups. For example, the incidence of TMPRSS2-ETS family gene fusion or PTEN inactivation has been reported to be lower in the Asian population.6,7
Recently, exome sequencing has discovered novel somatic mutations of considerable frequencies in PCa,8 which is a remarkable discovery considering that a recurrent mutation is a rare event in PCa. One of the frequently mutated genes was Mediator complex subunit 12 (MED12) gene located on Xq13.1 which encodes a subunit of a multi-protein complex called Mediator.9 The mediator plays an essential role in the transcription machinery and is composed of a set of distinct modules. MED12 participates in the formation of CDK8 kinase module of the mediator complex thereby engaging in the nuclear transduction of signals from several different oncogenic pathways10,11 and it was suggested that MED12 mutation may indirectly affect p53 or androgen signaling.8
However, one subsequent study failed to find the evidence for a frequent mutation in a Caucasian population, possibly due to the difference in the composition of the study cohort.12 Thus, in this study, we investigated the mutation status of MED12 in 102 radical prostatectomy specimen from the Korean men to explore its relevance in this distinct ethnic group testing the possibility that MED12 may represent another point of molecular difference between the Caucasian and Asian population.
MATERIALS AND METHODS
Clinicopathologic evaluation
One hundred and two cases of prostatic adenocarcinoma were selected for this study. All patients included underwent radical prostatectomy at Samsung Medical Center in Seoul, Korea between 1995 and 2006. Clinical data were retrieved through electronic medical records, and all relevant pathological features were separately reviewed by two pathologists (N Yoon and GY Kwon). Cases with incomplete resection or neoadjuvant treatment (radiotherapy or hormonal therapy) were excluded. The modified 2005 International Society of Urological Pathology criteria were utilized for Gleason grading. Tumor stage was determined according to the 7th Edition of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual. There were 75 cases (73.5%) of organ-confined tumors and 27 cases (26.5%) of nonorgan-confined ones.
Clinicopathological data including age (mean: 65; range: 44–77), Gleason score and pathologic stage are summarized in Table 1. The study protocol was approved by the Samsung Medical Center Institutional Review Board (2015-10-012-01). Informed consents were waived.
Table 1.
Clinicopathologic parameters of the 102 patients

Extraction of DNA
Two unstained 5-μm-thick sections were obtained from the representative blocks of formalin-fixed, paraffin-embedded (FFPE) samples. Original H and E slides were reviewed to identify the region of tumor and tumor tissues were macrodissected from corresponding areas of unstained slides. Additional H and E slides were prepared after obtaining the slides for DNA extraction and reviewed to confirm the area of tumors. Genomic DNA was isolated from FFPE tumor samples using a ReliaPrep™ FFPE gDNA extraction kit (Promega, Madison, WI, USA). The concentration of DNA was determined by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Sequencing of MED12 exons 2 and 26
Polymerase chain reaction (PCR) was performed in a total volume of 20 μl mixture containing 100 ng of template DNA, 1X PCR reaction buffer, 2.5 mM each of deoxynucleoside triphosphates (dNTPs), 10 μM of primers, and 2.5 U Taq DNA polymerase. Primers were designed to detect all known hot spots12,13 using Primer3 Plus software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) set to melting temperature of 60°C. Primer pairs were used to amplify the complete coding sequences of MED12 exons 2 and 26 (Table 2. The thermal cycling was 5 min at 95°C, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and an extension at 72°C for 30 s. PCR products were subjected to electrophoresis on 2% agarose gels and were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany).
Table 2.
MED12 Primer Sequences and PCR conditions
Sanger sequencing was performed with the same forward and reverse primers as used in PCR amplification. Reaction mixtures were composed of 4 μl of 2.5X Ready Reaction mix, 2 μl of 5X BigDye sequencing buffer, 2 μl of template (purified PCR product), and 10 μM of primer in a final volume made up to 20 μl with deioinized water. The thermal cycling was 1 min at 96°C for 1 cycle followed by 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. The PCR sequence products were purified using the BigDye Terminator PCR Purification Kit protocol (Qiagen) according to the manufacturer's instructions.14 The BigDye Terminator v1.1 kit (Applied Biosystems, Waltham, MA, USA) and the ABI 3130XL genetic analyzer (Applied Biosystems) were utilized for bidirectional sequencing. Sequence analysis was performed with sequencher version 4.10.1 (Gene Codes Corporation, Ann Arbor, MI, USA) and manual review of the chromatograms. Ambiguous sequences were subjected to repeated experiment with replicate PCR amplification reactions, and any sequences that were deviated from wild type were also re-sequenced. A case was considered as positive for mutation if abnormal sequences were detected on both the forward and reverse DNA strand.
Limit of detection (LOD) test for MED12 mutation
LOD test was performed to establish the threshold of tumor content for the detection of MED12 mutation. FFPE-tumor tissue from phyllodes tumor harboring heterozygous MED12 G44D mutation at exon 2 was diluted with varying amount of nonneoplastic FFPE-breast tissue to final tumor concentrations of 0%, 5%, 10%, 20%, 50%, and 100%. The mixtures were subjected to PCR amplification and Sanger sequencing in the same method as the test cases.
RESULTS
The average size of tumor on re-examination of recut slides after DNA extraction was 1.4 cm (range from 0.5 to 3 cm) in the largest dimension. After microdissection, the tumor content relative to the intervening stroma was estimated to be approximately 70% on average. The concentration of extracted DNA quantified by a spectrophotometer was 50 ng μl−1 on average. PCR product from all cases demonstrated clear bands at 244 bp for exon 2 and 202 bp for exon 26 on 2% agarose gels electrophoresis confirming that PCR was successfully performed.
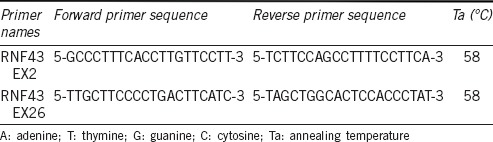
Six cases (three cases in exons 2 and three cases in exon 26) displayed irregular baseline with low double peaks in the sequence signal and thus were considered inadequate for interpretation. Microscopic slides for those cases were re-examined and revealed the adequate size of the tumors, five cases of which measured longer than 1.0 cm in the largest dimension. Repeat test was performed for those cases and showed clear sequence signals without any noise. Consequently, all cases were regarded as successfully sequenced and mutation of MED12 in either exon 2 or 26 was not found in any case of PCa (Figure 1a).
Figure 1.
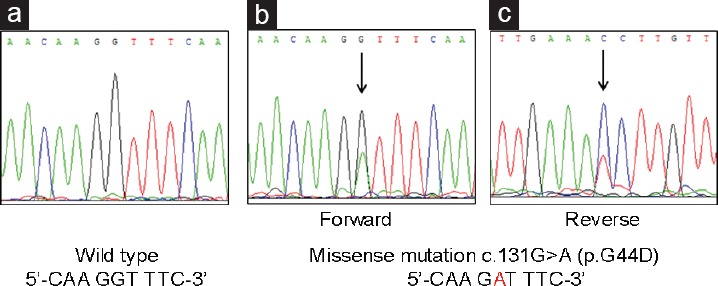
Representative sequencing results of MED12 in exon 2. All cases of prostatic adenocarcinoma showed wild type sequence (a), while considerable proportion of phyllodes tumors had missense mutation in exon 2, detected in both forward (b) and reverse strand (c).
In the limit of detection test using phyllodes tumor of the breast, mutation of MED12 at exon 2 was detectable from the tumor concentration of 10% relative to nonneoplastic breast tissue with wild-type MED12 (Figure 2).
Figure 2.
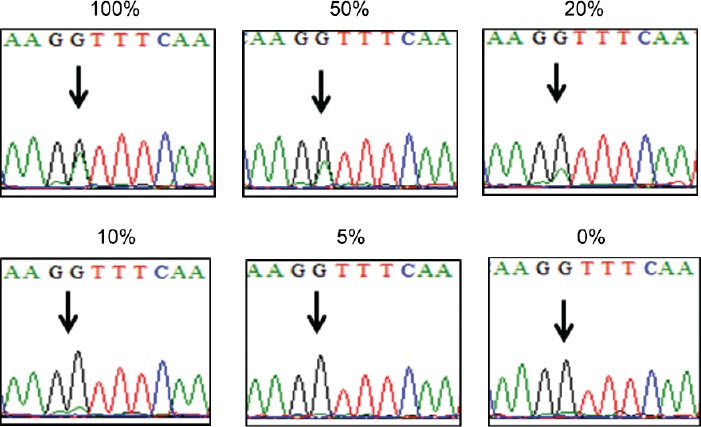
Limit of detection (LOD) test for MED12 mutation in exon 2. MED12 mutation was detectable from the concentration of 10% of mutation-positive phyllodes tumor diluted with nonneoplastic breast tissue. Percentage above each figure denotes the concentration of tumor relative to the nonneoplastic tissue with wild type MED12.
DISCUSSION
In this study, mutation of MED12 in exon 26 was not identified in any of the 102 cases of radical prostatectomy. This finding is in accordance with the results from the study by Stoehr et al.12 while in contrast to those from the work by Barbieri et al.8 and, therefore, requires careful interpretation.
First, also considering that the data from The Cancer Genome Atlas Project identified the presence of the same mutation through whole exome sequencing,15 MED12 mutation appears to be a recurrent finding in at least a certain group of PCa patients. The explanations for the failure to detect that mutation in our study and the studies by Stoehr et al. can be sought either in technical aspect or in population characteristics of the study cohorts. In the technical aspect, the methods employed by the researchers were not the same; the original work by Barbieri et al. and the Cancer Genome Atlas Project were conducted through exome sequencing which is generally perceived as more sensitive than Sanger sequencing performed in the study by Stoehr et al. and ours.
However, Stoehr et al. believe that they could exclude technical failure in their study as the reason that they could not detect the mutation, and we also share their confidence. In our study, PCR amplification was successfully performed in every case yielding PCR product of optimal size (244 bp for exon 2 and 202 bp for exon 26) and cases with ambiguous reading on sequencing was resolved on repeated experiment. Furthermore, we carefully reviewed the H and E slides, which were prepared directly after the recut for the DNA extraction and confirmed the adequate microdissection.
One drawback of our study was that it was performed exclusively with FFPE tissue and it is a well-established fact that formalin-fixation and/or prolonged storage can elicit damage to nucleic acids.16,17,18 However, DNA damage in such situation usually manifests as DNA fragmentation or sequence artifacts19 and we believe that utilization of FFPE did not confer considerable limitation on our results considering that the sequencing was successfully performed in every case. Moreover, it is recently reported that MED12 mutation in exon 2 is frequently found in fibroadenoma of breast13 and we could demonstrate the presence of a mutation in a considerable proportion of breast tumors in FFPE tissue of a comparable period of storage in our department (Figure 1b and 1c).
Sanger sequencing is usually considered as having a threshold of detection at 20% of a mutant allele relative to the wild type and tumor cell counts of 300–500 cells.20,21 According to our results of Limit of Detection (LOD) study, the threshold for detection is estimated at approximately 10% for MED12 mutation in our laboratory. Also considering that our cases had an adequate tumor in terms of total size and relative area as compared to stromal tissue, we believe that MED12 mutation is absent in PCa of our study cohort as analyzed by this traditional “gold standard.”
MED12 mutation in exon 2 reported in stromal component of fibroadenoma13 and uterine smooth muscle tumors22,23 was also explored in this study, but we could not detect the mutation in that exon, either.
In view of the population characteristics, our study cohort represents a fairly homogenous group of patients consisting exclusively of Korean male since in Korea, the patients are mostly domestic and occasional foreigners, especially non-Asian, can be readily identified by the letters for their names (Korean alphabet is used for Korean patients). This genetic composition of study cohort is a considerable advantage in the study of diseases which display remarkable ethnic variation such as PCa, although it can also be a limitation which precludes addressing the question in its entirety. PCa is the most common male cancer in predominantly Western developed countries while relatively uncommon in Asian region24,25 and recently increasing in incidence in Far East Asian countries, possibly due to combined effect of the introduction of PSA screening and westernization of the lifestyle.2,4,5 In aforementioned two studies of MED12, the patients appear to be mostly Caucasian and the study cohort of Stoehr et al. is considered more homogenous. However, ethnic background in those studies is assumed from the region of the residence and not clearly documented.
It has been pointed out that there may exist a difference in the molecular pathways of PCa between the Asian and Caucasian population. The incidence of TMPRSS2-ERG gene fusion has been reported to be lower in Korean,26,27 Japanese,6 and Chinese7 than in Caucasian population and PTEN inactivation has been less frequently detected in Chinese patients.7 The rate of SPOP mutation was similar26 or slightly lower28 in Korean population and the studies on KRAS or BRAF mutation have yielded inconsistent results regarding ethnic difference.29,30,31
Thus, it appears worthwhile to explore the presence of MED12 mutation in PCa from Korean patients, and we hypothesized that MED12 mutation may constitute one of the molecular alterations which are more frequent in PCa of Asian population considering that MED12-mediated pathway may affect androgen receptor signaling8,32 which is heavily dependent on ethnicity.33 The results of our study are not supportive of this hypothesis, and it remains to be unveiled what molecular mechanism is more actively operating in PCa of Korean and Asian patients than in PCa of Western patients.
AUTHOR CONTRIBUTIONS
NY participated in the experiment, drafting, and revision of the manuscript. SL drafted the manuscript. SYK performed the experiment and participated in the revision. GYK conceived of the study and coordinated the manuscript and the revision. HGJ and BCJ participated in the financing of the study and retrieval of clinical data. SIS, SSJ, HML, and HYC contributed to the collection of data and refinement of the manuscript in clinical aspect. All authors have read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
This work was supported by a grant from the Korean Foundation for Cancer Research (CB-2011-04-01).
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Daniyal M, Siddiqui ZA, Akram M, Asif HM, Sultana S, et al. Epidemiology, etiology, diagnosis and treatment of prostate cancer. Asian Pac J Cancer Prev. 2014;15:9575–8. doi: 10.7314/apjcp.2014.15.22.9575. [DOI] [PubMed] [Google Scholar]
- 3.Nath A, Singh JK, Vendan SE, Priyanka, Sinha S. Elevated level of prostate specific antigen among prostate cancer patients and high prevalence in the Gangetic zone of Bihar, India. Asian Pac J Cancer Prev. 2012;13:221–3. doi: 10.7314/apjcp.2012.13.1.221. [DOI] [PubMed] [Google Scholar]
- 4.Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, et al. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–92. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 5.Jung KW, Park S, Kong HJ, Won YJ, Lee JY, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012;44:11–24. doi: 10.4143/crt.2012.44.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyagi Y, Sasaki T, Fujinami K, Sano J, Senga Y, et al. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010;23:1492–8. doi: 10.1038/modpathol.2010.149. [DOI] [PubMed] [Google Scholar]
- 7.Mao X, Yu Y, Boyd LK, Ren G, Lin D, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70:5207–12. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampjarvi K, Makinen N, Kilpivaara O, Arola J, Heinonen HR, et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107:1761–5. doi: 10.1038/bjc.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turunen M, Spaeth JM, Keskitalo S, Park MJ, Kivioja T, et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014;7:654–60. doi: 10.1016/j.celrep.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–22. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoehr R, Taubert H, Gaisa NT, Smeets D, Kneitz B, et al. Lack of evidence for frequent MED12 p. L1224F mutation in prostate tumours from Caucasian patients. J Pathol. 2013;230:453–6. doi: 10.1002/path.4208. [DOI] [PubMed] [Google Scholar]
- 13.Lim WK, Ong CK, Tan J, Thike AA, Ng CC, et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46:877–80. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 14.Heikens E, Fleer A, Paauw A, Florijn A, Fluit A. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol. 2005;43:2286–90. doi: 10.1128/JCM.43.5.2286-2290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genomics Hub (CGHub). Sample UUIDs: 8d91b55d-0a51-44d3-bcc7-d6d95027683a, 787d4d9f-1c69-406f-8e1c-2458d77085e7, 986964bc-b680-4f c1-8f93-20772f0a8949, 104e14d3-f1d5-443f-88fd-6d67c98b1eff. Santa Cruz, CA: University of California, Santa Cruz; [Last cited on 2014 Mar 09]. Available from: https://www.cghub.ucsc.edu . [Google Scholar]
- 16.Inoue T, Nabeshima K, Kataoka H, Koono M. Feasibility of archival non-buffered formalin-fixed and paraffin-embedded tissues for PCR amplification: an analysis of resected gastric carcinoma. Pathol Int. 1996;46:997–1004. doi: 10.1111/j.1440-1827.1996.tb03580.x. [DOI] [PubMed] [Google Scholar]
- 17.Gallegos Ruiz MI, Floor K, Rijmen F, Grunberg K, Rodriguez JA, et al. EGFR and K-ras mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. 2007;29:257–64. doi: 10.1155/2007/568205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solassol J, Ramos J, Crapez E, Saifi M, Mange A, et al. KRAS mutation detection in paired frozen and Formalin-Fixed Paraffin-Embedded (FFPE) colorectal cancer tissues. Int J Mol Sci. 2011;12:3191–204. doi: 10.3390/ijms12053191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 20.Nikiforov YE, Biddinger PW, Thompson LD. Diagnostic Surgical Pathology and Molecular Genetics of the Thyroid. Baltimore, MD: Lippincott William and Wilkins; 2009. p. 413. [Google Scholar]
- 21.Kulkarni S. Clinical Genomics. Amsterdam: Academic Press; 2014. p. 7. [Google Scholar]
- 22.Halder SK, Laknaur A, Miller J, Layman LC, Diamond M, et al. Novel MED12 gene somatic mutations in women from the Southern United States with symptomatic uterine fibroids. Mol Genet Genomics. 2015;290:505–11. doi: 10.1007/s00438-014-0938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwetye KE, Pfeifer JD, Duncavage EJ. MED12 exon 2 mutations in uterine and extrauterine smooth muscle tumors. Hum Pathol. 2014;45:65–70. doi: 10.1016/j.humpath.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–64. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 25.Sim HG, Cheng CW. Changing demography of prostate cancer in Asia. Eur J Cancer. 2005;41:834–45. doi: 10.1016/j.ejca.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Blattner M, Lee DJ, O’Reilly C, Park K, MacDonald TY, et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia. 2014;16:14–20. doi: 10.1593/neo.131704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K, Chae JY, Kwak C, Ku JH, Moon KC. TMPRSS2-ERG gene fusion and clinicopathologic characteristics of Korean prostate cancer patients. Urology. 2010;76:1268.e7–13. doi: 10.1016/j.urology.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. Apmis. 2013;121:626–33. doi: 10.1111/apm.12030. [DOI] [PubMed] [Google Scholar]
- 29.Zong Y, Goldstein AS, Huang J. The molecular basis for ethnic variation and histological subtype differences in prostate cancer. Sci China Life Sci. 2013;56:780–7. doi: 10.1007/s11427-013-4522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu M, Zhang W, Shan L, Song J, Shang D, et al. Mutation status of somatic EGFR and KRAS genes in Chinese patients with prostate cancer (PCa) Virchows Arch. 2014;464:575–81. doi: 10.1007/s00428-014-1566-x. [DOI] [PubMed] [Google Scholar]
- 31.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, et al. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosome Cancer. 2012;51:1014–23. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 32.Shaikhibrahim Z, Offermann A, Braun M, Menon R, Syring I, et al. MED12 overexpression is a frequent event in castration-resistant prostate cancer. Endocr Relat Cancer. 2014;21:663–75. doi: 10.1530/ERC-14-0171. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman CM, Lowe LP, Lee H, Hayes MG, Dyer AR, et al. Ethnic variation in allele distribution of the androgen receptor (AR) (CAG) n repeat. J Androl. 2012;33:210–5. doi: 10.2164/jandrol.111.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]


