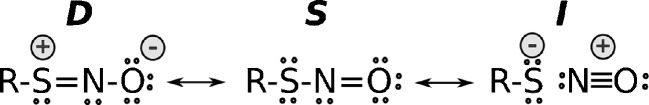Abstract
Thiol redox chemical reactions play a key role in a variety of physiological processes, mainly due to the presence of low-molecular-weight thiols and cysteine residues in proteins involved in catalysis and regulation. Specifically, the subtle sensitivity of thiol reactivity to the environment makes the use of simulation techniques extremely valuable for obtaining microscopic insights. In this work we review the application of classical and quantum–mechanical atomistic simulation tools to the investigation of selected relevant issues in thiol redox biochemistry, such as investigations on (1) the protonation state of cysteine in protein, (2) two-electron oxidation of thiols by hydroperoxides, chloramines, and hypochlorous acid, (3) mechanistic and kinetics aspects of the de novo formation of disulfide bonds and thiol−disulfide exchange, (4) formation of sulfenamides, (5) formation of nitrosothiols and transnitrosation reactions, and (6) one-electron oxidation pathways.
Keywords: Thiols, Oxidation, Redox homeostasis, Computer simulations
Introduction
Thiol biochemistry is mainly related to cysteine (Cys), which is one of the least abundant amino acids incorporated into proteins. However, due to its peculiar physico-chemical properties, Cys reactivity is unique. Specifically, the large size of the sulfur atom and the relatively low dissociation energy of the S−H bond render Cys with the ability to perform both nucleophilic and redox-active functions that are unfeasible for the other naturally occurring amino acids. Most commonly, the acid dissociation constant (pKa) values of thiol groups in Cys residues are relatively close to the physiological pH, and the ionization state of Cys is therefore highly sensitive to the environment. The thiol ionization state governs Cys nucleophilicity and redox susceptibility, thereby facilitating the unique functions of Cys: nucleophilic and redox catalysis, allosteric regulation, site of posttranslational modification, metal coordination, and structural stabilization (Fomenko et al. 2008). Additionally, the reactivity of specific Cys residues can be largely affected by protein environment, increasing thiol reactivity in specific reactions. For example, the reduction of hydroperoxides by peroxidatic thiols in Cys-based peroxidases is increased by factors of 103–107 compared with free Cys (Ferrer-Sueta et al. 2011). Similarly, thiol–disulfide exchange reactions are catalyzed by specific Cys-based oxidoreductases (Jensen et al. 2009). Protein factors affecting Cys reactivity are only starting to be unraveled, and computer simulations provide important tools to the understanding of the molecular basis of catalysis.
Computational techniques for modeling biochemical problems have emerged during the last decades as an important tool to complement experimental information (Dror et al. 2012). The in silico-generated models and the information which can be obtained throughout their study have been shown to be very useful for analyzing the structural, spectroscopic, and kinetic data provided by the experimental methods. In particular, computer simulations are a systematic and economical tool that allow the analysis of the dependence of a property of interest on static (e.g., amino acid sequence) and dynamical factors (Dror et al. 2012). In recent years, the increase in the computing power and in the accuracy of the models made possible in many cases to draw biologically relevant conclusions and propose new hypotheses based mainly on simulation data (van der Kamp and Mulholland 2013). In this context, redox biochemistry of thiols, a complex research area in which relevant reactions may occur in both solution and protein environments, constitutes an ideal benchmark for many computational techniques.
The modeling of phenomena which do not involve the formation or breaking of chemical bonds may be achieved, in principle, without resorting to quantum mechanics (QM), by employing classical force fields (even though such force fields often contain parameters based on QM calculations). Among the most widely used force fields for biomolecules are AMBER (Cornell et al. 1995), CHARMM (MacKerell et al. 2004), and GROMOS (Schmid et al. 2011). The time scale accessible in atomistic simulations with modern hardware technology is limited to the microsecond range, yet the predictive power of simulation schemes has increased enormously in the last years by the development of advanced sampling techniques, such as accelerated dynamics (Hamelberg et al. 2004), replica exchange (Zhang and Chen 2013), metadynamics (Laio and Parrinello 2002a), and multiple steered molecular dynamics (MD) (Park et al. 2003).
Classical simulation techniques are not able to deal with reactive processes where covalent chemical bonds are being formed or broken, in which cases it is necessary to employ QM schemes. There are two main strategies to tackle chemical processes in biochemistry. One is based on electronic structure calculations using appropriate model systems of moderate size, typically including the active site plus eventually the most relevant region of the environment, and/or adding a continuum representation of the environment. The second strategy is to employ hybrid QM–molecular mechanical (QM-MM) methods, which were introduced in the 1970s by Warshel and Levitt (1976). These are adequate for the investigation of chemical events that take place in a limited region of a large system (the QM region), to be modeled using QM at a certain level of theory, which may range from simplified valence bond schemes, to semiempirical methods, to Hartree–Fock and density functional theory (DFT) (Laio et al. 2002b; Crespo et al. 2003, 2005; Senn and Thiel 2009). The remainder of the system (MM region) is treated at the less expensive molecular mechanics, or classical level. The use of this kind of technique for the investigation of chemical reactivity in solution and in enzymes has become very popular during the last years (Lebrero et al. 2005; Crespo et al. 2006; van der Kamp and Mulholland 2013).
In this work we discuss results on classical and QM computer simulations of selected processes related to thiol redox biochemistry (Fig. 1). In section 1, we review investigations on the protonation state of Cys in protein environments, and in section 2 we examine and discuss the two-electron oxidation of thiols by hydroperoxides, chloramines, and hypochlorous acid. We discuss mechanistic and kinetics aspects of the de novo formation of disulfide bonds and thiol−disulfide exchange in section 3. Section 4 is devoted to the formation of sulfenamides, in section 5 we present results on the formation of nitrosothiols, and transnitrosation reactions, and, finally, in section 6 we focus on a number of one-electron oxidation pathways.
Fig. 1.
Most relevant thiol redox reaction pathways. Different routes of oxidation of thiol/thiolate are shown in a schematic way. Thiol acid–base behavior is presented in reaction (I). Reaction (II) shows the one-electron oxidation of thiolates to thiyl radical. Thiolates can be oxidized by hydroperoxides, hypochlorous acid, and chloramines in two-electron oxidation processes that yield the corresponding sulfenic acid (III), which in turn can be further oxidized to sulfinic and sulfonic acids if oxidant is in excess (IV). Inter- and intramolecular disulfides are de-novo formed from the reaction of a sulfenic acid with other thiol (V) and, subsequently, thiol−disulfide exchanges can occur (VI). Protein or protein-like thiolates can also rearrange to a cyclic product called sulfenamide that involves the reaction of the sulfur atom with the backbone’s NH moiety of the preceding residue in the sequence (VII). Thiyl radicals can react with molecular oxygen/superoxide or other thiol, yielding sulfinic acid and disulfides as final products, respectively (reactions VIII and IX). Nitrosothiols can be produced both by radical and non-radical pathways: both thiol–thiolate oxidation by reactive nitrogen species such as dinitrogen trioxide, or thiyl radical recombination with nitric oxide are called S-nitrosation and S-nitrosylation (reactions X and XI). Nitrosothiols can react with other thiols, exchanging the nitroso group in the so-called trans-S-nitrosations (XII) or releasing HNO to form a new disulfide (XIII)
Section 1: pKa of thiol
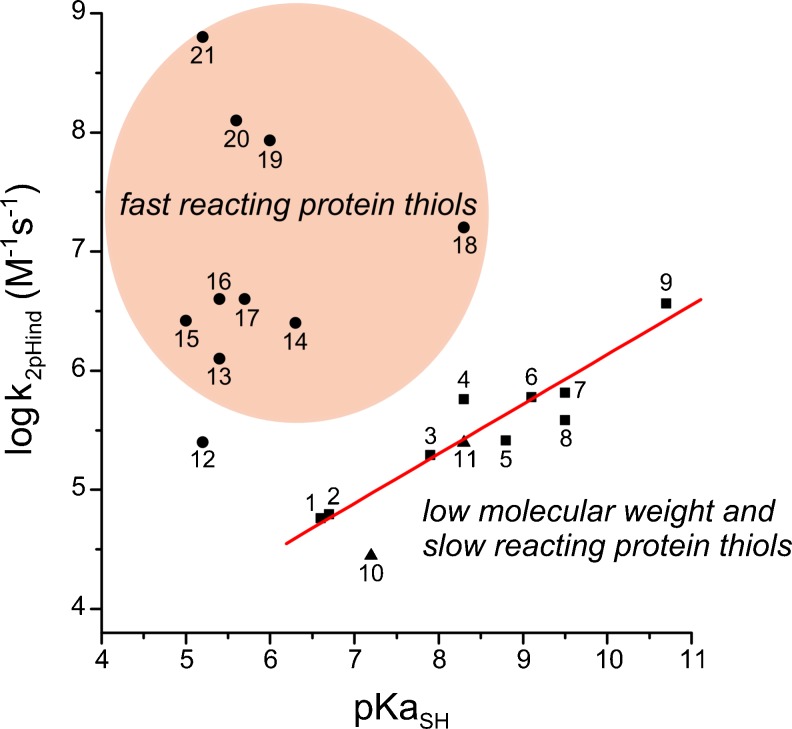
Many enzymes depend upon redox-active Cys. Since thiolates are much better nucleophiles than the neutral form of thiols (Winterbourn and Metodiewa 1999), their pKas are an essential factor for the understanding of their reactivity. While pH-dependent reactivities involving thiolates as nucleophiles at close to physiological pH are usually higher for those thiols with the lower thiol pKa values, due to a higher thiolate availability, pH-independent reactivities (intrinsic reactivity of thiolates) show the opposite trend (Gilbert 1990; Trujillo et al. 2007). Indeed, for low-molecular-weight thiols and a great number of protein Cys residues, the pKa value is a useful marker to predict thiolate reactivity, with low-molecular-weight thiols being more reactive than those with higher pKas (as shown for peroxynitrite in Fig. 2). However, it must be taken into account that in specific reactions, the Brønsted coefficient fails in this prediction for some protein thiols, the so-called “fast reacting protein thiols”, as shown in Fig. 2 (Trujillo et al. 2007).
Fig. 2.
Intrinsic thiolate reactivities with peroxynitrite (k 2pHind) as a function of thiol pKa (pKa SH). Low-molecular-weight thiols (filled squares 1–6) show a positive Brønsted correlation, as indicated by the solid red line, consistent with the thiols with a higher pKa being better nucleophiles. Some protein thiols (filled triangles 10–11) react with peroxynitrite, as anticipated according to their thiol pKa. Other protein thiols (filled circles 12–21), react much faster than expected, indicating that protein factors other than thiol pKa are determining this reactivity. 1 L-cysteine (Cys) ethyl ester, 2 Cys methyl ester, 3 penicillamine, 4 Cys, 5 glutathione, 6 mercapto ethyl guanidine, 7 homocysteine, 8 N-acetyl Cys, 9 dihidrolipoic acid, 10 Trypanosoma brucei tryparedoxin, 11 human serum albumin, 12 human arylamine N-acetyltransferase 1, 13 DJ1, 14 TSA2, 15 Mycobacterium tuberculosis AhpC, 16 creatinine kinase, 17 TSA1, 18 GAPDH, 19 red blood cell peroxiredoxin 2 (Prx2), 20 protein-tyrosine phosphatase 1B (PTP1B), 21 human Prx5. Modified from Trujillo et al. (2007) and Ferrer-Sueta et al. (2011)
Factors that control Cys pKa in proteins
Factors that affect the stability of charged species play a crucial role in pKa shifts. Therefore, aqueous solvation, H-bonds, charges of nearby residues, and helix dipole effects may influence pKa values (Roos et al. 2013a). Cys residues are very sensitive to their environment; consequently, a wide range of pKaSH values has been measured/calculated. For example, in different proteins of the thioredoxin superfamily, pKaSH in the range 3.5–8 range have been found (see references in Table 1).
Table 1.
Predicted pKa of L-cysteine residues in different proteins by different computational methodsa
| Protein | Protein Data Bank ID | L-cysteine residue number | Experimental | FDPB (electrostatic classical approach) | PROPKA3 (minimized structure) (empirical approach) | Quantum mechanical calculations |
|---|---|---|---|---|---|---|
| Escherichia coli Trx1 | 1xob | 32 | 7.1b | 7.6 | 6.6d | 6.5d |
| Human glutharedoxin | 1jhb | 22 | 3.5c | 5.5–7.2c | ||
| Human serum albumin | 1n5u | 34 | 5.0d | 7.7 | ||
| Oryctolagus cuniculus glyceraldehide-3-phosphate-dehydrogenase | 1j0x | 149 | 5.5d | 4.2 | ||
| Bacillus subtilis thiol-disulfide oxidoreductase resA | 1su9 | 76 | 8.2d | 10.0d | 8.1d | |
| Staphylococcus aureus thiorredoxin | 2o89 | 29 | 6.4d | 4.0d | 6.5d |
The role of charged side-chains and long-range electrostatics seems to be important (Lim et al. 2012). However, despite some exceptions, computational and experimental results indicate that the surrounding charged chains are not sufficient to explain large pKa shifts, possible due to the fact that side chains have to be long and flexible to establish strong interactions with the thiolate. Hence, a stable ionic contact would imply an entropic cost. It must be taken into account that when charges are exposed to a high dielectric medium, such as water, the electrostatic interaction between charges is diminished (Mossner et al. 1998; Jao et al. 2006).
In contrast, hydrogen bonds seem to have a strong influence on the pKa. Compared to oxygen or nitrogen, sulfur has lower electronegativity and a larger radius, both of which reduce its ability to participate in hydrogen bonds. In spite of this, there is strong evidence supporting the notion that the thiol group can act as a hydrogen bond donor or acceptor and that thiolate can act as hydrogen bond acceptor (Colebrook and Tarbell 1961). The associated bond distances are longer than those with nitrogen or oxygen, but some QM calculations have suggested that, in some cases, sulfur may be almost as strong a hydrogen bond acceptor as oxygen (Wennmohs et al. 2003). It has been reported that a correlation exists between the number of H bonds to the sulfur and lower predicted pKa values (Li et al. 2005; Foloppe and Nilsson 2007). It is important to remark that small structural rearrangements can form or destroy hydrogen bonds, thereby regulating reactivity through pKa shifts.
Another factor which affects the pKa of Cys residues is the helical effect, which has been attributed to the vector sum of the microdipole moments of the individual peptide units (Wada 1976). Finite difference Poisson–Boltzmann (FDPB) calculations have also suggested that the helix dipole depends on the geometry and solvent exposure of the termini (Sengupta et al. 2005).
pKa experimental determinations in proteins are far from trivial due to the often abundant number of protonable residues and the dependence of the pKa on the three-dimensional protein structure, as well as the solvation environment. Such determinations are usually made by different spectroscopic methods, nuclear magnetic resonance (NMR) and determination of the pH profiles of reaction rates. However, different experiments may lead to different results (Thurlkill et al. 2006; Roos et al. 2013a). Due to these difficulties, many computer methodologies have been developed in the last years in order to predict the pKa more accurately. Although great advances have been made in this area and very promising models have been created and tested for a variety of systems, only semi-quantitative tendencies have been observed to date, and very careful interpretations of the data are required (Alexov et al. 2011).
Electrostatic classical approach
Many pKa calculations are done by using the implicit solvent FDPB approach (Bashford and Karplus 1990; Yang et al. 1993; Honig and Nicholls 1995). This way of modeling the aqueous environment implies considering the protein as a set of spheres with a different dielectric constant (ε) than the solvent. The pKa value is calculated from the relative shift to the reference intrinsic pKa of the same residue free in aqueous solution, as previously described (Fitch and García-Moreno 2007). For free Cys, a pKa of around 8.3 is generally accepted. Many software packages are available for calculating pKa shifts, including the MEAD package and the PCE web tool (Bashford and Karplus 1990; Sanchez et al. 2008; Salsbury et al. 2012a). Using these approximations, various research groups have more recently focused on accounting for the effect of protein dynamics on pKaSH calculations. In different peroxiredoxin (Prx) systems. Salsbury and co-workers found that minimal changes in the surrounding environment of the Cys could lead to major changes in the pKa value and also that these effects could be related to conformational changes and long-range electrostatic communication paths between different Cys residues (Yuan et al. 2010; Salsbury et al. 2012b).
Empirical approach
Another method that has been used to predict the pKa of reactive Cys is the empirical pKa predictor PROPKA, which is available online (http://propka.ki.ku.dk/). The pKa shift for titratable residues is calculated as a function of the sum of energy contributions of the surrounding residues. Although this approach is simplistic, it is a very fast method with a fair correlation to the experimental determinations of pKa in some reported cases (Søndergaard et al. 2011). PROPKA has been updated three times. While in PROPKA1 (Li et al. 2005) no pKa shifts due to ligands, ions, and structural water molecules were considered, these effects were incorporated in PROPKA2 (Bas et al. 2008). In PROPKA3 and PROPKA3.1 (Søndergaard et al. 2011), the residues are no longer classified as either buried or surface residues, rather a linear interpolation between the two is used.
QM calculations
The pKa of free Cys in water has been calculated by DFT (Canle et al. 2005). These calculations have also been performed using a DFT-based approach developed by Roos and co-workers (Roos et al. 2009). Basically, these methods use the information obtained by an electronic structure calculation (free energy, atomic charges, etc.) to calculate or extrapolate pKaSH. For example, Roos and co-workers use the linear correlation between the atomic charge calculated on the sulfur atom of thiols (as natural population analysis: NPA) and their experimentally determined pKa to quantitatively calculate the pKaSH. These methods seem to be very accurate (linear correlation between theoretical and experimental values, R 2 = 0.96) (Marino and Gladyshev 2012), yet computationally very demanding. A brief comparison on the performance of different pKaSH prediction methods is presented in Table 1. In summary, although the ability to predict the pKa has improved considerably, further development is needed before computational calculations can be used to realistically calculate the pKa in protein Cys residues. To date, the best correlation with experimental results that may be achieved is the method based in DFT (Roos et al. 2009). However, this approach is computationally expensive. The PROPKA and FDPB methods show a fair correlation with experimental results and can be used to obtain a general idea about the Cys pKa, but should not yet be used to predict precise values.
Section 2: Thiol two-electron oxidation
Hydroperoxides
The reduction of hydroperoxides via thiol oxidation is a key event that has been implicated in a great variety of biological processes, such as antioxidant responses, redox signaling and transduction, regulation of different enzymes and channels, among others. (Barford 2004; Jones 2008; Winterbourn and Hampton 2008; Flohe 2010). Although Fig. 1 presents a generalized picture of the first step of this oxidation reaction, the actual reaction for each of the different hydroperoxides may vary somewhat. Here, we describe the hydrogen peroxide (H2O2) and peroxynitrous acid (ONOOH) cases as illustrative examples.
Thiols react with peroxides (Edwards 1962), chloramines, and hypochlorous acid (Peskin and Winterbourn 2001) in a bimolecular fashion. As shown in Fig. 1, the process includes two reaction steps; the first one (which is the topic of this section) involves the two-electron oxidation of the thiol moiety to yield sulfenic acid, and the second one involves de novo disulfide formation by the conjugation of the sulfenic acid generated in the first step with the other thiol (see section 3).
H2O2 is known to be produced in many cell types as a response to a variety of extracellular stimuli and could work as an intracellular messenger (Rhee et al. 2005). The oxidation of thiols by H2O2 has been reported to proceed through a SN2 mechanism (Edwards 1962). Using state of the art hybrid quantum-classical (QM-MM) MD simulations (QM-treated solute embedded in a MM-treated water box), we elucidated the reaction mechanism of thiol oxidation by H2O2 for a model methanethiolate system (Zeida et al. 2012). Our results show that there is a proton transfer between H2O2 oxygen atoms that occurs after the transition state (TS) has been reached (Fig. 3), proceeding directly to unprotonated sulfenic acid and water; these observations are in agreement with previous reported theoretical data (Chu and Trout 2004; Cardey and Enescu 2005; 2007a).
| 1 |
Fig. 3.
Free energy profile obtained by a quantum mechanics–molecular mechanical (QM-MM) umbrella sampling simulation. Free energy (kcal/mol) is plotted versus the reaction coordinate (Å). Illustrative models of the reaction mechanism steps are also depicted. Modified from Zeida et al. (2012)
We also found that the solvent plays a key role in positioning the reactants and that there is significant charge redistribution in the first stages of the reaction. The reaction is driven by the tendency of the slightly charged peroxidatic oxygen to become even more negative in the product via an electrophilic attack on the negative sulfur atom. These results are inconsistent with the SN2 mechanism, which predicts a protonated sulfenic acid and hydroxyl anion as stable intermediates (Edwards 1962).
Oxidation second order pH-independent rate constants of the reaction shown in Eq. 1 are approximately 20 M-1 s-1 for low-molecular-weight thiols in aqueous solution and exhibit almost no dependence on the thiol pKa value (Winterbourn and Metodiewa 1999). Remarkably, the rate constants for peroxidatic thiols in Cys-dependent peroxidases, such as Prxs, are several orders of magnitude larger, in the approximately 104–108 M-1 s-1 range (Parsonage et al. 2005; Navrot et al. 2006; Manta et al. 2009). It has recently been proposed that the environment of each Prx’s active site could account for a H-network and substrate placing such as to provide an alternative mechanism to the one found in aqueous solution (Hall et al. 2010; Ferrer-Sueta et al. 2011). Moreover, in a very insightful work, Nagy and co-workers modeled this environment on the human Prx2 and performed QM calculations for these model systems in order to gain an understanding of the influence of the residues nearby the peroxidatic Cys. These authors reported that both active site Arg residues are fundamental to explain the H-network that could in turn explain TS stabilization (Nagy et al. 2011). This study represents the first attempt to comprehend the extraordinary reactivity of Prxs via molecular modeling, and much more work is needed to shed light on this subject.
Peroxynitrite (as the sum of ONOO- and ONOOH) is formed in the cell by the reaction between the superoxide anion (O2 •-) and nitric oxide (•NO) radicals (Goldstein and Czapski 1995; Pryor and Squadrito 1995; Kissner et al. 1997). Both ONOOH and ONOO- are strong oxidants which have been implicated in numerous biologically relevant processes associated with protein function modification and cellular signaling, among others [for comprehensive reviews, see Pacher et al. (2007) and Ferrer-Sueta and Radi (2009)]. The specific case of ONOOH reduction by thiols has been studied very profoundly from a kinetic viewpoint (Radi et al. 1991; Koppenol et al. 1992; Trujillo and Radi 2002).
In a very recent work, we conducted an integrated kinetic and theoretical study of the oxidation of Cys by peroxynitrite:
| 2 |
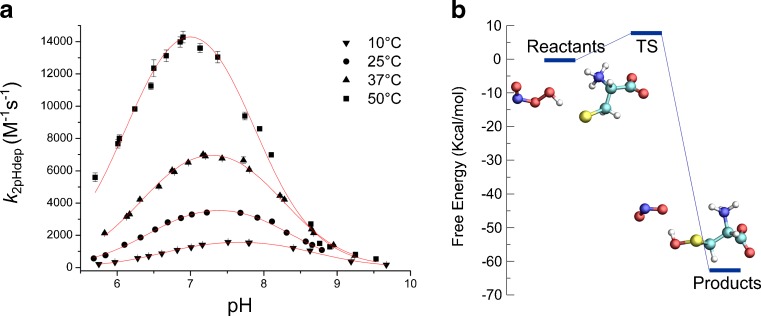
We determined the pH-independent thermodynamic activation parameters from kinetics experiments and also explored the reaction mechanism and system properties on the basis of hybrid QM-MM MD simulations (Zeida et al. 2013). This work represents the first theoretical study of this important reaction. Our results underline the pH dependency of the process and the significance of the solvent in assisting in the orientation of ONOOH and allowing the charge reorganization to take place. Although we found that the TS structure for this reaction is very similar to that observed for H2O2, unlike what was proposed for that case, we did not observe the proton transfer and the process concludes in the protonated Cys sulfenic acid (CysSOH) and nitrite ion (NO2 ¯) as products, which is then perfectly consistent with a substitution-like mechanism (Fig. 4).
Fig. 4.
Kinetic and QM-MM study of Cys oxidation by peroxynitrite (ONOO-). a “Bell-shaped” plots of the dependence of k 2pHdep (M-1 s-1) as a function of pH, at T = 10, 25, 37, and 50 °C. b Estimated free energies of the reactants, transition states (TS), and products. Illustrative pictures of reactants and products are depicted. Modified from Zeida et al. (2013)
Taking these results into account, it is essential to obtain atomistic detailed information on the mechanism of hydrogen peroxide and peroxynitrite reduction by different protein thiols, such as like peroxidatic Cys in Prxs and thiol-dependent glutathione peroxidases (GPxs), in order to understand how the protein environment can modify so drastically thiol’s capability to scavenge these oxidants. Although scarce, available kinetic data indicate that the reactivities of thiol-dependent peroxidases, such as Prxs, with fatty acid hydroperoxides and amino acid-derived hydroperoxides are also high (Peskin et al. 2010; Reyes et al. 2011).
Chloramines and hypochlorite
Thiol oxidations by hypohalous acids, such as hypochlorous acid (HOCl), and by chloramines are favorable reactions which have been implicated with alterations in regulatory and/or signaling pathways in cells exposed to neutrophile-derived oxidants (Peskin and Winterbourn 2001). In the case of HOCl, the reaction has been proposed to proceed via the formation of a very unstable sulfenyl halide (RS-Cl), which readily rearranges to sulfenic acid (Nagy and Ashby 2007). On the other hand, the oxidation of thiols by chloramines would not form a sulfenic acid, and the condensation with other thiol to form a disulfide would proceed directly from RS-Cl (Peskin and Winterbourn 2001; Stacey et al. 2012). All of these mechanistic hypotheses are based on the results of kinetic experiments (see Lo Conte and Carroll 2012 for a complete review); nevertheless, a complete atomistic description of these processes is needed for a thorough understanding. In this context, computer simulations appear to be the appropriate tool to shed light on this issue.
Section 3: Disulfide bonds
The analysis per se of the disulfide bond has been the subject of multiple studies, both experimental and computational. The combination of computational and spectroscopic tools has been a very useful duo approach for determining the physicochemical properties of the disulfide bond in small XSSX systems (X = H, Cl, F, CH3, −CH2CH2CH2−) (Boyd et al. 1983; Dixon et al. 1985; Honda and Tajima 1986; Honda and Tajima 1990; Cárdenas-Jirón et al. 1993), various organic molecules (Suzuki et al. 1990; Benassi et al. 1997; Ackermann et al. 2009), diglycosyl-disulfides (Fehér et al. 2011) and even complex protein models, such as disulfide-bridged cyclic tetrapeptides (Rao et al. 1983; Li et al. 2013) and the 4,5-ditiaheptano-7-lactam (Hudáky et al. 2004), and the famous antioxidant and GPx mimic cyclic selenamide Ebselen and its derivatives (Pearson and Boyd 2006; Pearson and Boyd 2007; Heverly-Coulson and Boyd 2009). Beyond being one of the most significant chemical bonds of sulfur, the disulfide bond is also one of the most important protein structural motifs found in nature. Formed through the oxidative binding of two cysteines (Eq. 3), disulfide bonds constitute a major chemical modification in the structure of proteins which leads to increased conformational stability of the native state, mainly by reducing the conformational entropy of the unfolded state (Zhang et al. 1994; Abkevich and Shakhnovich 2000).
| 3 |
The introduction of disulfide bonds into proteins by site-directed mutagenesis with the aim to stabilize the native folded state has been explored for “engineering” S–S bridges into unstable proteins (Perry and Wetzel 1984; Katz and Kossiakoff 1986; Wetzel et al. 1988). The same computational prediction approaches as those used in the as yet unsolved biochemical problem of the prediction of the correct disulfide bonding pattern in proteins can also be used for achieving the best combination of disulfide bonds when the aim is stabilization (Srinivasan et al. 1990; Marino and Gladyshev 2011; Savojardo et al. 2011).
As observed from Eq. 3, the reversible formation of disulfide bonds is modulated by the redox potential (Sevier and Kaiser 2002) and the pH of the environment (Roos et al. 2013a) and is often catalyzed by the action of specific proteins (Sevier and Kaiser 2002; Kadokura et al. 2003; Depuydt et al. 2011). At the present time, only two main chemical pathways through which disulfide bonds are formed in proteins have been described: (1) a sulfenic acid form of a Cys reacts with other Cys residues (Eq. 4); (2) a mechanism which involves the nucleophilic cleavage and thiol–disulfide exchange (Eq. 5) between a nucleophilic thiolate and a pre-formed disulfide bond. Both reactions are of paramount relevance in cellular physiology. There are of course a few known alternative reactions, such as the reaction of S-nitrosothiols with thiols (see section 5) or the encounter of two thiyl radicals (section 6), but these are less likely to occur in vivo.
| 4 |
| 5 |
Regarding the disulfide formation reaction via Eq. 4, the rate at which this reaction occurs has recently become an important discussion topic. While the sulfenic acid formation reaction was described at a much earlier date (Barton et al. 1973; Winterbourn and Metodiewa 1999), Luo et al. (2005) were the first to include not only the analysis of the formation of sulfenic acid, but also the formation of cystine through the reaction depicted in Eq. 4. Using an experimental design that involved reverse-phase high performance liquid chromatography (HPLC) and a two-step nucleophilic model for interpretation of the data, these authors proposed that the reaction of Eq. 4 evolved with a specific rate constant of 720 ± 70 M-1 s-1 (25 °C and pH 6). However, the following year Ashby and Nagy (2006a) published a comment on this paper arguing that the kinetic model chosen by the authors was not suitable. In the same journal issue, Anderson and Luo replied defending their choice of the kinetic model used for describing the system reactions (Anderson and Luo 2006). Finally, Ashby and Nagy decided to publish two independent new studies (Ashby and Nagy 2006b, 2007) where they showed that the value of the rate constant for the reaction of Eq. 4 is greater than 105 M-1 s-1, clearly much larger than that determined by Luo and co-workers in their original study. To add more controversy to the issue, there is also some discrepancy between some of the computational results. Two computational studies support the mechanism proposed by Luo and co-workers: in one of these, the authors combined MD and quantum chemical simulations to understand the mechanism of activation of the OxyR transcription factor by hydrogen peroxide (Kóňa and Brinck 2006). A very strong solvent effect in the disulfide formation reaction step was observed, resulting in a free energy of activation value of 14.4 kcal/mol in water. In the second study, Bayse (2011) used DFT and solvent-assisted proton exchange (SAPE) and reported a value of −27.5 kcal/mol for reaction free energy change and a free energy of activation in water of 12.5 kcal/mol, consistent with the mechanism proposed by Luo et al. (2005) at low [H2O2]/[Cys] ratios. In a recent work that deals with the oxidation of zinc−thiolate complexes by hydrogen peroxide, Kassim et al. (2011) calculated the reaction between the model systems CH3SOH and CH3S- to give dimethyl sulfide and hydroxide. The authors predicted that the reaction would be extremely fast since the corresponding potential energy barrier was lower than 1 kcal/mol, supporting, conversely, the results obtained by Ashby and Nagy (2006b, 2007). A similar tendency was observed in a study reported by Dokainish and Gauld (2013), where despite the complete reaction of study being different from the ones reviewed here, in the final step, a disulfide bond is formed between a thiolate and a sulfenic acid. In this case, the authors observed that the reduction of the sulfenic acid to give a disulfide bond can effectively occur without a barrier. A number of experimental studies of this particular reaction have been carried out within proteins (Trujillo et al. 2007; Turell et al. 2008; Hugo et al. 2009; Peskin et al. 2013), this reaction has not been intensively studied through computer simulations. Differences are observed when free Cys rate constants are compared with those obtained for the Cys residues in protein environments, as well as between the different protein models investigated to date. Clearly, despite the insights obtained through the use of computational tools, the reasons for all this variance in results remain unclear, and ultimately experimental characterization would appear to be necessary.
The reactions of disulfide bond cleavage have been studied for over 50 years (Parker and Kharasch 1959). One of these reactions, the thiol−disulfide exchange, has received particular attention over the years (Fava et al. 1957; Singh and Whitesides 1990; Keire et al. 1992; Bulaj et al. 1998). Many discussions have focused on determining the exact mechanism through which the thiol−disulfide exchange reactions occur, and it now appears that an agreement has been reached. Structural and energetic data suggest that as the central atom in disulfide becomes larger (−SS−H vs. −SS−CH3), the pathway tends to shift away from an addition–elimination (A–E) mechanism, and the SN2 pathway comes into operation (Bachrach and Mulhearn 1996; Bachrach et al. 2002; Bachrach and Pereverzev 2005; Hayes and Bachrach 2003). However, a precise partitioning of the mechanism into SN2 versus A–E has proved to be difficult because of the flatness of the potential energy surfaces (PESs) in the region of the TS and intermediates. A broad review of the kinetics and mechanisms of thiol−disulfide exchange and the challenges and advances associated with this type of investigation (small molecules vs. enzymatic systems) from a rigorous chemical point of view has been recently published (Nagy 2013). In addition, most of these results have been re-examined in two recent and comprehensive computational works (Fernandes and Ramos 2004; Bach et al. 2008). In the latter study, the authors characterized the mechanism and the transition state for the reaction of Eq. 5 when the substituents R are small. Using ab initio calculations (DFT and post-Hartree–Fock approaches) and corrections for solvation (COSMO and CPCM), these data suggest that thiolate–disulfide exchange involves a discrete intermediate resulting from the attack of the incoming thiolate on a disulfide bond, resulting in the formation of an anionic trisulfide structure [δ-S–S–Sδ-] that exists in an energy well sufficiently deep to allow for such an exchange process. Thus, the mechanism of the thiolate–disulfide exchange most likely proceeds by an SN2 process whereby the corresponding trisulfur anionic intermediate is formed.
What becomes clear from all of the above examples is that, given the large charge transfer along the reaction progress, the protein environment where these reactions may occur will play a key role in the mechanism and catalysis of the reaction. For this, an accurate description of the thiolate, disulfide, and leaving group solvation is essential for understanding the costs and benefits of carrying out this reaction in the interior of an enzyme. In recent studies (Hayes and Bachrach 2003; Geronimo et al. 2009), the microhydration of small disulfide molecules, such as dimethyldisulfide, either neutral or radical anionic, has been explored by means of ab initio calculations, revealing drastic changes in its electron affinity induced by only a few water molecules. These studies provide an important take-home message: it would seem mandatory that explicit quantum water molecules be included to achieve a complete and satisfactory picture of disulfide bond chemistry. This inclusion could be essential when these reactions are interpreted inside proteins.
The formation of disulfide bonds is not only important for the thermal stabilization of proteins, but also for proteins that carry out key redox functions through them. This has become evident in those proteins where their function is a consequence of the dynamic and sensitive exchange between two different redox states, reduced (2 CysH) and oxidized (Cys–Cys) (Sevier and Kaiser 2002; Marino and Gladyshev 2011). Another type of scenario is possible when the protein function is not intimately related with a redox Cys-mediated catalysis but still plays a physiological role in the response to oxidative stress through disulfide bond formation (Fomenko et al. 2008). In these cases, the disulfide bridge affects some protein properties, i.e., local structure, solubility, conformational entropy, among others, that ultimately impact on its not necessarily redox function. The Cys residues involved in this particular kind of modification [and also oxidation to CysSOH and S-nitrosylation) have been classified as “regulatory cysteines” (Fomenko et al. 2008). Computational tools to study these particular cases are a combination of the above. First, it is necessary to characterize the structure and dynamics of reduced and oxidized forms and to establish the impact of disulfide bond formation in the specific function of the protein in both oxidation states. Sometimes this is not easy to obtain because the structures of proteins in both oxidation states are very different, and computation requires having a starting structure (X-ray, NMR, etc.). In order to get a complete picture, an understanding of the physiological environment is necessary, as well as of the agents responsible for these changes (small oxidizing and reducing agents, traffic and mechanisms of protein secretion, proteins responsible for disulfide exchange, reversibility of these processes, etc.). Finally, a QM-MM calculation is the most appropriate approach for exploring and describing the chemical reaction pathway connected with protein function.
Section 4: Sulfenamides
Sulfenic acid (Cys−OH) may act as a metastable oxidized form or as an intermediate, giving rise to more stable products such as disulfide, sulfinic acid, or sulfonic acids (Giles et al. 2003; Paulsen and Carroll 2010). As oxidation can lead to the loss of protein activity, there are alternative mechanisms to recover the Cys residue that may be observed when disulfide formation is not possible. In this section we refer to an autorecovery mechanism by the protein itself that involves the formation of a sulfenamide, a cyclic product which involves the reaction of the sulfur atom with the backbone NH moiety of the preceding residue in the sequence (Fig. 5). This intermediate was seen for the first time in X-ray studies of protein tyrosine phosphatase 1B (PTP1B) (Salmeen et al. 2003). An increasing body of evidence suggests that the cellular redox states of the catalytic Cys are involved in determining tyrosine phosphatase activity. Peroxides can regulate cellular processes by the transient inhibition of protein tyrosine phosphatases through the reversible oxidization of their catalytic Cys, which suppresses protein dephosphorylation (Buhrman et al. 2005; Brandes et al. 2009; Miki and Funato 2012). The discovery of sulfenamide formation in PTP1B emerged as a possible mechanism to prevent overoxidation of functional Cys without the need of external regulators as organic molecules or proteins (Paulsen and Carroll 2010). Since the discovery of the formation of this intermediate in PTP1B (Salmeen et al. 2003), several proteins have been identified as forming sulfenamides, and this reaction is emerging as an important mechanism of auto-resolution of protein oxidation. In Bacillus subtilis, this phenomenon is involved in the control of the expression of Prx in response to reactive oxygen species (ROS): cyclic sulfenamide prevents the overoxidation of the organic hydroperoxide resistance regulator (OhrR) and acts as a slow switch to prevent DNA binding, allowing the transcription of the Prx genes (Lee et al. 2007; Eiamphungporn et al. 2009). Cyclic sulfenamide was also detected in the D2 distal domain of PTPalpha. This protein consists of two phosphatase domains, one proximal (D1) with phosphatase activity and one distal (D2), not directly involved in phosphatase activity. In this case the sulfenamide has been proposed as an allosteric regulator of the D1 domain, controlling its catalytic activity (Yang et al. 2007).
Fig. 5.
Energy profile for sulfenamide formation in protein tyrosine phosphatase 1B (PTP1B) for histidine protonated in the epsilon nitrogen (HIE). Cys 215 (as CysSOH) and Ser 216 were treated at the density functional theory level (PBE/dzvp), while the rest of the protein and water molecules were treated classically. Reactant, TS, and products are shown as ball and stick representations. Solid lines distances, dashed lines putative bonds. Modified from Defelipe (in preparation)
We have recently provided evidence that sulfenamide formation can be achieved in those proteins that adopt a constrained conformation of the Cys residue (Defelipe et al., in preparation) which has an specific value of the Phi angle, located in the forbidden region of the Ramachandran plot. This constrained conformation is present in almost all proteins reported to form the intermediate and in many proteins that have a reactive Cys. The constrained Cys was identified to be located in a conserved helix-loop-beta-sheet motif present in different folds, which positions the NH backbone that will form the cyclic product in a conformation needed for the reaction to occur (Table 2). The formation of sulfenamide in proteins has been proposed to occur through oxidation of the constrained Cys to sulfenic acid, followed by a nucleophilic attack of the backbone nitrogen atom of the following residue in the sequence to the sulfur atom of oxidized Cys. Synthetic thiols having ortho-amide substituents have been proposed as good models for proteins because they enforce proximity of the amide and Cys thiol groups (Sivaramakrishnan et al. 2005), highlighting the importance of specific conformations in enabling the reaction to occur. The proposed mechanism in the model organic compounds that have the S–OH moiety, as in sulfenic acid, involves a concerted mechanism that yields the cyclic sulfenamide and the loss of a water molecule (Sivaramakrishnan et al. 2005).
Table 2.
Statistical analysis of proteins in a given protein familya that display the strand-loop-helix motif with the corresponding Cys in a “forbidden” conformation
| Important families | Pfam accession | Percentage of proteins in the Pfam family with structural motif | Experimental Information oxidationb | Reference |
|---|---|---|---|---|
| Y_phosphatase | PF00102 | 70,27 | Yes | (Yang et al. 2007; Sivaramakrishnan et al. 2005) |
| Glutamine amidotransferase (GATase) | PF00117 | 69,56 | No | |
| Rhodanese-like domain | PF00581 | 37,50 | Yes | (Seo and Carroll 2009) |
| Dual specificity phosphatase | PF00782 | 30,77 | Yes | (Alonso et al. 2004; Eiamphungporn et al. 2009) |
| Carbon–nitrogen hydrolase | PF00795 | 91,67 | No | |
| SNO glutamine amidotransferase | PF01174 | 83,33 | Yes (ortholog) | (Wolf et al. 2008) |
| DJ-1/PfpI | PF01965 | 68,75 | Yes | (Canet-Avilés et al. 2004; Wilson et al. 2004; Wolf et al. 2008) |
aAs defined by the Pfam HMM model; for more information, see Sonnhammer et al. (1997)
bExperimental information on cysteine oxidation which has been reported in any member of the Pfam family
We performed QM–MM calculations starting with X-ray crystal of PTP1B; Cys 215 (as CySOH) and Ser 216 were treated at the DFT theory level (PBE/dzvp) while the rest of the protein and water molecules were treated classically using the AMBER force field. We found that the reactions occur in a concerted mechanism involving the abstraction of the amide hydrogen by the OH group from the sulfenic acid and the subsequent formation of the the S–N bond (Defelipe, in preparation) (Fig. 5). The S–O bond rupture is clearly homolytic based on the neutral charge in the S and OH groups in the apparent TS. The Psi dihedral angle does not change significantly during the reaction, again showing that backbone conformation is important. These results are consistent with the fact that the reaction is very slow, as measured in synthetic probes and based on previous calculations (Sarma and Mugesh 2007). A possible explanation of why the process actually happens, even if at slow rates, may be given by flaws in the computational methodology, such as the neglect of entropic effects. Relatively high activation energy barriers can be partially overcome as the reactants are constrained and located in the same molecule ready to react. By analyzing the conformation of a model peptide we found that proteins overcame a penalty of around 8 kcal/mol to achieve the constrained conformation; on the other hand, local environment around the reactive Cys seemed not to be important as we failed to find any conserved residues or amino acid type with respect to sequence or structure around the reactive Cys in all proteins that have the helix-loop-beta-sheet motif.
The physiological relevance of sulfenamides is still a matter of discussion. The fact that some proteins which have a reactive Cys lack a regulatory mechanism to protect it from oxidative attack and that a constrained conformation found in many of these protein families may be relevant for its formation opens the possibility that this mechanism might be more general than previously thought when it was identified in PTP1B.
Section 5: S-nitrosothiols
S-nitrosothiols (RSNOs) were first synthesized in 1909 by (Tasker and Jones 1909). In 1974, Incze and co-workers showed that these compounds exert biological activity, such as antibacterial effects (Incze et al. 1974). The first studies in mammalian systems were performed in the early 1980s by Ignarro (1999). At that time, a plethora of investigations followed into the many different aspects of •NO biochemistry: the generation of nitric oxide, its function, and fate (Bredt and Snyder 1993; Marletta 1993; Nathan and Xie 1994; Gross and Wolin 1995; Kerwin et al. 1995; Crane et al. 1998). As part of this extensive research, the presence of RSNOs in biological systems was demonstrated (Rockett et al. 1991; Goldman et al. 1998). This led to a considerable interest into the physiological role of RSNOs, their formation, reactivity, and delivery, either as a small molecule adduct or as a post-translational chemical modification of a protein.
S-nitrosation and S-nitrosylation are thought to be a major mechanism of NO-mediated signaling in pathology and physiology (Hess et al. 2005). S-nitrosated proteins have been found in many tissues (Bryan et al. 2004), and these proteins are involved in a wide range of functions, including transcription, channel activity, response to hypoxia, and cell death (Hess et al. 2005). Although it is widely accepted that RSNOs occurs in vivo, mechanisms of S-nitrosation in biological systems are poorly understood. •NO is synthesized from nitric oxide synthase (NOS), but it does not directly react with thiols to form RSNOs (Hogg et al. 1996). Several mechanisms (1, 2, and 3, as follows) have been proposed:
the formation of nitrosating species from •NO oxidation by a molecular oxygen-dependent process (Wink et al. 1994; Goldstein and Czapski 1996);
thiol oxidation to form a thiyl radical, which can combine with •NO to form RSNO (Jourd’heuil et al. 2003; Madej et al. 2008);
direct addition of •NO to RSH to form the radical intermediate RSNOH followed by oxidation of this radical by oxygen or some other one-electron acceptor (Gow et al. 1997).
Regarding mechanism 1, there is little evidence that this effect is important in vivo (Liu et al. 1998; Moller et al. 2007). The reaction of •NO with the thiyl radical has been reported by some authors (Schrammel et al. 2003; Hofstetter et al. 2007), and some discrepancies about the rate constant have been observed. A pulse radiolysis study by Madej et al. (2008) has shed light on this issue, with these authors demonstrating that the rate constants for this reaction in a variety of Cys and glutathione (GSH)-derived thiols are (2–3) × 109 M-1 s-1—two orders of magnitude higher than those previously reported. Regarding mechanism 3, there has been significant interest in the role of metal ions and metalloproteins in RSNO formation (Stubauer et al. 1999): peroxidases and hemoglobin (Nagababu et al. 2006; Angelo et al. 2006) as well as in dinitrosyl iron complexes (Boese et al. 1995). Gow and co-workers proposed that electron acceptors could facilitate S-nitrosation by oxidizing the thionitroxyl radical (Gow et al. 1997), suggesting that single electron acceptors may facilitate RSNO formation. Broniowska et al.( 2012) recently observed that ferric cytochrome c can, under anaerobic conditions, efficiently promote glutathione S-nitrosation by acting as an electron acceptor (Basu et al. 2010).
Two aspects of these S-nitroso compounds are of particular interest in this review. One is their electronic structure and properties, and the other is their reactivity, in particular the reaction of RSNOs with other thiols.
Electronic structure description of RSNO by QM calculations
S-nitrosothiols, with certain exceptions, are unstable in aqueous solution. For example S-nitrosoglutatione (GSNO) undergoes decomposition over hours, while S-nitrosoCys has a half life of less than 2 min. The S–N bond in RSNOs is weak, with a bond dissociation energy (BDE) in the range of 25–30 kcal/mol (Bartberger et al. 2001; Lu et al. 2001; Grossi and Montevecchi 2002) and a long bond distance (1.75 Å to almost 2.00 Å) relative to a typical S–N bond (1.77–1.79 Å: range of values for available crystal structures of RSNOs) (Arulsamy et al. 1999; Wang et al. 2002; Baciu and Gauld 2003). At the same time, the evidence of cis and trans conformers of RSNOs separated by a barrier of approx. 10 kcal/mol (Arulsamy et al. 1999) suggests a partial S–N double bond character (Bartberger et al. 2000, 2001; Lu et al. 2001; Grossi and Montevecchi 2002; Wang et al. 2002). Hence, an attractive challenge for those researchers using the computational chemistry approach is to determine whether or not such variable optimized lengths are artifacts of the methods employed or indicative of the particular nature of the N–S bonds.
Quantum chemical calculations of RSNOs do not provide a consistent description of the RSNO structure and energetics since the results are highly dependent on the method and basis set used (see Table 1) (Baciu and Gauld 2003). For example, calculations employing a relatively high-level coupled-cluster with single and double excitations or quadratic configuration interaction with single and double excitations (QCISD) underestimate the BDE(S–N) by more than 10 kcal/mol (Baciu and Gauld 2003), whereas DFT and various composite methodologies, such as G3 and CBS-QB3, predict BDE(S–N) values close to those experimentally determined. Timerghazin et al. (2008a) suggested that this unusual and seemingly inconsistent properties of RSNOs indicates that the RSNO wave function possibly possesses a multireference character (Timerghazin et al. 2008b). Weinhold et al. (2005) demonstrated that natural resonance theory (NRT) analysis of the single determinant DFT electron density indicates that the electronic structure of RSNOs can be expressed as a combination of three resonance structures (S, D, I; Scheme 1) (Timerghazin et al. 2007).
Scheme 1.
Representation of S-nitrosothiols (RSNOs) in terms of three resonance structures—D, S, and I
The conventional structure S implies a single S–N bond. The zwitterionic structure D, with a double S–N bond, would account for the cis–trans isomerization of RSNOs, while the ionic structure I would explain the unusually long S–N bond. In addition, reactivity and stability of the RSNOs could be modulated via N-coordination to other molecules. It has recently been shown how the stability and reactivity of RSNOs can significantly change when coordinated to metal complexes such as pentachloroIridate(III) and pentacyanoferrate(II) (Szacilowski and Chmura 2005; Perissinotti et al. 2006, 2008).
RSNO reaction with thiols
One possible reaction between RSNOs and thiols is a simple transnitrosation reaction that results from the nucleophilic attack on the nitrogen of the RSNO (reaction 1, Scheme 2) and which contributes to the •NO transfer without involving free •NO during the process. However, this is not the only possible chemical outcome, as the reaction of RSNOs with other thiols is complex and leads to the generation of a variety of species, including ammonia (NH3), •NO, nitrous oxide (N2O), and nitrite (NO2 -) (Singh et al. 1996). The chemistry is complex and may involve a series of sequential reactions leading to a multitude of products. It has been proposed that many of the observed products may result from the disulfide formation reaction (also known as S-thiolation reaction) and thus the nuclephilic attack on the sulfur atom of the RSNO (reaction 2, Scheme 2), leading to disulfide and the initial generation of nitroxyl (HNO) (Wong et al. 1998).
Scheme 2.
The two reactions studied: transnitrosation and disulfide formation
The two resonance structures discussed above, D and I (see Scheme 2), with the opposite formal charge distribution and bonding patterns, can explain the duality of the reactivity of RSNOs with nucleophiles. While D favors a nucleophilic attack at the electrophilic S+ atom, I describes the •NO group as a nitrosonium ion NO+ and thus favors a nucleophilic attack at the N atom. Talipov and Timerghazin (2013) recently demonstrated that complexation with charged and polar residues modulate the reactivity of biological RSNOs through D and I effects. However, specific interactions with charged/polar residues are not the only way to control the RSNOs reactivity. For instance, catalytic effects of external electric fields created by the protein environment can modify the RSNO structure and reactivity (Timerghazin and Talipov 2013).
Transnitrosation
Transnitrosation is a second order reaction with a barrier height in the range of 13–18 kcal/mol (Perissinotti et al. 2005; Li et al. 2006). The reaction rate depends on pH, suggesting that the RS- is the reactive species (Barnett et al. 1994, 1995; Munro and Williams 2000; Wang et al. 2001). The experimental evidence is also supported by QM calculations, first by Houk and co-workers for the methylthiol (MeSH) model system (Houk et al. 2003), and later revisited for the same model system by Li et al. (2006). It is proposed that transnitrosation proceeds by the attack of a thiolate on an electrophilic RSNO through an anionic [RSN(O)SR]- intermediate. We have also proposed this intermediate and characterized it through 15 N, 1HNMR, and QM calculations for the Cys ethylester (CEE) system, a much more relevant model system from the biological point of view (Perissinotti et al. 2005).
To date, transnitrosation has been observed experimentally and supported by quantum calculations; whether it plays a key biological role needs further research but it may be a relevant pathway for •NO trafficking. We will present here a detailed analysis of the transnitrosation of the more relevant model system, CEE.
The CEE system
This reaction was studied experimentally and computationally by Perissinotti et al. (2005). Figure 6a shows the energetics for the reaction in gas phase and water (PCM), together with the optimized structures for all species. We observed similar environmental effects for the MeSH system, but the barrier was much higher, and the intermediate seemed to be more thermodynamically favored in the CEE system.The barrier was experimentally measured in water and the intermediate characterized by 15 N and 1H NMR techniques in methanol. Figure 6b shows the experimental characterization and the 15 N NMR assignment together with the the QM prediction for the reaction intermediate. The choice of methanol as solvent allowed characterization of the transient intermediate while all attempts in water failed. The potential structure that we suggested for the intermediate is shown in Fig. 6c and, in accordance to that proposed by Schlegel (Li et al. 2006), based on the 1H NMR and 2D NMR assignment it would appear that a thiol molecule assists the process.
Fig. 6.
a Energetics and optimized species for the transnitrosation reaction between the Cys ethylester (CEE) and the corresponding S-nitrosothiol in gas phase (black) and water (PCM, red) at the B3LYP/6-311 + G* level of theory. b Intermediate assignment from 15 N nuclear magnetic resonance (NMR) spectrum for the reaction in methanol (reference: nitromethane). c Suggested structure for the intermediate based on 1H NMR and 2-dimensional correlation spectroscopy (2D COSY), and heteronuclear correlation (HETCOR) NMR spectra. Modified from Perissinotti et al. 2005
Disulfide formation
In vivo disulfide formation occurs in competition with transnitrosation and may be associated with the release of •NO. It has been established that RSNOs are sensitive to both photolytic (Wang et al. 2002) and transition metal ion-dependent decomposition, two processes that could significantly interfere with transnitrosation as well as disulfide formation.
The MeSH system
Li et al. (2006) showed that the reaction of MeSH with MeSNO to generate HNO and disulfide has a prohibitively high barrier in the gas phase. Without considering an explicit molecule of water, the barrier is 12 kcal/mol higher than transnitrosation in gas phase. Inclusion of an explicit water molecule lowers the difference to 9 kcal/mol, and adding bulk solvent reduces the difference to 3–5 kcal/mol. Similar to transitrosation reactions, the corresponding reactions with the MeS radical were also studied. These authors found that the reaction for the MeS radical with RSNO to form the disulfide bond is barrierless. This observation is in accord with the autocatalytic effect of the MeS radical on the decomposition rate of MeSNO (Oliveira et al. 2002) (Table 3).
Table 3.
Energetics (kcal/mol) for the disulfide formation reaction between MeSH and MeSNO in gas phase in which one explicit water molecule in gas phase was included and using different solvating schemes to account for the effect of water and acetonitrilea
| Species | CBS-QB3/6-311 + G* | |||
|---|---|---|---|---|
| Gas | Gas + H2O | CPCM (water) + H2O | CPCM (CH3CN) + H2O | |
| 1 + 2 | 0 | 0 | 0 | 0 |
| TS | 50.4 | 36.1 | 33.2 | 30.3 |
| MeSSMe + HNO | 5.6 | 6.0 | 2.1 | 4.3 |
aData from Li et al. (2006)
General considerations
The relevance of the different reactive species, namely, the RSH, RS-, and RS radicals, is determined by the reaction conditions. The energy barriers of RSNO for neutral RSH are very high, but they can be lowered by the inclusion of explicit water molecules. The involvement of the RS- as a reactive species lowers the barrier heights even more. In each case, RS radicals reacting with RSNO have the lowest barrier for both transnitrosation and disulfide formation reactions.
The experimental work of Hogg (1999) shows that the S-nitrosation reaction is several orders of magnitude faster than disulfide formation at pH = 7 and room temperature. Since the mechanisms for both reactions involving RSH have very high barriers, the reactions involving RS- will contribute the most. From the other side, although the mechanisms involving thiyl radicals have the lowest barriers, the concentration of the radical in the solution is very low, and the contribution of these radicals will depend on temperature, the presence of transition metals, and light.
Finally, it is important to emphasize that the chemical reaction between RSNOs and thiols is complex due to the multiple secondary reactions that take place and the differing conditions which govern the favorableness of each reaction. It is also of crucial importance to consider the RSH/RSNO ratio, the environment, and oxygen availability in biological systems when speculating on the fate and reaction of RSNOs. In addition, the selectivity of S-nitrosation of Cys residues against disulfide formation might in part be determined by the local electric field exerted by nearby charged residues, alfa-helices dipoles, or membrane potentials.
Section 6: One-electron oxidation paths overview
Low-molecular-weight thiols, such as glutathione and Cys protein residues, are good antioxidant scavengers because along with other capabilities they react rapidly with a great variety of cellular free radicals (hydroxyl, phenoxyl, peroxyl radicals, among others) (Sjöberg et al. 1982; Swarts et al. 1989; Folkes et al. 2011). Concomitantly, these reactions produce thiyl radicals that need to be converted or removed from the cell. Thiolates, molecular oxygen (O2), O2 •-, and •NO can react subsequently with these thiyl radicals, yielding disulfides, sulfinyl radical, or nitrosothiols (as discussed in the previous section) as final products, respectively (Winterbourn and Hampton 2008; Winterbourn 2013). However, the other central role for protein radicals (such as glycyl and thiyl radicals) in biology should not be forgotten. These are formed during the catalysis of key metabolic reactions (Stubbe and van Der Donk 1998; Buckel 2013), as in the case of the pyruvate formate-lyase (Becker and Kabsch 2002) and benzylsuccinate synthase (Bharadwaj et al. 2013) enzymes. These new findings open the door to a new world of free radical mechanisms in protein, where the thiyl radical is one of the leading actors.
The thiyl radical has been the subject of a few theoretical QM studies in which the unusual spectroscopic and one-electron reduction potential properties measured in these kind of systems are explained in terms of their electronic structure and the surrounding environment, especially by the number of H bonds to the sulfur (Engström et al. 2000; van Gastel et al. 2004) and by the ligand polarity around the sulfur atom (Roos et al. 2013b).
After the reaction of a thiolate with a free radical, the fate of the generated thiyl radical is extremely sensitive to its surroundings. Although its major physiological decay route would imply the reaction with another thiolate to yield the disulfide radical anion (Buettner 1993), with the subsequent reaction being with O2 to produce O2 •- (reactions 6 and 7) (Winterbourn 2013), secondary pathways may be activated.
| 6 |
| 7 |
Particularly, O2 can react directly with thiyl radicals, yielding sulfynil radicals which can be converted to sulfinic and sulfenic acids (Winterbourn and Metodiewa 1994).
A biologically important and discussed issue of this radical chemistry is the direct reaction of O2 •- with thiols (Winterbourn and Metodiewa 1999). In this context, two QM investigations have confirmed the possibility for a three-electron-bonded superoxide–thiol complex and the likelihood of decomposition into the sulfinyl radical and hydroxide (Cardey et al. 2007, 2009).
Another significant issue of thiyl radical biochemistry is the intramolecular electron transfer (IET) between tyrosyl radicals (Tyr-O•) and Cys residues. This event has been studied in model Tyr/Cys-containing peptides (Prütz et al. 1989; Zhang et al. 2005) and some proteins (Romero et al. 2003; Bhattacharjee et al. 2007), and it represents a key factor in controlling the final sites and yields of protein oxidative modifications. Recently, Petruk et al. (2012) used MD and QM-MM calculations to analyze the molecular basis of IET. The results support a sequential, acid/base equilibrium-dependent and solvent-mediated, proton-coupled electron transfer from Tyr-O• to the thiyl radical in which not only energetic and kinetics of the reversible IET are key physico-chemical factors, but also the pKa values of the Tyr phenol and Cys thiol groups (Petruk et al. 2012).
All of these mechanisms are not only relevant in the context of antioxidant defense, but they are also components of the complex network of radical reactions that assist regulation of the oxidation state of cellular thiols (Winterbourn 2013). Clearly, the atomistic detailed description of these particular processes is not entirely unraveled, and computer simulation techniques appear to be a useful tool to contribute to this understanding.
Concluding remarks
Thiol redox biochemistry is a rich field in which classical and QM computer simulation techniques provide valuable tools to shed light onto the different possible reaction pathways. Specifically, the subtle sensitivity of thiol reactivity with the environment results in these techniques being extremely useful. In this context, the contribution of hybrid QM-MM schemes represents an ideal tool.
Different theoretical schemes based on classical MD and QM-MM, can complement each other by covering different time and space scales, thereby providing a complete picture of the molecular basis of thiol reactivity. The agreement with the experimental data, when available, constitutes a stringent test to the reliability of these approaches. Computational simulation has been shown to be a precious tool to validate, contrast, and complement experimental observations, offering a microscopic view, and providing new insights that sometimes are impossible to obtain with experimental approaches.
Acknowledgments
This work was supported in part by the University of Buenos Aires, ANPCyT (PICT-25667), and Consejo Nacional de Investigaciones Científicas y Tecnicas (CONICET). DAE and AT are CONICET staff researchers. AZ, CMG, and LAD acknowledge a CONICET fellowship. RR and MT acknowledge the financial support of the Howard Hughes Medical Institute, National Institutes of Health, Agencia Nacional de Investigación e Innovación (ANII, Uruguay) and Comisión Sectorial de Investigación Científica (CSIC), Universidad de la República.
Conflict of interests
None.
Animal study statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Abbreviations
- Cys
L-cysteine
- CysS-
Cysteinate
- CysSOH
Cysteine sulfenic acid
- DFT
Density functional theory
- GPx
Glutathione peroxidase
- GSH
Glutathione
- MD
Molecular dynamics
- MM
Molecular mechanics
- Prx
Peroxiredoxin
- QM
Quantum mechanics
- ROS
Reactive oxygen species
Footnotes
Special Issue Advances in Biophysics in Latin America
References
- Abkevich VI, Shakhnovich EI. What can disulfide bonds tell us about protein energetics, function and folding: simulations and bioninformatics analysis. J Mol Biol. 2000;300:975–985. doi: 10.1006/jmbi.2000.3893. [DOI] [PubMed] [Google Scholar]
- Ackermann KR, Koster J, Schlücker S. Conformations and vibrational properties of disulfide bridges: Potential energy distribution in the model system diethyl disulfide. Chem Phys. 2009;355:81–84. [Google Scholar]
- Alexov E, Mehler EL, Baker N, Baptista AM, Huang Y, Milletti F, Nielsen JE, Farrell D, Carstensen T, Olsson MHM, Shen JK, Warwicker J, Williams S, Word JM. Progress in the prediction of pKa values in proteins. Proteins. 2011;79:3260–3275. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Anderson BD, Luo D. Application of an exact mathematical model and the steady-state approximation to the kinetics of the reaction of cysteine and hydrogen peroxide in aqueous solution: a reply to the Ashby and Nagy commentary. J Pharm Sci. 2006;95:19–24. doi: 10.1002/jps.20539. [DOI] [PubMed] [Google Scholar]
- Angelo M, Singel DJ, Stamler JS. An S-nitrosothiol (SNO) synthase function of hemoglobin that utilizes nitrite as a substrate. Proc Natl Acad Sci USA. 2006;103:8366–8371. doi: 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulsamy N, Bohle DS, Butt JA, Irvine GJ. Interrelationships between conformational dynamics and the redox chemistry of S-nitrosothiols. J Am Chem Soc. 1999;121:7115–7123. [Google Scholar]
- Ashby MT, Nagy P. On the kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J Pharm Sci. 2006;95:15–18. doi: 10.1002/jps.20521. [DOI] [PubMed] [Google Scholar]
- Ashby MT, Nagy P. Revisiting a proposed kinetic model for the reaction of cysteine and hydrogen peroxide via cysteine sulfenic acid. Int J Chem Kinet. 2006;39:32–38. [Google Scholar]
- Bach RD, Dmitrenko O, Thorpe C. Mechanism of thiolate−disulfide interchange reactions in biochemistry. J Org Chem. 2008;73:12–21. doi: 10.1021/jo702051f. [DOI] [PubMed] [Google Scholar]
- Bachrach SM, Hayes JM, Dao T, Mynar JL. Density functional theory gas- and solution-phase study of nucleophilic substitution at di- and trisulfides. Theor Chem Acc. 2002;107:266–271. [Google Scholar]
- Bachrach SM, Mulhearn DC. Nucleophilic substitution at sulfur: SN2 or addition-elimination? J Phys Chem. 1996;100:3535–3540. [Google Scholar]
- Bachrach SM, Pereverzev A. Competing elimination and substitution reactions of simple acyclic disulfides. Org Biomol Chem. 2005;3:2095–2101. doi: 10.1039/b501370d. [DOI] [PubMed] [Google Scholar]
- Baciu C, Gauld JW. An assessment of theoretical methods for the calculation of accurate structures and SN bond dissociation energies of S-nitrosothiols (RSNOs) J Phys Chem A. 2003;107:9946–9952. [Google Scholar]
- Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Barnett DJ, McAnimly J, Williams DLH. Transnitrosation between nitrosothiols and thiols. J Chem Soc Perkin Trans. 1994;2:1131–1133. [Google Scholar]
- Barnett DJ, Rios A, Williams DLH. NO- group transfer (transnitrosation) between S-nitrosothiols and thiols II. J Chem Soc Perkin Trans. 1995;2:1279–1282. [Google Scholar]
- Bartberger MD, Houk KN, Powell SC, Mannion JD. Theory, spectroscopy, and crystallographic analysis of S-nitrosothiols: conformational distribution dictates spectroscopic behavior. J Am Chem Soc. 2000;122:5889–5890. [Google Scholar]
- Bartberger MD, Mannion JD, Powel SC, Stamler JS. SN dissociation energies of S-nitrosothiols: on the origins of nitrosothiol decomposition rates. J Am Chem Soc. 2001;123:8868–8869. doi: 10.1021/ja0109390. [DOI] [PubMed] [Google Scholar]
- Barton JP, Packer JE, Sims RJ. Kinetics of the reaction of hydrogen peroxide with cysteine and cysteamine. J. Chem. Soc., Perkin Trans. 1973;2:1547–1549. [Google Scholar]
- Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- Bashford D, Karplus M. pKa’s of Ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, Broniowska KA, Hogg N, Kim-Shapiro DB. A novel role for cytochrome c: efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–263. doi: 10.1016/j.freeradbiomed.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J Am Chem Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- Bayse CA. Transition states for cysteine redox processes modeled by DFT and solvent-assisted proton exchange. Org Biomol Chem. 2011;9:4748–4751. doi: 10.1039/c1ob05497j. [DOI] [PubMed] [Google Scholar]
- Becker A, Kabsch W. X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA—How the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. J Biol Chem. 2002;277:40036–40042. doi: 10.1074/jbc.M205821200. [DOI] [PubMed] [Google Scholar]
- Benassi R, Fiandri GL, Taddei F. A theoretical MO ab initio approach to the conformational properties and homolytic bond cleavage in aryl disulphides. J Mol Struc-Theochem. 1997;418:127–138. [Google Scholar]
- Bharadwaj VS, Dean AM, Maupin CM. Insights into the glycyl radical enzyme active site of benzylsuccinate synthase: a computational study. J Am Chem Soc. 2013;135:12279–12288. doi: 10.1021/ja404842r. [DOI] [PubMed] [Google Scholar]
- Boese M, Mordvintcev P, Vanin AF, Busse R, Mulsch A. S-nitrosation of serum albumin by dinitrosyl-iron complex. J Biol Chem. 1995;270:29244–29249. doi: 10.1074/jbc.270.49.29244. [DOI] [PubMed] [Google Scholar]
- Boyd RJ, Perkyns JS, Ramani R. Conformations of simple disulfides and L-cystine. Can J Chem. 1983;61:1082–1085. [Google Scholar]
- Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antiox Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH. Nitric oxide: a physiologic messenger molecule. Annu Rev Biochem. 1993;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- Broniowska KA, Keszler A, Basu S, Kim-Shapiro DB, Hogg N. Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem J. 2012;442:191–197. doi: 10.1042/BJ20111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan NS, Rassaf T, Maloney RE, Rodriguez CM, Saijo F, Rodriguez JR, Feelish JR. Cellular targets and mechanisms of nitros (yl) ation: an insight into their nature and kinetics in vivo. Proc Natl Acad Sci USA. 2004;101:4308–4313. doi: 10.1073/pnas.0306706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W. Bacterial methanogenesis proceeds by a radical mechanism. Angew Chem Int Ed. 2013;52:2–5. doi: 10.1002/anie.201304593. [DOI] [PubMed] [Google Scholar]
- Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, a-tocopherol, and ascorbate. Arch Biochem Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Parker B, Sohn J, Rudolph J, Mattos C. Structural mechanism of oxidative regulation of the phosphatase Cdc25B via an intramolecular disulfide bond. Biochemistry. 2005;44:5307–5316. doi: 10.1021/bi047449f. [DOI] [PubMed] [Google Scholar]
- Bulaj G, Kortemme T, Goldenberg DP. Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37:8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- Canet-Avilés RM, Wilson M, Miller DW, Ahmad R, McLendon C, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canle López M, Ramos DR, Santaballa JA. A DFT study on the microscopic ionization of cysteine in water. Chem Phys Lett. 2005;417:28–33. [Google Scholar]
- Cárdenas-Jirón GI, Cárdenas-Lailhacar C, Toro-Labbé A. Theoretical analysis of the internal rotation, molecular structures and electronic properties of the XSSX series of molecules (X = H, F, Cl) J Mol Struc-Theochem. 1993;282:113–122. [Google Scholar]
- Cardey B, Enescu M. A computational study of thiolate and selenolate oxidation by hydrogen peroxide. ChemPhysChem. 2005;6:1175–1180. doi: 10.1002/cphc.200400568. [DOI] [PubMed] [Google Scholar]
- Cardey B, Enescu M. Selenocysteine versus cysteine reactivity: A theoretical study of their oxidation by hydrogen peroxide. J Phys Chem A. 2007;111:673–678. doi: 10.1021/jp0658445. [DOI] [PubMed] [Google Scholar]
- Cardey B, Enescu M. Cysteine oxidation by the superoxide radical: a theoretical study. ChemPhysChem. 2009;10:1642–1648. doi: 10.1002/cphc.200900010. [DOI] [PubMed] [Google Scholar]
- Cardey B, Foley S, Enescu M. Mechanism of thiol oxidation by the superoxide radical. J Phys Chem A. 2007;111:13046–13052. doi: 10.1021/jp0731102. [DOI] [PubMed] [Google Scholar]
- Chu JW, Trout BL. On the mechanisms of oxidation of organic sulfides by H2O2 in aqueous solutions. J Am Chem Soc. 2004;126:900–908. doi: 10.1021/ja036762m. [DOI] [PubMed] [Google Scholar]
- Colebrook LD, Tarbell DS. Evidence for hydrogen bonding in thiols from NMR measurements. Proc Natl Acad Sci USA. 1961;47:993–996. doi: 10.1073/pnas.47.7.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Trainer JA. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- Crespo A, Marti MA, Estrin DA, Roitberg AE. Multiple-steering QM-MM calculation of the free energy profile in chorismate mutase. J Am Chem Soc. 2005;127:6940–6941. doi: 10.1021/ja0452830. [DOI] [PubMed] [Google Scholar]
- Crespo A, Marti MA, Roitberg AE, Amzel M, Estrin DA. The catalytic mechanism of peptidylglycine a-hydroxylating monooxygenase investigated by computer simulation. J Am Chem Soc. 2006;128:12817–12828. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]
- Crespo A, Scherlis DA, Martí MA, Ordejón P, Roitberg AE, Estrin DA. A DFT-based QM-MM approach designed for the treatment of large molecular systems: application to chorismate mutase. J Phys Chem B. 2003;107:13728–13736. [Google Scholar]
- Depuydt M, Messens J, Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Zeroka D, Wendoloski JJ, Wasserman ZR. The molecular structure of H2S2 and barriers to internal rotation. J Phys Chem. 1985;89:5334–5336. [Google Scholar]
- Dokainish HM, Gauld JW. A molecular dynamics and quantum mechanics/molecular mechanics study of the catalytic reductase mechanism of methionine sulfoxide reductase A: formation and reduction of a sulfenic acid. Biochemistry. 2013;52:1814–1827. doi: 10.1021/bi301168p. [DOI] [PubMed] [Google Scholar]
- Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: A computational microscope for molecular biology. Annu Rev Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Jeng MF, Tennant LL, Slaby I, Lindell M, Cui DS, Kuprin S, Holmgren A. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- Edwards JO. In: Peroxide reaction mechanisms. 1. Edwards JO, editor. New York: Interscience; 1962. pp. 67–106. [Google Scholar]
- Eiamphungporn W, Soonsanga S, Lee J-W, Helmann JD. Oxidation of a single active site suffices for the functional inactivation of the dimeric Bacillus subtilis OhrR repressor in vitro. Nucleic Acids Res. 2009;37:1174–1181. doi: 10.1093/nar/gkn1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström M, Vahtras O, Ågren H. MCSCF and DFT calculations of EPR parameters of sulfur centered radicals. Chem Phys Lett. 2000;328:483–491. [Google Scholar]
- Fava A, Iliceto A, Camera E. Kinetics of the thiol-disulfide exchange. J Am Chem Soc. 1957;79:833–838. [Google Scholar]
- Fehér K, Matthews RP, Kövér KE, Naidoo KJ, Szilágyi L. Conformational preferences in diglycosyl disulfides: NMR and molecular modeling studies. Carbohydr Res. 2011;346:2612–2621. doi: 10.1016/j.carres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Fernandes PA, Ramos MJ. Theoretical insights into the mechanism for thiol/disulfide exchange. Chem Eur J. 2004;10:257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- Fitch CA, García-Moreno EB (2007) Structure-based pKa calculations using continuum electrostatics methods. Curr Protoc Bioinf Chapter 8:Unit 8.11. doi:10.1002/0471250953.bi0811s16. [DOI] [PubMed]
- Flohe L. Changing paradigms in thiology from antioxidant defense toward redox regulation. Methods Enzymol. 2010;473:1–39. doi: 10.1016/S0076-6879(10)73001-9. [DOI] [PubMed] [Google Scholar]
- Folkes LK, Trujillo M, Bartesaghi S, Radi R, Wardman P. Kinetics of reduction of tyrosine phenoxyl radicals by glutathione. Arch Biochem Biophys. 2011;506:242–249. doi: 10.1016/j.abb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Foloppe N, Nilsson L. Stabilization of the catalytic thiolate in a mammalian glutaredoxin: structure, dynamics and electrostatics of reduced pig glutaredoxin and its mutants. J Mol Biol. 2007;372:798–816. doi: 10.1016/j.jmb.2007.05.101. [DOI] [PubMed] [Google Scholar]
- Fomenko DE, Marino SM, Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells. 2008;26:228–235. [PMC free article] [PubMed] [Google Scholar]
- Geronimo I, Chéron N, Fleurat-Lessard P, Dumont E. How does microhydration impact on structure, spectroscopy and formation of disulfide radical anions? An ab initio investigation on dimethyldisulfide. Chem Phys Lett. 2009;481:173–179. [Google Scholar]
- Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- Giles N, Watts A, Giles G, Fry F. Metal and redox modulation of cysteine protein function. Chem Biol. 2003;10:677–693. doi: 10.1016/s1074-5521(03)00174-1. [DOI] [PubMed] [Google Scholar]
- Goldman RK, Vlessis AA, Trunkey DD. Nitrosothiol quantification in human plasma. Anal Biochem. 1998;259:98–103. doi: 10.1006/abio.1998.2651. [DOI] [PubMed] [Google Scholar]
- Goldstein S, Czapski G. The reaction of NO. with O2.- and HO2.: a pulse radiolysis study. Free Radic Biol Med. 1995;19:505–510. doi: 10.1016/0891-5849(95)00034-u. [DOI] [PubMed] [Google Scholar]
- Goldstein S, Czapski G. Mechanism of the nitrosation of thiols and amines by oxygenated no solutions: the nature of the nitrosating intermediates. J Am Chem Soc. 1996;118:3419–3425. [Google Scholar]
- González Lebrero MC, Perissinotti LL, Estrin DA. Solvent effects on peroxynitrite structure and properties from QM/MM simulations. J Phys Chem A. 2005;109:9598–9604. doi: 10.1021/jp054224l. [DOI] [PubMed] [Google Scholar]
- Gow AJ, Buerk DG, Ischiropoulos H. A novel reaction mechanism for the formation of S-nitrosothiol in vivo. J Biol Chem. 1997;272:2841–2845. doi: 10.1074/jbc.272.5.2841. [DOI] [PubMed] [Google Scholar]
- Gross SS, Wolin MS. Nitric oxide: pathophysiological mechanisms. Annu Rev Physiol. 1995;57:737–769. doi: 10.1146/annurev.ph.57.030195.003513. [DOI] [PubMed] [Google Scholar]
- Grossi L, Montevecchi PC. A kinetic study of S–nitrosothiol decomposition. Chem Eur J. 2002;8:380–387. doi: 10.1002/1521-3765(20020118)8:2<380::AID-CHEM380>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Hall A, Parsonage D, Poole LB, Karplus PA. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J Mol Biol. 2010;402:194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- Hayes JM, Bachrach SM. Effect of micro and bulk solvation on the mechanism of nucleophilic substitution at sulfur in disulfides. J Phys Chem A. 2003;107:7952–7961. [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nature Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Heverly-Coulson GS, Boyd RJ. Reduction of hydrogen peroxide by glutathione peroxidase mimics: reaction mechanism and energetics. J Phys Chem A. 2009;114:1996–2000. doi: 10.1021/jp910368u. [DOI] [PubMed] [Google Scholar]
- Hofstetter D, Nauser T, Koppenol WH. The glutathione thiyl radical does not react with nitrogen monoxide. Biochem Biophys Res Commun. 2007;360:146–148. doi: 10.1016/j.bbrc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Hogg N. The kinetics of S-transnitrosation: a reversible second-order reaction. Anal Biochem. 1999;272:257–262. doi: 10.1006/abio.1999.4199. [DOI] [PubMed] [Google Scholar]
- Hogg N, Singh RJ, Kalyanaraman B. The role of glutathione in the transport and catabolism of nitric oxide. FEBS Lett. 1996;382:223–228. doi: 10.1016/0014-5793(96)00086-5. [DOI] [PubMed] [Google Scholar]
- Honda M, Tajima M. Ab initio study of disulfide bond: Part I. Contribution of d polarization functions. J Mol Struc-Theochem. 1986;136:93–98. [Google Scholar]
- Honda M, Tajima M. Ab initio study of disulphide bond: Part II. The change in atomic distance between sulphur atoms on the reduction of disulphide to dithiol. J Mol Struc-Theochem. 1990;204:247–252. [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Houk NK, Hietbrink BH, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, Stamler JS, Toone EJ. Nitroxyl disulfides, novel intermediates in transnitrosation reactions. J Am Chem Soc. 2003;125:6972–6976. doi: 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- Hudáky I, Gáspári Z, Carugo O, Čemažar M, Pongor S, Perczel A. Vicinal disulfide bridge conformers by experimental methods and by ab initio and DFT molecular computations. Proteins. 2004;55:152–168. doi: 10.1002/prot.10581. [DOI] [PubMed] [Google Scholar]
- Hugo M, Turell L, Manta B, Botti H, Monteiro G, Netto LES, Alvarez B, Radi R, Trujillo M. Thiol and sulfenic acid oxidation of AphE, the one-cystein peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry. 2009;48:9416–9426. doi: 10.1021/bi901221s. [DOI] [PubMed] [Google Scholar]
- Ignarro LJ. Nitric oxide: a unique endogenous signaling molecule in vascular biology (Nobel lecture) Angew Chem Int Ed. 1999;38:1882–1892. doi: 10.1002/(SICI)1521-3773(19990712)38:13/14<1882::AID-ANIE1882>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Incze K, Farkas J, Mihalys V, Zukal E. Antibacterial effect of cysteine-nitrosothiol and possible percursors thereof. Appl Microbiol. 1974;27:202–205. doi: 10.1128/am.27.1.202-205.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao SC, English Ospina SM, Berdis AJ, Starke DW, Post CB, Mieyal JJ. Computational and mutational analysis of human glutaredoxin (thioltransferase): probing the molecular basis of the low pKa of cysteine 22 and its role in catalysis. Biochemistry. 2006;45:4785–4796. doi: 10.1021/bi0516327. [DOI] [PubMed] [Google Scholar]
- Jensen KS, Hansen RE, Winther JR. Kinetic and thermodynamic aspects of cellular thiol-disulfide redox regulation. Antioxid Redox Signal. 2009;11:1047–1058. doi: 10.1089/ars.2008.2297. [DOI] [PubMed] [Google Scholar]
- Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:849–868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourd’heuil D, Jourd’heuil FL, Feelish M. Oxidation and nitrosation of thiols at low micromolar exposure to nitric oxide evidence for a free radical mechanism. J Biol Chem. 2003;278:15720–15726. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- Kassim R, Ramseyer C, Enescu M. Oxidation of zinc-thiolate complexes of biological interest by hydrogen peroxide: a theoretical study. Inorg Chem. 2011;50:5407–5416. doi: 10.1021/ic200267x. [DOI] [PubMed] [Google Scholar]
- Katz BA, Kossiakoff A. The crystallographically determined structures of atypical strained disulfides engineered into subtilisin. J Biol Chem. 1986;261:15480–15485. [PubMed] [Google Scholar]
- Keire DA, Strauss E, Guo W, Noszal B, Rabenstein DL. Kinetics and equilibria of thiol/disulfide interchange reactions of selected biological thiols and related molecules with oxidized glutathione. J Org Chem. 1992;57:123–127. [Google Scholar]
- Kerwin JF, Lancaster JR, Feldman PL. Nitric oxide: a new paradigm for second messengers. J Med Chem. 1995;38:4343–4362. doi: 10.1021/jm00022a001. [DOI] [PubMed] [Google Scholar]
- Kissner R, Nauser T, Bugnon P, Lye PG, Koppenol WH. Formation and properties of peroxynitrite as studied by laser flash photolysis, high-pressure stopped-flow technique, and pulse radiolysis. Chem Res Toxicol. 1997;10:1285–1292. doi: 10.1021/tx970160x. [DOI] [PubMed] [Google Scholar]
- Kóňa J, Brinck T. A combined molecular dynamics simulation and quantum chemical study on the mechanism for activation of the OxyR transcription factor by hydrogen peroxide. Org Biomol Chem. 2006;4:3468–3478. doi: 10.1039/b604602a. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, Beckman JS. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem Res Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002a;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A, VandeVondele J, Rothlisberger U. A Hamiltonian electrostatic coupling scheme for hybrid Car–Parrinello molecular dynamics simulations. J Chem Phys. 2002b;116:6941–6947. [Google Scholar]
- Lee J, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Bravo-Rodriguez K, Phillips C, Seidel RW, Wieberneit F, Stoll R, Doltsinis NL, Sanchez-Garcia E, Sander W. Conformation and dynamics of a cyclic disulfide-bridged peptide: effects of temperature and solvent. J Phys Chem B. 2013;117:3560–3570. doi: 10.1021/jp4007334. [DOI] [PubMed] [Google Scholar]
- Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- Li J, Wang PH, Schlegel HB. A computational exploration of some transnitrosation and thiolation reactions involving CH3SNO, CH3ONO and CH3NHNO. Org Biomol Chem. 2006;4:1352–1364. doi: 10.1039/b600177g. [DOI] [PubMed] [Google Scholar]
- Lim JC, Gruschus JM, Kim G, Berlett BS, Tjandra N, Levine RL. A low pKa cysteine at the active site of mouse methionine sulfoxide reductase A. J Biol Chem. 2012;287:25596–25601. doi: 10.1074/jbc.M112.369116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Miller MJS, Joshi MS, Thomas DD, Lancaster JR. Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc Natl Acad Sci USA. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte M, Carroll KS. The chemistry of thiol oxidation and detection. In: Jakob U, editor. Oxidative stress and redox regulation. 1. New York: Springer; 2012. pp. 1–51. [Google Scholar]
- Lu JM, Wittbrodt JM, Wang K, Wen Z. NO affinities of S-nitrosothiols: a direct experimental and computational investigation of RS-NO bond dissociation energies. J Am Chem Soc. 2001;123:2903–2904. doi: 10.1021/ja000384t. [DOI] [PubMed] [Google Scholar]
- Luo D, Smith SW, Anderson BD. Kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J Pharm Sci. 2005;94:304–316. doi: 10.1002/jps.20253. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S. Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic Biol Med. 2008;44:2013–2018. doi: 10.1016/j.freeradbiomed.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Manta B, Hugo M, Ortiz C, Ferrer-Sueta G, Trujillo M, Denicola A. The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch Biochem Biophys. 2009;484:146–154. doi: 10.1016/j.abb.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid Redox Signal. 2011;15:135–146. doi: 10.1089/ars.2010.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. Analysis and functional prediction of reactive cysteine residues. J Biol Chem. 2012;287:4419–4425. doi: 10.1074/jbc.R111.275578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marletta MA. Nitric oxide synthase structure and mechanism. J Biol Chem. 1993;268:12231–12234. [PubMed] [Google Scholar]
- Miki H, Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem. 2012;151:255–261. doi: 10.1093/jb/mvs006. [DOI] [PubMed] [Google Scholar]
- Moller MN, Li Q, Vitturi DA, Robinson JM, Lancaster JR, Denicola A. Membrane “lens” effect: focusing the formation of reactive nitrogen oxides from the NO/O2 reaction. Chem Res Toxicol. 2007;20:709–714. doi: 10.1021/tx700010h. [DOI] [PubMed] [Google Scholar]
- Mossner E, Huber-Wunderlich M, Glockshuber R. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxido- reductases. Protein Sci. 1998;7:1233–1244. doi: 10.1002/pro.5560070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro AP, Williams DLH. Reactivity of sulfur nucleophiles towards S-nitrosothiols. J Chem Soc Perkin Trans. 2000;2:1794–1797. [Google Scholar]
- Nagababu E, Ramasamy S, Rifkind JM. S-Nitrosohemoglobin: A mechanism for its formation in conjunction with nitrite reduction by deoxyhemoglobin. Nitric Oxide. 2006;15:20–29. doi: 10.1016/j.niox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Nagy P, Ashby MT. Reactive sulfur species: kinetics and mechanisms of the oxidation of cysteine by hypohalous acid to give cysteine sulfenic acid. J Am Chem Soc. 2007;129:14082–14091. doi: 10.1021/ja0737218. [DOI] [PubMed] [Google Scholar]
- Nagy P, Karton A, Betz A, Peskin AV, Pace P, O’Reilly RJ, Winterbourn CC. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide a kinetic and computational study. J Biol Chem. 2011;286:18048–18055. doi: 10.1074/jbc.M111.232355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. Kinetics and mechanisms of thiol-disulfide exchange covering direct substitution and thiol oxidation-mediated pathways. Antioxid Redox Signal. 2013;18:1623–1641. doi: 10.1089/ars.2012.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Xie Q. Regulation of biosynthesis of nitric oxide. J Biol Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- Navrot N, Collin V, Gualberto J, Gelhaye E, Hirasawa M, Rey P, Rouhier N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006;142:1364–1379. doi: 10.1104/pp.106.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MG, Shishido SM, Seabra AB, Morgon NH. Thermal stability of primary S-nitrosothiols: roles of autocatalysis and structural effects on the rate of nitric oxide release. J Phys Chem A. 2002;106:8963–8970. [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kahlili-Araghi F, Tajkhorshid E, Schulten K. Free energy calculation from steered molecular dynamics simulations using Jarzynski’s equality. J Chem Phys. 2003;119:3559–3567. [Google Scholar]
- Parker AJ, Kharasch N. The scission of the sulfur-sulfur bond. Chem Rev. 1959;59:583–628. [Google Scholar]
- Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JK, Boyd RJ. Modeling the reduction of hydrogen peroxide by glutathione peroxidase mimics. J Phys Chem A. 2006;110:8979–8985. doi: 10.1021/jp0615196. [DOI] [PubMed] [Google Scholar]
- Pearson JK, Boyd RJ. Density functional theory study of the reaction mechanism and energetics of the reduction of hydrogen peroxide by ebselen, ebselen diselenide, and ebselen selenol. J Phys Chem A. 2007;111:3152–3160. doi: 10.1021/jp071499n. [DOI] [PubMed] [Google Scholar]
- Perissinotti LL, Estrin DA, Leitus G, Doctorovich FJ. A surprisingly stable S-nitrosothiol complex. J Am Chem Soc. 2006;128:2515–2513. doi: 10.1021/ja0565976. [DOI] [PubMed] [Google Scholar]
- Perissinotti LL, Leitus G, Shimon L, Estrin DA, Doctorovich F. A unique family of stable and water-soluble S-nitrosothiol complexes. Inorg Chem. 2008;47:4723–4733. doi: 10.1021/ic7024999. [DOI] [PubMed] [Google Scholar]
- Perissinotti LL, Turjanski AG, Estrin DA, Doctorovich F. Transnitrosation of nitrosothiols: characterization of an elusive intermediate. J Am Chem Soc. 2005;127:486–487. doi: 10.1021/ja044056v. [DOI] [PubMed] [Google Scholar]
- Perry LJ, Wetzel R. Disulfide bond engineered into T4 lysozyme: stabilization of the protein toward thermal inactivation. Science. 1984;226:555–557. doi: 10.1126/science.6387910. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Cox AG, Nagy P, Morgan PE, Hampton MB, Davies MJ, Winterbourn CC. Removal of amino acid, peptide and protein hydroperoxides by reaction with peroxiredoxins 2 and 3. Biochem J. 2010;432:313–321. doi: 10.1042/BJ20101156. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Dickerhof N, Poynton RA, Paton LN, Pace PE, Hampton MB, Winterbourn CC. Hyperoxidation of peroxiredoxins 2 and 3: rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. J Biol Chem. 2013;288:14170–14177. doi: 10.1074/jbc.M113.460881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin AV, Winterbourn CC. Kinetics of the reactions of hypochlorous acid and amino acid chloramines with thiols, methionine, and ascorbate. Free Radic Biol Med. 2001;30:572–579. doi: 10.1016/s0891-5849(00)00506-2. [DOI] [PubMed] [Google Scholar]
- Petruk AA, Bartesaghi S, Trujillo M, Estrin DA, Murgida D, Kalyanaraman B, Marti M, Radi R. Molecular basis of intramolecular electron transfer in proteins during radical-mediated oxidations: Computer simulation studies in model tyrosine–cysteine peptides in solution. Arch Biochem Biophys. 2012;525:82–91. doi: 10.1016/j.abb.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prütz WA, Butler J, Land EJ, Swallow AJ. The role of sulphur peptide functions in free radical transfer: a pulse radiolysis study. Int J Radiat Biol. 1989;55:539–556. doi: 10.1080/09553008914550591. [DOI] [PubMed] [Google Scholar]
- Pryor WA, Squadrito GL. The chemistry of peroxynitrite: a product from the reaction of nitric oxide with superoxide. Am J Physiol. 1995;268:699–722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- Rao BNN, Kumar A, Balaram H, Ravi A, Balaram P. Nuclear Overhauser effects and circular dichroism as probes of β-turn conformations in acyclic and cyclic peptides with pro-X sequences. J Am Chem Soc. 1983;105:7423–7428. [Google Scholar]
- Reyes AM, Hugo M, Trostchansky A, Capece L, Radi R, Trujillo M. Oxidizing substrate specificity of Mycobacterium tuberculosis alkyl hydroperoxide reductase E: kinetics and mechanisms of oxidation and overoxidation. Free Radic Biol Med. 2011;51:464–473. doi: 10.1016/j.freeradbiomed.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo H. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Rockett KA, Awburn MM, Cowden WB, Clark IA. Killing of Plasmodium falciparum in vitro by nitric oxide derivatives. Infect Immun. 1991;59:3280–3283. doi: 10.1128/iai.59.9.3280-3283.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N, Radi R, Linares E, Augusto O, Detweiler CD, Mason RP, Denicola A. Reaction of human hemoglobin with peroxynitrite isomerization to nitrate and secondary formation of protein radicals. J Biol Chem. 2003;278:44049–44057. doi: 10.1074/jbc.M305895200. [DOI] [PubMed] [Google Scholar]
- Roos G, Foloppe N, Messens J. Understanding the pKa of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- Roos G, De Proft F, Geerlings P. Electron capture by the thiyl radical and disulfide bond: ligand effects on the reduction potential. Chem Eur J. 2013;19:5050–5060. doi: 10.1002/chem.201203188. [DOI] [PubMed] [Google Scholar]
- Roos G, Foloppe N, Van Laer K, Wyns L, Nilsson L, Geerlings P, Messens J. How thioredoxin dissociates its mixed disulfide. PloS Comp Biol. 2009;5:1000461. doi: 10.1371/journal.pcbi.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng T-C, Hinks J, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- Salsbury FR, Poole LB, Fetrow JS. Electrostatics of cysteine residues in proteins: parameterization and validation of a simple model. Proteins. 2012;80:2583–2591. doi: 10.1002/prot.24142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsbury FR, Yuan Y, Knaggs MH, Poole LB, Fetrow JS. Structural and electrostatic asymmetry at the active site in typical and atypical peroxiredoxin dimers. J Phys Chem B. 2012;116:6832–6843. doi: 10.1021/jp212606k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Riddle M, Woo J, Momand J. Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein Sci. 2008;17:473–481. doi: 10.1110/ps.073252408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma BK, Mugesh G. Redox regulation of protein tyrosine phosphatase 1B (PTP1B): a biomimetic study on the unexpected formation of a sulfenyl amide intermediate. J Am Chem Soc. 2007;129:8872–8881. doi: 10.1021/ja070410o. [DOI] [PubMed] [Google Scholar]
- Savojardo C, Fariselli P, Martelli PL, Shukla P, Casadio R. Prediction of the bonding state of cysteine residues in proteins with machine-learning methods. Lect Notes Comput Sc. 2011;6685:98–111. [Google Scholar]
- Schmid N, Eichenberger A, Choutko A, Riniker S, Winger M, Mark A, van Gunsteren W. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- Schrammel A, Gorren ACF, Schmidt K, Pfeiffer S, Mayer B. S-nitrosation of glutathione by nitric oxide, peroxynitrite, and.NO/O2-. Free Radical Biol Med. 2003;34:1078–1088. doi: 10.1016/s0891-5849(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Behera RN, Smith JC, Ullmann GM. The alpha helix dipole: screened out? Structure. 2005;13:849–855. doi: 10.1016/j.str.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- Seo YH, Carroll KS. Facile synthesis and biological evaluation of a cell-permeable probe to detect redox-regulated proteins. Bioorg Med Chem Lett. 2009;19:356–359. doi: 10.1016/j.bmcl.2008.11.073. [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- Singh R, Whitesides GM. Comparisons of rate constants for thiolate−disulfide interchange in water and in polar aprotic solvents using dynamic proton NMR line shape analysis. J Am Chem Soc. 1990;112:1190–1197. [Google Scholar]
- Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. The chemistry of the S-nitrosoglutathione/glutathione system. Proc Natl Acad Sci USA. 1996;93:14448–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Keerthi K, Gates KS. A chemical model for redox regulation of protein tyrosine phosphatase 1B (PTP1B) activity. J Am Chem Soc. 2005;127:10830–10831. doi: 10.1021/ja052599e. [DOI] [PubMed] [Google Scholar]
- Sjöberg L, Eriksen TE, Révész L. The reaction of the hydroxyl radical with glutathione in neutral and alkaline aqueous solution. Radiat Res. 1982;89:255–263. [PubMed] [Google Scholar]
- Søndergaard CR, Olsson HMM, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Srinivasan N, Sowdhamini R, Ramakrishnan C, Balaram P. Conformations of disulfide bridges in proteins. Int J Pept Protein Res. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]
- Stacey MM, Vissers MC, Winterbourn CC. Oxidation of 2-cys peroxiredoxins in human endothelial cells by hydrogen peroxide, hypochlorous acid, and chloramines. Antioxid Redox Signal. 2012;17:411–421. doi: 10.1089/ars.2011.4348. [DOI] [PubMed] [Google Scholar]
- Stubauer G, Guffre A, Sarti P. Mechanism of S-nitrosothiol formation and degradation mediated by copper ions. J Biol Chem. 1999;274:28128–28133. doi: 10.1074/jbc.274.40.28128. [DOI] [PubMed] [Google Scholar]
- Stubbe J, van Der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Fukushi K, Ikawa S, Konaka S. Vibrational spectra and conformation of diallyl disulphide in the liquid state. J Mol Struc. 1990;221:141–148. [Google Scholar]
- Swarts SG, Becker D, DeBolt S, Sevilla MD. Electron spin resonance investigation of the structure and formation of sulfinyl radicals: reaction of peroxyl radicals with thiols. J Phys Chem. 1989;93:155–161. [Google Scholar]
- Szacilowski A, Chmura SZ. Interplay between iron complexes, nitric oxide and sulfur ligands: structure, (photo) reactivity and biological importance. Coord Chem Rev. 2005;249:2408–2436. [Google Scholar]
- Talipov MR, Timerghazin QK. Protein control of S-nitrosothiol reactivity: interplay of antagonistic resonance structures. J Phys Chem B. 2013;117:1827–1837. doi: 10.1021/jp310664z. [DOI] [PubMed] [Google Scholar]
- Tasker HS, Jones HO. The action of mercaptans on acid chlorides. Part II. The acid chlorides of phosphorus, sulphur, and nitrogen. J Chem Soc. 1909;95:1910–1918. [Google Scholar]
- Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timerghazin QK, Talipov MR. Unprecedented external electric field effects on S-nitrosothiols: possible mechanism of biological regulation? J Phys Chem Lett. 2013;4:1034–1038. doi: 10.1021/jz400354m. [DOI] [PubMed] [Google Scholar]
- Timerghazin QK, Pelsherbe GH, English AM. Resonance description of S-nitrosothiols: insights into reactivity. Org Lett. 2007;9:3049–3052. doi: 10.1021/ol0711016. [DOI] [PubMed] [Google Scholar]
- Timerghazin QK, English AM, Peslherbe GH. On the multireference character of S-nitrosothiols: a theoretical study of HSNO. Chem Phys Lett. 2008;454:24–29. [Google Scholar]
- Timerghazin QK, Peslherbe GH, English AM. Structure and stability of HSNO, the simplest S-nitrosothiol. Phys Chem Chem Phys. 2008;10:1532–1539. doi: 10.1039/b715025c. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Clippe A, Manta B, Ferrer-Sueta G, Smeets A, Declercq JP, Knoops B, Radi R. Pre-steady state kinetic characterization of human peroxiredoxin 5: taking advantage of Trp84 fluorescence increase upon oxidation. Arch Biochem Biophys. 2007;467:95–106. doi: 10.1016/j.abb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Radi R. Peroxynitrite reaction with the reduced and the oxidized forms of lipoic acid: new insights into the reaction of peroxynitrite with thiols. Arch Biochem Biophys. 2002;397:91–98. doi: 10.1006/abbi.2001.2619. [DOI] [PubMed] [Google Scholar]
- Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Durán R, Freeman BA, Radi R, Alvarez B. Reactivity of sulfenic acid in human serum albumin. Biochemistry. 2008;47:358–367. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52:2708–2728. doi: 10.1021/bi400215w. [DOI] [PubMed] [Google Scholar]
- van Gastel M, Lubitz W, Lassmann G, Neese F. Electronic structure of the cysteine thiyl radical: a DFT and correlated ab initio study. JAm Chem Soc. 2004;126:2237–2246. doi: 10.1021/ja038813l. [DOI] [PubMed] [Google Scholar]
- Wada A (1976) The alpha-helix as an electric macro-dipole. Adv Biophys 1–63 [PubMed]
- Wang K, Wen Z, Zhang W, Xian M, Cheng JP, Wang PG. Equilibrium and kinetics studies of transnitrosation between S-nitrosothiols and thiols. Bioorg Med Chem Lett. 2001;11:433–436. doi: 10.1016/s0960-894x(00)00688-0. [DOI] [PubMed] [Google Scholar]
- Wang PG, Xian M, Tang X, Wu X. Nitric oxide donors: chemical activities and biological applications. Chem Rev. 2002;102:1091–1134. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- Warshel A, Levitt M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Weinhold F, Landis CR, Valency, Bonding . A natural bond orbital donor-aceptor perspective. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Wennmohs F, Staemmler V, Schindler M. Theoretical investigation of weak hydrogen bonds to sulfur. J Chem Phys. 2003;119:3208–3218. [Google Scholar]
- Wetzel R, Perry LJ, Baase WA, Becktel WJ. Disulfide bonds and thermal stability in T4 lysozyme. Proc Natl Acad Sci U S A. 1988;85:401–405. doi: 10.1073/pnas.85.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Amour CVS, Collins JL, Ringe D, Petsko GA. The 1.8-Å resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily. Proc Natl Acad Sci USA. 2004;101:1531–1536. doi: 10.1073/pnas.0308089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink DA, Nims RW, Darbyshire JF, Christodoulou D, Hanbauer I, Cox GW, Laval F, Laval J, Cook JA. Reaction kinetics for nitrosation of cysteine and glutathione in aerobic nitric oxide solutions at neutral pH. Insights into the fate and physiological effects of intermediates generated in the NO/O2 reaction. Chem Res Toxicol. 1994;7:519–525. doi: 10.1021/tx00040a007. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. In: Oxidative stress and redox regulation. 1. Jakob U, Reichmann D, editors. Netherlands: Springer; 2013. pp. 43–58. [Google Scholar]
- Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Metodiewa D. The reaction of superoxide with reduced glutathione. Arch Biochem Biophys. 1994;314:284–290. doi: 10.1006/abbi.1994.1444. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, et al. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics. 2008;8:3139–3153. doi: 10.1002/pmic.200701062. [DOI] [PubMed] [Google Scholar]
- Wong PSY, Hyun J, Fukuto JM, Shirota FN, DeMaster EG, Shoeman DW, Nagasawa HT. Reaction between S-nitrosothiols and thiols: generation of nitroxyl (HNO) and subsequent chemistry. Biochemistry. 1998;37:5362–5371. doi: 10.1021/bi973153g. [DOI] [PubMed] [Google Scholar]
- Yang A-S, Gunner MR, Sampogna R, Sharp K, Honig B. On the calculation of pKas in proteins. Proteins. 1993;15:252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- Yang J, Groen A, Lemeer S, Jans A, Slijper M, et al. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Knaggsa MH, Poolec LB, Fetrowab JS, Salsbury FR. Conformational and oligomeric effects on the cysteine pK(a) of tryparedoxin peroxidase. J Biomol Struct Dyn. 2010;28:51–70. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeida A, Babbush R, González Lebrero MC, Trujillo M, Radi R, Estrin DA. Molecular basis of the mechanism of thiol oxidation by hydrogen peroxide in aqueous solution: challenging the SN2 paradigm. Chem Res Toxicol. 2012;25:741–746. doi: 10.1021/tx200540z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeida A, González Lebrero MC, Trujillo M, Radi R, Estrin DA. Mechanism of cysteine oxidation by peroxynitrite: An integrated experimental and theoretical study. Arch Biochem Biophys. 2013;539:81–86. doi: 10.1016/j.abb.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Zhang T, Bertelsen E, Alber T. Entropic effects of disulphide bonds on protein stability. Nat Struct Biol. 1994;1:434–438. doi: 10.1038/nsb0794-434. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen J. Efficiency of adaptive temperature-based replica exchange for sampling large-scale protein conformational transitions. J Chem Theory Comp. 2013;9:2849–2856. doi: 10.1021/ct400191b. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu Y, Joseph J, Kalyanaraman B. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides treated with myeloperoxidase, H2O2, and NO2-EPR spin trapping studies. J Biol Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]