Abstract
Arginine kinase is an essential enzyme which is closely related to energy metabolism in marine invertebrates. Arginine kinase provides a significant role in quick response to environmental change and stress. In this study, we simulated a tertiary structure of Sepia pharaonis arginine kinase (SPAK) based on the gene sequence and conducted the molecular dynamics simulations between SPAK and Zn2+. Using these results, the Zn2+ binding sites were predicted and the initial effect of Zn2+ on the SPAK structure was elucidated. Subsequently, the experimental kinetic results were compared with the simulation results. Zn2+ markedly inhibited the activity of SPAK in a manner of non-competitive inhibitions for both arginine and ATP. We also found that Zn2+ binding to SPAK resulted in tertiary conformational change accompanying with the hydrophobic residues exposure. These changes caused SPAK aggregation directly. We screened two protectants, glycine and proline, which effectively prevented SPAK aggregation and recovered the structure and activity. Overall, our study suggested the inhibitory effect of Zn2+ on SPAK and Zn2+ can trigger SPAK aggregation after exposing large extent of hydrophobic surface. The protective effects of glycine and proline against Zn2+ on SPAK folding were also demonstrated.
Keywords: Aggregation, arginine kinase, Zn2+, inhibition, molecular dynamics simulation, Sepia pharaonis
1. INTRODUCTION
The cuttlefish Sepia pharaonis belongs to cephalopods, and it is an important marine resource for human consumption [1]. Sepia pharaonis has excellent breeding prospects based on its characteristics of larger body type, fast growth and suitability for high-density cultivation. At present, research into the artificial breeding of Sepia pharaonis is being carried out in China. It has shown that Sepia pharaonis is particularly sensitive to environmental changes in its breeding environment [2]. Changes in salinity and heavy metal concentration cause a severe stress reaction, including stress-induced inking behavior, and can even lead to massive death. Thus, it is important to study the stress response of cephalopods and their defense mechanisms under different environmental conditions.
Previous studies have indicated that energy metabolism-related enzymes in invertebrates are importantly responsible for adapting the environmental stress. In this regard, arginine kinase (AK, EC 2.7.3.3) is distinctively found in marine invertebrates to be associated with adapting the environmental troubles caused from physical and chemical factors [3-6]. AK is a phosphagen kinase that catalyzes the reversible reaction of phosphoryl group’s transfer from various sources of phosphagens [7, 8]. It has been generally recognized that AK plays a pivotal role in ATP buffering in invertebrates both temporal and extreme conditions where muscle and nerves cells require immediate and high fluctuating energy demands via catalyzing Mg2+ cofactor dependent phosphoryl transfer [9, 10]. The evolution of a marine organism’s metabolic patterns generally involves further adaptation to the aquatic environment and this was supported by the homologous sequence alignments of various origins of AKs [11-13]. Some of divalent metal ions (i.e. Cd2+, Cu2+ and Pb2+) were found to have significant inhibitory effects on AK catalysis, which could directly affect the body’s energy balance and destroy energy homeostasis in invertebrates [5, 14-15].
Cuttlefish muscle is a good food source of some important essential minerals [16-18]. However, the increasing pollution of seawater by metal ions often leads to excessive intracellular unfavorable accumulation. Excessive Zn2+ can be toxic to marine organisms regardless of that Zn2+ is a basic ingredient of several important enzymes and displays multiple functions in the catalytic reaction [19-20]. It is evident that Zn2+ binding directly mediates hydrophobic exposure of enzyme active site pocket and structural unfolding, as well as unfavorable aggregation in a broad variety of metallic/non-metallic enzymes, including creatine kinase and AK [21,22]. The roles of Zn2+ in ocean invertebrates have not been well elucidated, but it has been found that Zn2+ concentrations in the body of marine organisms, especially for marine invertebrates, are more excessive than other organisms’ metabolisms [23-25]. The reason of Zn2+ can exist excess concentration in tissues of marine invertebrates is that it is prone to bind to macromolecules or present as insoluble metal inclusions in tissues [26]. It has rarely been reported the role of Zn2+ on enzymes’ structure and function in aquatic organisms and thus, it could be a topic of interest to investigate the role of excess Zn2+ on enzyme derived from aquatic organism such as Sepia pharaonis that is one of food source for human. Accordingly, we elucidated the inhibitory effect of Zn2+ on arginine kinase from Sepia pharaonis (SPAK) in this study and the protective functional role of some osmolytes against Zn2+ in controlling changes in enzyme structure and function and combined these studies with ORF gene sequence analysis and computational simulations. Exploration the basic characteristics of the energy metabolism related enzymes might provide significant message on the metabolic adaptation mechanisms during environmental stress.
2. MATERIALS AND METHODS
2.1. Materials
Magnesium acetate, thymol blue, ATP, arginine, ANS, zinc acetate dehydrate were obtained from Sigma-Aldrich (Shanghai, China). Sephacryl S-200 and Cellulose DE-52 were purchased from GE Healthcare (Piscataway, NJ, USA) and Whatman (GE Healthcare), respectively. The other chemicals were the analytical grade and locally obtained.
2.2. cDNA Cloning of SPAK and RACE
Total RNA was extracted from tissue samples of healthy adult S. Pharaonis using a Trizol RNA extraction reagent (Invitrogen, Carlsbad, USA). Oligo (dT) primer was used to initiate the reverse transcription process, and cDNA synthesis was conducted by M-MLV reverse transcriptase (Takara, Shiga, Japan). Partial fragments of the AK gene from the prepared cDNA were obtained with primers AK-F and AK-R (Table 1). The purified PCR products were subcloned into pMD18-T vector (Takara). Positive clones were sequenced.
Table 1.
The Primers Used for RACE and cDNA Cloning of SPCK.
| Primer | Sequence (5’—3’) |
|---|---|
| AK-F | TTGCTGAAGTCCTTGATGCYGT |
| AK-R | TCATGGTRGTACCCAAGTTGC |
| 3’RACE1 | TGGCAGAGGAATTTACTTCAACCAGGACA |
| 3’RACE2 | GAAGAAATTGACCTTTGCCAAGAAGGATAACA |
| 5’RACE1 | CAGTCAGCTTCTGTTGAGTGTCTGGGGT |
| 5’RACE2 | GCCTGCAGAGCTTCAACTGAGACCTTC |
| 5’RACE Outer Primer | GCCACGCGTCGACTAGTACGGGGGGGGGG |
| 5’RACE Inner Primer | GGCCACGCGTCGACTAGTAC |
The missing cDNA sequences of the SPAK gene were then obtained by 3’-RACE and 5’-RACE using RACE Kit in accordance with the manufacturer’s instructions (Takara). For 3’ RACE, RT product was amplified with nested forward PCR primers (3’ RACE1 and 3’ RACE2) to obtain the partial sequence and an adaptor primer. PCR was performed under hot-start conditions (95ºC, 3 min) for 33 cycles of 94ºC 30 s, 58ºC 30 s, and 72ºC 60 s, and then 7 min at 72ºC. The oligo-dC primer and terminal deoxynucleotidyl transferase was used to amplify the 5’-half of the SPAK cDNA. Outer PCR was carried out by 5’RACE1 and 5’RACE Outer Primer. Inner PCR was conducted by 5’RACE2 and 5’RACE Inner Primer. PCR amplification was performed for 33 cycles of 94ºC 30 s, 68ºC 30 s, and 72ºC 60 s, and then 7 min at 72ºC. Finally, the amplified products were subcloned and sequenced, respectively. The sequences were spliced and analyzed using BLAST in the GenBank database.
2.3. Computational Simulations of SPAK
The pseudo quadratic restraint with simulated annealing method (PQR-SA) [27] was used to establish a SPAK homology model. A knowledge-based statistical energy potential (STAP) was used [28] for higher protein like scores. All simulations and analyses were used in the CHARMM program. To validate the structure model, the radius of gyration of the protein and the associated protein-like scores were measured. A functional study of the structure, dockable pocket site prediction (DPSP) was used and identified the appropriate ligand-binding pocket. To understand the role of Zn2+ in SPAK, molecular dynamics simulations (MD) were prepared with the action of Zn2+. The experimental concentration was calculated, and eight Zn2+ ions were randomly added in the simulation. Ten-nanosecond (ns) MD simulations were carried out and the atomic coordinates were saved every 1 picosecond (ps) for further analyses. The protein stabilities were checked by root mean square deviation of the alpha carbon relative to its initiating structure. The bound Zn2+ ions were counted as a function of time. Protein flexibilities were checked based on the feature of residue number by root mean square fluctuation (RMSF). The final structures after 10 ns MD simulations were applied to determine the secondary structure and to compare the two structures with and without Zn2+. The secondary structure ratio was tested by DSSP [29].
2.4. SPAK Purification and Activity Assay
SPAK enzyme samples were extracted and purified from the muscle of cuttlefish by homogenate, cellulose DE-52 ion-exchange chromatography and Sephacryl S-200 gel chromatography according the previous procedures [30,31]. AK activity was determined by monitoring H+ generation in the catalytic reaction of ATP and arginine with an indicator thymol blue as the previous reported methods [31,32]. The absorption change value per minute was measured at 575 nm by a Shimadzu UV-1800 spectrophotometer. The OD value at 280 nm was measured to calculate the protein content using the corresponding standard curve based on bull serum albumin.
2.6. Kinetic Analysis
The Lineweaver-Burk equation of non-competitive inhibition in double-reciprocal form is as follows, and it was used to analysis the inhibition type and kinetics:
| (1) |
The secondary replot can be established from the following equation:
Y-intercept
| (2) |
The values including Vmax, Km and Ki, can be calculated from the two equations and the corresponding figures. When the obtained data in the secondary replot shown in Eq. (2) is linearly well fitted, it assumes that the inhibitor binding site is a single class.
2.7. Protein Unfolding Measurement Induced by Zn2+
For the protein unfolding measurement, various concentrations of Zn2+ was incubated with SPAK for 2 h at 20 °C. Fluorescence emission spectra were measured using a F-4500 spectrofluorometer (Hitachi, Japan) according the previous procedures [33]. Intrinsic fluorescence was measured following excitation at 280 nm. The emission wavelength was from 300 to 400 nm. Zn2+-treated SPAK was incubated with 40 μM ANS for 30 min in the dark to probe the hydrophobic surface. The excitation and emission wavelengths for ANS-binding fluorescence of SPAK were 380 nm and 420-600 nm, respectively. The Zn2+ mediated aggregation of SPAK was followed by the absorbance record at 400 nm. All above measurements were performed in the buffer (20 mM Tris-acetic acid, pH 8.0).
RESULTS
3.1. RACE and Gene Cloning of SPAK
The SPAK gene sequences were successfully amplified and submitted to GenBank under the accession number 1799738. The SPAK cDNA comprises 1251 bp with an 1050 bp open reading frame. The BLAST results displayed more than 80% similarity to AKs of Sepiella maindroni (AEK26855.1), Sepioteuthis lessoniana (BAA95610.1), Octopus vulgaris (BAA95609.1) and Amphioctopus fangsiao (AEK65120.1) (Fig. 1a). Phylogenetic analysis result displayed that SPAK sequences closely clustered with the homologous AKs from mollusca followed by cuttlefish, octopus, chiton, oyster, conch and clam (Fig. 1b).
Figure 1.
Multiple alignment and neighbor-joining tree of SPAK. (A) Multiple sequence alignments of SPAK with five other homologous arginine kinase amino acid sequences. (B) Phylogenetic relationship of the AK amino acid sequences. MEGA4 software was used to construct the neighbor-joining tree.
3.2. Homology Modeling and Molecular Dynamics Simulations of SPAK
A SPAK structure model was created using the homology modeling method (Fig. 2a). The template structures used were 1m15, 1qh4, 3jpz, 3l2f, 2k9i, 2ba3, 1p94, and 2cpg (PDB entry codes). The best template was 1m15 with a sequence identity of 55%. The radius of gyration of SPAK was lower than the standard value of the same size of SPAK (green dot in Fig. 2b). This indicates that the model was more tightly packed than the X-ray structures. The protein like scores were measured and marked on the high resolution X-ray structure space (Fig. 2c). The clash and Ramachandran related scores (WhatRama, MolRama, ProRama, and rotamer) had good indicators comparable to the X-ray experimental structures. From these results, the developed homology model was shown to be a useful protein model for further study, such as docking and simulations.
Figure 2.
Homology protein model of SPAK. (A) The developed homology model of SPAK. The protein structure is drawn as a cartoon image colored red (α helix) and yellow (β sheet). (B) The radius of gyration (Rg) is indicated by total residue number. The SPAK structure is indicated by the green dot. The best fitting curve for X-ray structures is shown in blue. (C) The protein like scores of the SPAK homology model structure in X-ray structure space. The shaded grey circle represents the high 50% X-ray structure scores.
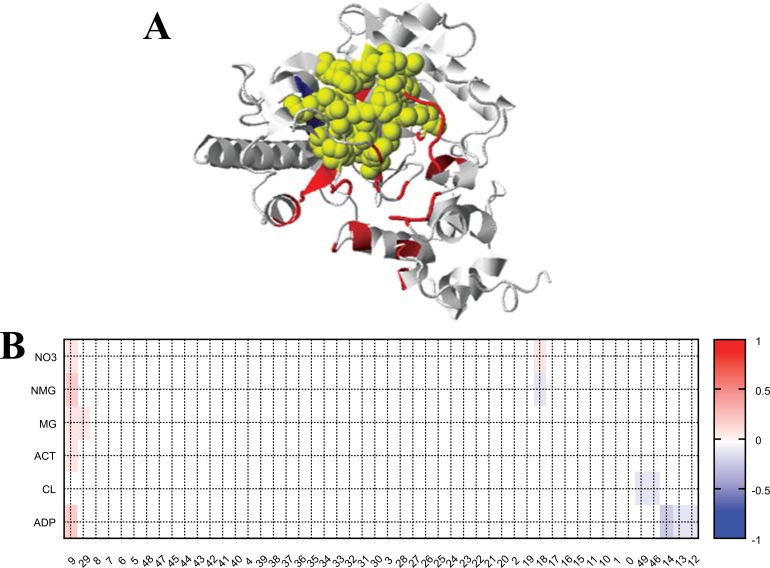
The DPSP server was used to predict plausible ligands for SPAK. DPSP identified an ADP binding site with high DPSP score (Fig. 3). The interacting residues are shown in Figure 3a as yellow spheres (residue numbers: 118 120 176 179 181 215 218 223 227 274 278 302 304 305 306 307 317).
Figure 3.
Predicted ADP binding site of SPAK. (A) SPAK protein structure with plausible ADP binding sites (shown as yellow spheres in the structure view and yellow circles in the heat map figure). (B) The heat map of DPSP scores. The darker red color indicates a higher affinity binding site. The X- and Y-axes represent pocket numbers and found ligand lists. (The color version of the figure is available in the electronic copy of the article).
After 10 ns simulations, protein flexibility and Zn bound ligand residues were sought (Fig. 4a). The Zn interacting residues are highlighted in red at the bottom of Fig. 4a. The ADP binding sites are shown marked with blue at the bottom of Fig. 4a. Two Zn2+ overlapped in the ADP binding sites, while the other five Zn2+ didn’t overlap (Fig. 4b). Thus the Zn2+ can interfere with SPAK, and may cause inactivity of
Figure 4.
Structural analyses of SPAK with Zn2+. (A) RMSF plot with (red line)/without (green line) Zn2+. The Zn2+ bound sites are marked by squares in the bottom plot (red: final structure, green: initial structure, and blue: plausible ADP binding site predicted by DPSP). (B) The final structure with Zn2+. ADP binding sites and Zn2+ are indicated by blue and brown spheres, respectively. (C) The final structures with (red) and without (blue) bound Zn2+. (D) The secondary structure schemes showing the differences between the final structures. Str1/str2 represent with/without Zn2+, respectively. The α-helix, β-sheet and coil are indicated by red, yellow and blue patterns, respectively. (The color version of the figure is available in the electronic copy of the article).
SPAK at a high concentration of Zn2+. The final two structures were superimposed (Fig. 4c) and the secondary structures were aligned according to the residues (Fig. 4d). The backbone similarity of the two structures is 3.2 Å. The
secondary structure ratios of the two structures are very similar, and no great structural change was found although the first simulation was in the presence of Zn2+. In subsequent kinetic studies, we attempted to validate the simulation results with respect to activity modulation and conformational changes due to Zn2+ binding.
3.3. SPAK Activity Inhibition by Zn2+
SPAK was mixed with different concentrations of Zn2+ for 2 h at 20 °C and measured the relative activity. The results showed that Zn2+ inhibited AK activity with a concentration related pattern (Fig. 5). Relative SPAK activity remained about 15% when the Zn2+ concentration was greater than 0.03 mM (Fig. 5a). Measurement of the IC50 gave a value of 10.6 ± 0.6 µM (n = 3). SPAK could be refolded due to the dilution effect in the Zn2+ absent substrate system (Fig. 5b). The relative SPAK activity remained about 25% when the Zn2+ concentration was greater than 0.03 mM. The IC50 value was 11.2 ± 0.8 µM (n = 3).
Figure 5.
The activity inhibition of Zn2+ on SPAK. Different concentrations of Zn2+ was incubated with SPAK for 2 h in buffer (20 mM Tris-acetic acid, pH 8.0) at 20°C, followed the relative activity measurement with corresponding Zn2+ concentrations (A) or without Zn2+ (B). The final concentration of SPAK was 0.06 μM. Data are presented as average values, n=3.
3.4. Noncompetitive Inhibition Induced by Zn2+
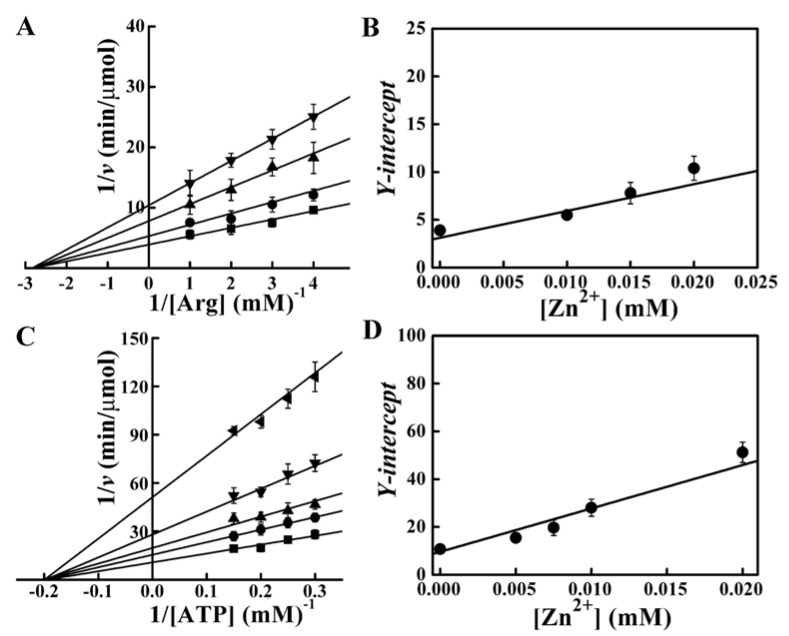
Double-reciprocal Lineweaver–Burk plots were used to determine the inhibition type and mechanisms of Zn2+. The results showed changes in the values of apparent Vmax. The inhibition induced by Zn2+ was non-competitive mode for arginine (Fig. 6a) and ATP (Fig. 6c). The secondary plot (Y-intercept versus [Zn2+]) was linear (Fig. 6b and Fig. 6d), indicating that the SPAK inhibition site of Zn2+ was single or single class. The Ki values were 11.17 ± 0.87 mM for arginine and 5.31 ± 0.61 mM for ATP. The dynamic constants of SPAK were Km = 0.36 ± 0.08 mM and Vmax = 57.81 ± 6.72 U/mg for arginine and Km = 5.11 ± 0.75 mM and Vmax = 46.78 ± 4.96 U/mg for ATP. The result of noncompetitive inhibition indicated that Zn2+ could bind SPAK, and that Zn2+ might reduce the enzyme activity by impacting its tertiary structure. To verify our supposition, the tertiary structure changes of SPAK incuced by Zn2+ were measured.
Figure 6.
Double-reciprocal plot in the presence of Zn2+. (A) Lineweaver-Burk plot for arginine. Zn2+ concentrations were 0 (■), 0.01 (●), 0.015 (▲) and 0.02 (▼) mM, respectively. The concentration of ATP was 5 mM, and the final concentration of the enzyme was 0.12 μM. (B) The secondary plot for arginine. Data was derived from (A). (C) Lineweaver-Burk plot for ATP. Zn2+ concentrations were 0 (■), 0.005 (●), 0.075 (▲), 0.01 (▼) and 0.02 (◄) mM, respectively. The concentration of arginine was 5.7 mM, and the final concentration of the enzyme was 0.06 μM. (D) The secondary plot for ATP. Data was derived from (C).
3.5. Zn2+-Induced Tertiary Structural Change and Aggregation of SPAK
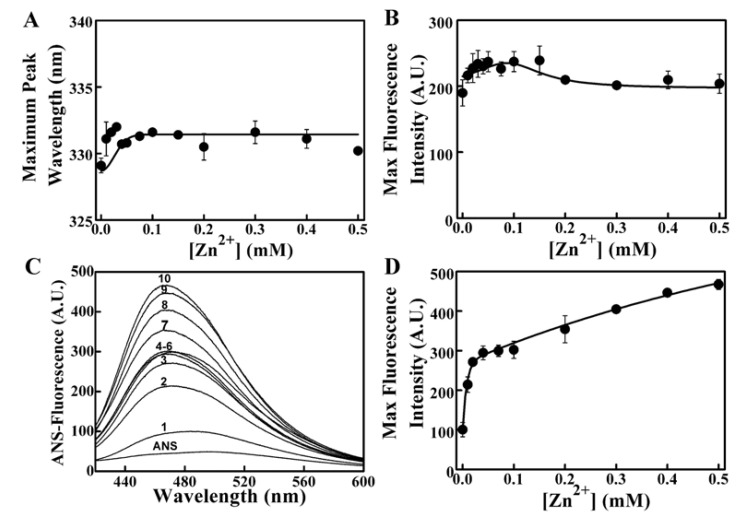
The intrinsic and ANS-binding fluorescence measurements were conducted to determine the tertiary structural changes of SPAK treated with different concentrations of Zn2+. Addition of Zn2+ at different concentrations resulted in a slight red-shift from 329 to 332 nm according to the maximum emission peak wavelength of intrinsic fluorescence (Fig. 7a). The intensity changed from about 190 A.U. to 240 A.U., while the concentration of Zn2+ changed from 0 to 0.15 mM, and then declined to about 200 A.U. (Fig. 7b). This finding indicated that the binding of Zn2+ did loosen the structure of SPAK and slightly induced the tertiary structural change. The ANS-binding fluorescence spectra measurements showed that Zn2+ significantly enhanced the ANS-binding fluorescence of the enzyme with the increase of concentration. The resulted indicated that the combination of enzyme with Zn2+ induced hydrophobic surface exposure (Figs. 7c and 7d). The hydrophobic disruption of SPAK could directly lead to protein misfolding and aggregation with the loss of enzymatic activity. Next, Zn2+-mediated SPAK aggregation was tested and the results showed that SPAK aggregation increased remarkably as the Zn2+ concentration increased (Fig. 8).
Figure 7.
Fluorescence changes of SPAK induced by Zn2+. (A) Maximum peak wavelength of intrinsic fluorescence versus [Zn2+]. (B) Maximum intrinsic fluorescence intensity versus [Zn2+]. (C) ANS-binding fluorescence changes of SPAK. The labels 1 to 10 indicate 0, 0.01, 0.02, 0.04, 0.07, 0.1, 0.2, 0.3, 0.4 and 0.5 mM Zn2+. (D) Maximum ANS-fluorescence intensity changes. Data was derived from (C). The final concentration of SPAK was 5 μM.
Figure 8.
SPAK aggregation induced by Zn2+. Labeled 1-6 represented 0, 0.05, 0.1, 0.2, 0.3 and 0.5 mM Zn2+, respectively. The final concentration of SPAK was 17 μM and the measurement was carried out at 20 °C.
3.6. Screening for Protective Osmolytes on the Inactivation of SPAK
Osmolytes can act as folding chaperones and protect the enzyme activity by facilitating the correct folding of protein. We screened the protective agents for Zn2+ mediated inactivation from five common osmolytes (glycine, proline, glycerol, liquaemin and sucrose). The experiment results showed that glycine and proline were efficient protective agents for SPAK activity among these osmolytes. SPAK was mixed with 0.03 mM Zn2+ (an effective SPAK-inactivated concentration) and the different concentrations of osmolytes, respectively. The activity of SPAK was significantly restored by glycine and proline in a concentration dependent manner (Figs. 9a and 9b). At a glycine concentration of 50 mM, 86% of the activity was restored. Surprisingly, approximately 100% of the enzyme activity was restored after incubation with 120 mM proline. However, glycerol, sucrose and liquaemin showed no protective function for SPAK activity in Zn2+ mediated inactivation (Figs. 9c to 9e).
Figure 9.
Osmolytes screening for SPAK reactivation. Different concentrations of glycine (A), proline (B), glycerol (C), sucrose (D) and liquaemin (E) were incubated with SPAK (inactivated by 0.03 mM Zn2+) for 2 h at 20 °C, respectively. The relative activity was tested, and the final concentration of SPAK was 0.06 μM.
3.7. Effect of Glycine and Proline on the SPAK Tertiary Structure and Aggregation
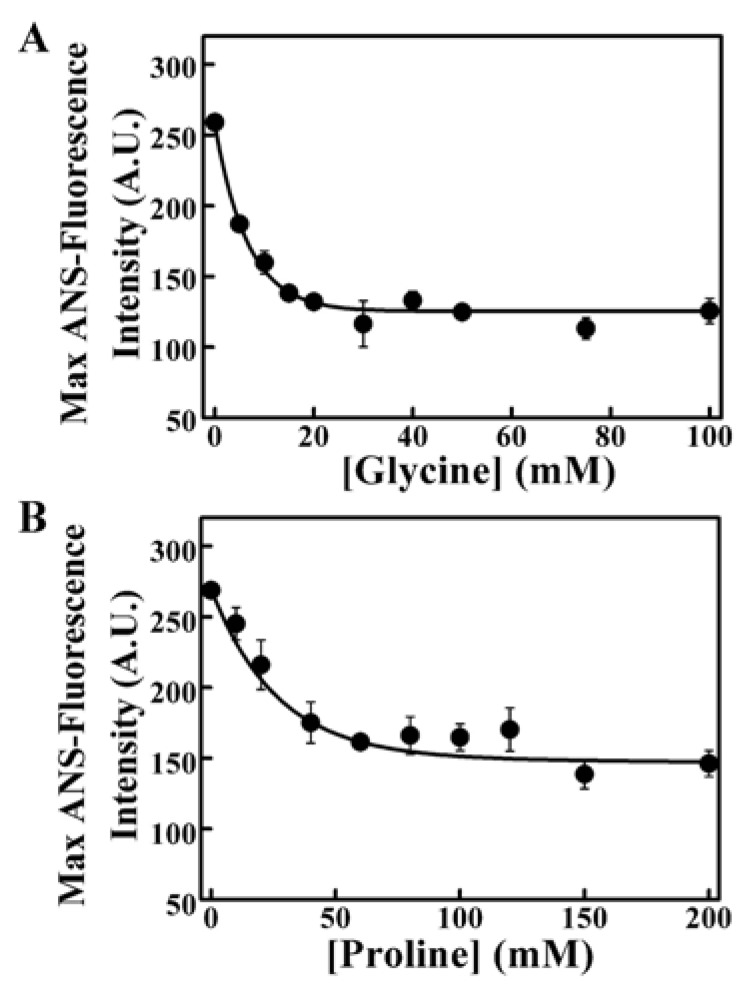
The structure protective effect of glycine and proline was tested through the ANS-binding fluorescence of 0.02 mM Zn2+-treated SPAK. The two osmolytes both obviously reduced the Zn2+-induced surface hydrophobicity with the increase of concentrations (Fig. 10). It showed that the recover of SPAK activity could depend mainly on the restoration of the structure. Zn2+ induced aggregation of SPAK was effectively reduced by glycine or proline (Fig. 11). Either 5 mM glycine or 100 mM proline could thoroughly prevent SPAK aggregation. Glycine and proline could act as folding chaperones by the structural protection and aggregation prevention of SPAK.
Figure 10.
Prevention of Zn2+-induced SPAK hydrophobic surface changes. The effects of glycine (A) and proline (B) were assayed by the ANS-fluorescence of SPAK (treated with 0.02 mM Zn2+). The enzyme concentration was 5 μM.
Figure 11.
Prevention of Zn2+-induced SPAK aggregation. (A) The protective effect of glycine. Labeled 1 to 5 represented 0, 1, 2, 4 and 5 mM glycine, respectively. (B) The protective effect of proline. Labeled 1 to 6 represented 0, 50, 60, 70, 80 and 100 mM proline, respectively. The concentrations of SPAK and Zn2+ were 17 μM and 0.5 mM, respectively.
4. DISCUSSION
The SPAK gene sequence obtained using the RACE technique was first revealed in this study. On the basis of the gene sequence results, we obtained a reasonable 3D structure of SPAK and carried out molecular dynamics simulation for SPAK with the binding of Zn2+. The overall predicted results implied that Zn2+ was a tight binding ligand and could directly affect the catalytic function of SPAK. The subsequent kinetic experiments confirmed that Zn2+ inhibited SPAK with a non-competitive inhibition mode. Zn2+ generated significant hydrophobic surface exposure and tertiary structure change of SPAK along with aggregation occurrence. The exposure of the hydrophobic groups caused by Zn2+ binding could directly result in misfolding of SPAK and thus modulated enzyme activity.
We found that among the osmolytes tested, glycine and proline not only prevented aggregation but also restored the activity and tertiary structure of SPAK that were destroyed due to the unfolding induced by Zn2+. These results implied that glycine and proline may regulate the activity and function of arginine kinase on unsuitable environmental conditions. The function of osmolytes in heavy metal poisoning preventment could be an interesting topic for marine invertebrates’ research.
Since the applied concentration of Zn2+ in this study was comparable to physiological Sepia pharaonis body condition, our results could be useful to elucidate the function of excessive Zn2+ in SPAK as a negative effecter. To illuminate more detail physiological functions of Zn2+ and of the osmolytes in vivo for SPAK and the relationship during SPAK unfolding, including the Zn2+-mediated SPAK aggregation problem, more systematic subsequent and sequential experiments should be conducted in the further studies.
5. CONCLUSION
We revealed the inhibitory effects of Zn2+ on SPAK by using computational MD simulation integrating enzymatic kinetics. Our results suggest concrete in vitro evidence of the toxicity and negative regulation of Zn2+ for arginine kinase in Sepia pharaonis and also suggest a putative protective role of osmolytes against Zn2+. The methodological approach for investigate target enzyme integrating with simulations could be applicable for the other enzymes from various ocean invertebrates. This study elucidates the mechanisms underlying Zn2+-related functions in cephalopods, and will help further clarify the stress mechanisms of cuttlefish under inhospitable environmental conditions.
ACKNOWLEDGEMENTS
This study was supported by the Ningbo Special Project of Agricultural Science and Technology (2014C11001) and the Zhejiang Provincial Top Key Discipline of Biological Engineering (KF2015002). Yue-Xiu Si was supported by the Zhejiang Provincial Top Key Discipline of Aquaculture (XRZSC1520). Dr. Jinhyuk Lee was supported through grants from the KOBIC Research Support Program, KRIBB Research Initiative Program, and the Pioneer Research Center Program through the National Research Foundation of Korea funded through the Ministry of Science, ICT & Future Planning (2013M3C1A3064780). Dr. Yong-Doo Park was supported through a grant from the Zhejiang Provincial Natural Science Foundation of China, “Towards studying the function of C3dg protein and elucidating its role in the pathogenesis of atopic dermatitis” (Grant No. LY14H110001) and through a fund from the Science and Technology Planning Project of Jiaxing (No. 2014AY21026).
LIST OF ABBREVIATIONS
- SPAK
arginine kinase from Sepia pharaonis
- ANS
1-anilinonaphthalene-8-sulfonate
- ATP
adenosine triphosphate
- MD
molecular dynamics
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Sikorski Z.E., Kolodziejska I. The composition and properties of squid meat. Food Chem. 1986;20:213–224. [Google Scholar]
- 2.Le K.X., Jiang X.M., Peng R.B., Luo J., Tang F., Wang C.L. Effects of four ecological factors on the growth and survival of Sepia pharaonis larvae. J. Biol. 2014;31:33–37. [Google Scholar]
- 3.Kinsey S.T., Lee B.C. The effects of rapid salinity change on in vivo arginine kinase flux in the juvenile blue crab, Callinectes sapidus. Comp. Biochem. Physiol. B. 2003;135:521–531. doi: 10.1016/s1096-4959(03)00121-0. [DOI] [PubMed] [Google Scholar]
- 4.Abe H., Hirai S., Okada S. Metabolic responses and arginine kinase expression under hypoxic stress of the kuruma prawn Marsupenaeus japonicus. Comp. Biochem. Physiol. A. 2007;146:40–46. doi: 10.1016/j.cbpa.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 5.Silvestre F., Dierick J.F., Dumont V., Dieu M., Raes M., Devos P. Differential protein expression profiles in anterior gills of Eriocheir sinensis during accl imation to cadmium. Aquat. Toxicol. 2006;76:46–58. doi: 10.1016/j.aquatox.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Morris S., van Aardt W.J., Ahern M.D. The effect of lead on the metabolic and energetic status of the Yabby, Cherax destructor, during environment al hypoxia. Aquat. Toxicol. 2005;75:16–31. doi: 10.1016/j.aquatox.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Ellington W.R. Evolution and physiological roles of phosphagen systems. Annu. Rev. Physiol. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- 8.Buttlaire D.H., Cohn M. Interaction of manganous ion, substrates, and anions with arginine kinase. Magnetic relaxation rate studies of water protons and kinetic anion effects. J. Biol. Chem. 1974;249:5733–5740. [PubMed] [Google Scholar]
- 9.Shofer S.L., Willis J.A., Tjeerdema R.S. Effects of hypoxia and toxicant exposure on arginine kinase function as measures by 31P-NMR magnetization transfer in living abalone. Comp. Biochem. Physiol. C. 1997;117:283–289. [Google Scholar]
- 10.Voncken F., Gao F., Wadforth C., Harley M., Colasante C. The phosphoarginine energy-buffering system of trypanosoma brucei involves multiple arginine kinase isoforms with different subcellular locations. PLoS One. 2013;8:e65908. doi: 10.1371/journal.pone.0065908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T., Furukohri T. Evolution of phosphagen kinase. Primary structure of glycocyamine kinase and arginine kinase from invertebrates. J. Mol. Biol. 1994;237:353–357. doi: 10.1006/jmbi.1994.1237. [DOI] [PubMed] [Google Scholar]
- 12.Furukohri T., Okamoto S., Suzuki T. Evolution of phosphagen kinase (III). Amino acid sequence of arginine kinase from the shrimp Penaeus japonicus. Zoolog. Sci. 1994;11:229–234. [PubMed] [Google Scholar]
- 13.Azzi A., Clark S.A., Ellington W.R., Chapman M.S. The role of phosphagen specificity loops in arginine kinase. Protein Sci. 2004;13:575–585. doi: 10.1110/ps.03428304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silvestre F., Dierick J.F., Dumont V., Dieu M., Raes M., Devos P. Differential protein expression profiles in anterior gills of Eriocheir sinensis during acclimation to cadmium. Aquat. Toxicol. 2006;76:46–58. doi: 10.1016/j.aquatox.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q., Wu C.G., Dong B., Li F.H., Liu F.Q., Xiang J.H. Proteomic analysis of acute responses to copper sulfate stress in larvae of the brine shrimp, Artemia sinica. Chin. J. Oceanology Limnol. 2010;28:224–232. [Google Scholar]
- 16.Ichihashi H., Kohno H., Kannan K., Tsumura A., Yamasaki S. Multielement analysis of purpleback flying squid using high resolution inductively coupled plasma-mass spectrometry (HR ICP-MS). Environ. Sci. Technol. 2001;35:3103–3108. doi: 10.1021/es010653v. [DOI] [PubMed] [Google Scholar]
- 17.Bustamante P., Grigiono S., Boucher-Rodoni R., Caurant F., Miramand P. Bioaccumulation of 12 trace elements in the tissues of the Nautilus Nautilus mae-romphalus from New Caledonia. Mar. Pollut. Bull. 2000;40:688–696. [Google Scholar]
- 18.Craing S., Overnell J. Metal in squid, Loligo forbesi, adults, eggs and hatchings. No evidence for a role for Cu- or Zn-methallothionein. Comp. Biochem. Physiol. C. 2003;134:311–317. doi: 10.1016/s1532-0456(02)00274-0. [DOI] [PubMed] [Google Scholar]
- 19.Jacobsen J.A., Major Jourden J.L., Miller M.T., Cohen S.M. To bind zinc or not to bind zinc: an examination of innovative approaches to improved metalloproteinase inhibition. 2010. [DOI] [PubMed]
- 20.Fukada T., Yamasaki S., Nishida K., Murakami M., Hirano T. Zinc homeostasis and signaling in health and diseases: Zinc signaling. J. Biol. Inorg. Chem. 2011;16:1123–1134. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong X., Zeng X., Zhou H.M. Effects of zinc on creatine kinase: activity changes, conformational changes, and aggregation. J. Protein Chem. 2000;19:553–562. doi: 10.1023/a:1007142117037. [DOI] [PubMed] [Google Scholar]
- 22.Liu T., Wang X. Zinc induces unfolding and aggregation of dimeric arginine kinase by trapping reversible unfolding intermediate. Acta Biochim. Biophys. Sin. (Shanghai) 2010;42:779–786. doi: 10.1093/abbs/gmq092. [DOI] [PubMed] [Google Scholar]
- 23.Shyn A., Chalk S.J., Smith K., Charnock N.L., Bielmyer G.K. Zinc distribution in the organs of adult Fundulus heteroclitus after waterborne zinc exposure in freshwater and saltwater. Arch. Environ. Contam. Toxicol. 2012;63:544–553. doi: 10.1007/s00244-012-9805-0. [DOI] [PubMed] [Google Scholar]
- 24.Núñez-Nogueira G., Fernández-Bringas L., Ordiano-Flores A., Gómez-Ponce A., de León-Hill C.P., González-Farías F. Accumulation and regulation effects from the metal mixture of Zn, Pb, and Cd in the tropical shrimp Penaeus vannamei. Biol. Trace Elem. Res. 2012;150:208–213. doi: 10.1007/s12011-012-9500-z. [DOI] [PubMed] [Google Scholar]
- 25.Mitra A., Chowdhury R., Banerjee K. Concentrations of some heavy metals in commercially important finfish and shellfish of the River Ganga. Environ. Monit. Assess. 2012;184:2219–2230. doi: 10.1007/s10661-011-2111-x. [DOI] [PubMed] [Google Scholar]
- 26.Eisler R. Zinc Hazards to Fish, Wildlife, and Invertebrates: A Synoptic Review. Contaminant Hazard Reviews Report. 1993;26:1–126. [Google Scholar]
- 27.Kim T.R., Oh S., Yang J.S., Lee S., Shin S., Lee J. A simplified homology-model builder toward highly protein-like structures: an inspection of restraining potentials. J. Comput. Chem. 2012;33:1927–1935. doi: 10.1002/jcc.23024. [DOI] [PubMed] [Google Scholar]
- 28.Kim T.R., Yang J.S., Shin S., Lee J. Statistical torsion angle potential (STAP) energy functions for protein structure modeling: a bicubic interpolation approach. Proteins. 2013;81:1156–1165. doi: 10.1002/prot.24265. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 30.Si Y.X., Lee J., Yin S.J., Gu X.X., Park Y.D., Qian G.Y. The inhibitory effects of Cu2+ on Exopalaemon carinicauda arginine kinase via inhibition kinetics and molecular dynamics simulations. Appl. Biochem. Biotechnol. 2015;176:1217–1236. doi: 10.1007/s12010-015-1641-z. [DOI] [PubMed] [Google Scholar]
- 31.Si Y.X., Lee J., Zhao F., Yin S.J., Park Y.D., Qian G.Y., Jiang X.M. Effects of cadmium on the cuttlefish Sepia pharaonis’ arginine kinase: unfolding kinetics integrated with computational simulations. J. Biomol. Struct. Dyn. 2015;19:1–15. doi: 10.1080/07391102.2015.1091747. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z., Pan J., Zhou H.M. A direct continuous pH-spectrophotometric assay for arginine kinase activity. Protein Pept. Lett. 2002;9:545–552. doi: 10.2174/0929866023408382. [DOI] [PubMed] [Google Scholar]
- 33.Yin S.J., Si Y.X., Chen Y.F., Qian G.Y., Lu Z.R., Oh S., Lee J., Lee S., Yang J.M., Lee D.Y., Park Y.D. Mixed-Type Inhibition of Tyrosinase from Agaricus bisporus by Terephthalic Acid: Computational Simulations and Kinetics. Protein J. 2011;30:273–280. doi: 10.1007/s10930-011-9329-x. [DOI] [PubMed] [Google Scholar]