Abstract
Salt-inducible kinases (SIKs) are a family of related serine-threonine kinases and are involved in controlling various metabolisms such as liver glucose homeostasis, hepatic lipogenesis, steroidogenesis, and adipogenesis. Here we investigated the regulatory role of SIK proteins in Toll-like receptor 4 (TLR4)-mediated signaling. Overexpression of SIK1 and SIK3, but not SIK2, significantly inhibited nuclear factor-κB activity in response to lipopolysaccharide stimulation and affected the expression of proinflammatory cytokines. In contrast, both SIK1KD and SIK3KD Raw 264.7 cells exhibit dramatic elevations of nuclear factor-κB activation and activations of downstream signaling molecules, such as TGF-β-activated kinase 1, p38, and c-Jun N-terminal kinase, in response to TLR4 stimulation, indicating that SIK1 and SIK3 are negatively involved in the TLR4-mediated signaling. Through biochemical studies, we found that SIK1 and SIK3 interact with TGF-β-activated kinase 1-binding protein 2 (TAB2), and interrupt the functional complex of TAB2-TNF receptor-associated factor 6 (TRAF6). Interestingly, the molecular interruption is induced to suppress the ubiquitination of TRAF6 in response to TLR4 stimulation. These result suggest that SIK1 and SIK3 negatively regulate TLR4-mediated signaling through the interruption of TAB2-TRAF6 complex and thereby the inhibition of ubiquitination of TRAF6. The present findings can be useful for a better understanding of multilevel interactions between the metabolic and immune systems.
It has become increasingly evident that immune cells reciprocally interact with metabolic diseases and that certain nonimmune pathologies result in the mobilization of the innate and adaptive immune systems (1–3). Recent studies of gene expression networks on adipose tissue from obese mice and human subjects have revealed extensive inflammatory gene networks associated with incidence of obesity (4, 5). In particular, cells that are involved in metabolic and immune responses show evidence of coordination and coevolution. For example, macrophages and adipocytes share many functional features (6). Thus, immunometabolism, which is the interplay between immunological and metabolic processes, has already provided insights on the pathogenic mechanism of obesity and substantial therapeutics (7). Recently, studies have been reported that AMP-activated protein kinase, which is an important serine/threonine kinase, plays a role in both the metabolic and immune systems (8, 9). In addition, a number of studies have emerged that the transcription factor nuclear factor-κB (NF-κB) is implicated in the development of metabolic diseases through the production of proinflammatory cytokines (3, 10–12). These studies strongly suggest that inflammatory responses via NF-κB activation may be associated with the development of metabolic diseases.
There is accumulating evidence that different macrophage populations play roles in the elimination of invaded pathogens and the resolution of inflammation (13, 14). Macrophages can classify into the classical activated macrophages, named by M1, and regulatory macrophages, named by M2 in terms of functional aspects. M1 macrophages are actively involved in the elimination of invaded pathogens by producing proinflammatory cytokines mediated by innate signaling including Toll-like receptors (TLR), whereas M2 macrophages is associated with the resolution of inflammation by producing antiinflammatory mediators such as IL-10, IL-4, and IL-13 (13–15).
Because of the distinct role of macrophages, current studies have been focused and interested on the specific regulation of these macrophages. In line with the interest, a recent report has suggested that the kinase inhibition of salt-inducible kinase (SIK) proteins strikingly increase many of the characteristic markers of regulatory macrophage such as IL-10 and other antiinflammatory molecules (16). They found that the inhibition of SIKs leads to the dephosphorylation of cAMP response element-binding protein-regulated transcriptional coactivator (CRTC)-3 at Ser62, Ser162, Ser329, and Ser370 in macrophages and stimulating the translocation of CRTC3 to the nucleus in which it promotes cAMP response element-binding protein-dependent gene transcription, including IL-10 gene transcription in TLR-stimulated macrophages (16). Nevertheless, the molecular and cellular mechanisms by which recruited monocytes differentiate into either classical M1 macrophages or regulatory M2 macrophages are fully remained to be investigated. Furthermore, how or which intercellular signals transmitting into the M2 macrophages induce the production of antiinflammatory cytokines and simultaneously suppress the production of proinflammatory cytokines is also unclear.
TLRs in innate immune cells including dendritic cells, macrophages, and neutrophils recognize and respond to both pathogen-associated molecular patterns and endogenous signals associated with danger (17, 18). All TLRs, with the exception of TLR3, recruit MyD88 to their receptor complex, as do members of the IL-1 receptor family. MyD88 recruits IL-1 receptor-associated kinase 1, IL-1 receptor-associated kinase 4 and then TNF receptor-associated factor 6 (TRAF6), which activate TGF-β-activated kinase 1 (TAK1), leading to the activation of NF-κB for the production of proinflammatory cytokines such as IL-1, IL-6, and TNF-α. Several binding proteins of TAK1, including TAK1-binding protein (TAB)-1, TAB2, TAB3, and TAB4, have been implicated to play a role in the regulation of TAK1 activity (19–21). A recent study has shown that TAB2 acts as a multifunctional protein that facilitates TRAF6 ubiquitination and assembly of an IL-1 signaling complex containing TRAF6, TAK1, and IκB kinase (22).
Here we report the functional regulation of SIK proteins in the TLR4-mediated signaling pathway. SIK1 and SIK3 interact with the TAB2 protein, and that critically inhibits the interaction of TRAF6 with the TAB2 protein. The inhibitory effect has a critical effect on the ubiquitination of TRAF6, and that results in the inhibition of NF-κB activation by TLR4 stimulation.
Materials and Methods
Cell culture
Human embryonic kidney (HEK) 293 cells were maintained in DMEM (Sigma) containing a high concentration of glucose (4.5 mg/mL) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin in a 5% CO2 atmosphere at 37°C. Raw 264.7 cells were maintained in DMEM containing 5% FBS and antibiotics (100 U/mL penicillin-G and 100 μg/mL streptomycin) at 37°C in a humidified incubator with 5% CO2. All Raw cells were initially seeded at 1 × 104 cells/well and cultured for 24 hours. SIK1- or SIK3-knockdown Raw 264.7 cells were maintained in DMEM medium containing 5% FBS, antibiotics, and puromycin (8 μg/mL).
Transfection methods
For the transfection of plasmids to be used in this work, we used two different systems: the Neon transfection system, which is a next-generation electroporation device (Invitrogen), for the transfection to Raw 264.7 cells, according to the respective manufacturer's instructions, and Lipofectamine LRX (Invitrogen) reagent for the transfection to HEK 293 cells. According to the respective manufacturer's instructions of the Neon transfection system, we tested the transfection efficacy by using a pcDNA-GFP vector and that was less than 50% in Raw 264.7 cells.
NF-κB-dependent luciferase reporter assay
Raw 264.7 cells were plated in 12-well culture plates. On the following day, the cells were transiently transfected with the indicated expression vectors using Neon transfection system (Invitrogen), according to the respective manufacturer's instructions. The total amount of DNA was kept constant by supplementation with empty vector, pcDNA3 (Invitrogen). Each transfection step included 300 ng of reporter plasmid together with 100 ng of Renilla for normalization of transfection efficiency. As a reporter plasmid, κ-luciferase reporter plasmid (pBIIx) was used (9). After 24–30 hours, the cells were lysed in luciferase lysis buffer (Promega). Where indicated, the cells were treated with 500 ng/mL lipopolysaccharide (LPS) for 6 hours. The lysates were divided and analyzed for firefly luciferase and Renilla activities using a luminometer. All the luciferase experiments were performed in triplicate. To confirm an appropriate expression of transfected expression plasmids, aliquots of whole-cell lysates were subjected to Western blot analysis and then analyzed by immunoblotting with anti-Flag, antihemagglutinin (HA), and antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH; Roche Applied Science) antibodies.
p65 and p50 (NF-κB) DNA binding assay by ELISA
Nuclear proteins from transfectants treated with or without LPS were prepared with the CelLytic NuCLEAR extraction kit in accordance with the manufacturer's protocol (Sigma). Activities of the transcription factors p65 and p50 were determined with the TransAM NF-κB transcription factor assay kit according to the manufacturer's instructions (Active Motif North America) (9). Binding of the transcription factors NF-κB-p65 and -p50 to the related consensus sequence on the plate-bound oligonucleotide were studied in the nuclear extracts following the manufacturer's procedure.
Analysis by nano-liquid chromatography, electrospray ionization, and tandem mass spectrometry
In-gel digestion was carried out with 12.5 ng/μL sequencing grade-modified trypsin (Promega) in 50 mM NH4HCO3 buffer (pH 7.8) at 37°C overnight. Produced tryptic peptides were extracted with 5% formic acid in 50% acetonitrile solution at room temperature for 20 minutes. The supernatants were collected and dried with SpeedVac (Thermo Scientific). Resuspended samples in 0.1% formic acid were purified and concentrated using C18 ZipTips (Millipore) before mass spectrometry analysis. The tryptic peptides were loaded onto a fused silica microcapillary column (12 cm × 75 μm) packed with C18 reversed-phase resin (5 μm, 200 Å). Peptide separation was conducted with a series of step gradients composed of initial isobaric flow for 5 minutes with 3% solvent B (0.1% formic acid in acetonitrile) and then linear gradient from 3% to 40% for 40 minutes. At the end of each running, 90% of solvent B was eluted for 10 minutes with the flow rate 250 nL/min. The percentage gradient of solvent B was against solvent A (0.1% formic acid in H2O). The column was directly connected to an LTQ linear ion-trap mass spectrometer (Finnigan) equipped with a nanoelectrospray ion source. The electrospray voltage was set at 1.95 kV, and the threshold for switching from mass spectrometry to tandem mass spectrometry was 500. The normalized collision energy for tandem mass spectrometry was 35% of main radio frequency amplitude and the duration of activation was 30 milliseconds. All spectra were acquired in data-dependent scan mode. Each full mass spectrometry scan was followed by five tandem mass spectrometry scans corresponding from the most intense to the fifth intense peaks of full mass spectrometry scan.
For data analysis, the acquired liquid chromatography, electrospray ionization, and tandem mass spectrometry fragment spectra were searched in the BioWorksBrowser (version Rev. 3.3.1 SP1; Thermo Fisher Scientific Inc) with the SEQUEST search engines against the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) nonredundant human database. The searching conditions were trypsin enzyme specificity, a permissible level for two missed cleavages, peptide tolerance of ±2 amu, a mass error of ±1 amu on fragment ions, and fixed modifications of carbamidomethylation of cysteine (+57 Da) and oxidation of methionine (+16 Da) residues.
Gene expression analysis by quantitative PCR
Total RNA was extracted using Trizol (Invitrogen). cDNAs were generated by the amfiRivert reverse transcription system (GenDEPOT). Quantitative PCR analysis against TNF-α, IL-1β, and IL-6 was performed using Roter-GeneQ (QIAGEN) according to the manufacturers' protocol. Values were normalized to GAPDH.
Measurement of IL-6 production
The quantity of mouse IL-6 in the culture supernatant fractions was measured with an ELISA (ELISA; eBioscience) according to the manufacturer's instructions.
Generation of SIK1- and SIK3-knockdown Raw 264.7 cells
Lentiviral particles containing short hairpin RNA (shRNA)-targeted SIK1 (sc-270371-V) or SIK3 (sc-141570-V) were purchased from Santa Cruz Biotechnology Inc. The 1 × 105 cells were cultured in a 24-well plate and infected with each lentiviral particle according to the manufacturer's protocol. Cells were cultured in puromycin (8 μg/mL)-containing medium for 2 weeks for selection of stable clones.
Plasmids and truncated mutant plasmids
Flag-tagged SIK1, HA-tagged SIK2, and Flag-tagged SIK3 plasmids were kindly provided by Dr Koo from Sungkyunkwan University School of Medicine (Seoul, Korea). For the generation of HA-tagged TAB2 truncated mutant plasmids, HA-TAB2 1–518, HA-TAB2 518–657, and HA-TAB2 518–693 truncated mutant plasmids were generated by PCR using HA-TAB2 wild-type as a template and then inserted into pcDNA3. The primers used for PCR are as follows: HA-TAB2 1–518, forward (5′-GCGAATTCATGGCCCAAG-3′) and reverse (5′-TATGTCGACTCAACT TATTCTATCCACATGTGCTAA-3′); HA-TAB2 518–657, forward (5′-ACGAATTCATGAGTGAAACACGGAAACTGAGTATG-3′) and reverse (5′-TATGTCGACTCAAGTCTTTGGTGTTTTGATGATGG-3); HA-TAB2 518–693, forward (5′-ACGAATTCATGAGTGAAACACGGAAACTGAGTATG-3′) and reverse (5′-TATGTCGACTCAGAAATGCCTTGGCAT C-3′).
Immunoprecipitation and Western blot analysis
To examine protein interactions in HEK 293 cells, the transfected cells were lysed in lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), 10 mM EDTA, 1% Triton X-100, 1% deoxycholate, 1.5% aprotinin, and 1 mM phenylmethylsulfonyl fluoride. Cellular debris was removed by centrifugation. Coimmunoprecipitation procedures were followed as previously described (22–25). For immunoprecipitation and Western blotting, we used anti-HA (Roche Applied Science), anti-Myc (Calbiochem), anti-FLAG (Sigma), antiphospho-TAK1 (Thr-187) (Cell signaling), anti-TAK1 (Cell Signaling), antiphospho-c-Jun N-terminal kinase (JNK; Cell Signaling), anti-JNK (Cell Signaling), antiphospho-p38 (Cell Signaling), anti-p38 antibodies (Cell Signaling), anti-TAB2 (Cell Signaling), antiubiquitin (Cell Signaling), anti-GAPDH (Cell Signaling), and normal rabbit IgG (Santa Cruz Biotechnology). The proteins were detected by the enhanced chemiluminescence system (Amersham Biosciences).
Statistical analyses
Results shown are the average of minimum of three independent experiments with error bars denoting standard deviation (±SD), as indicated in the figure legends. Comparison of different groups was carried out using the two-tailed unpaired Student t test. Differences were considered statistically at P < .05 or P < .01.
Results
The overexpression of SIK1 and SIK3, but not SIK2, inhibits NF-κB activation in response to TLR4 stimulation
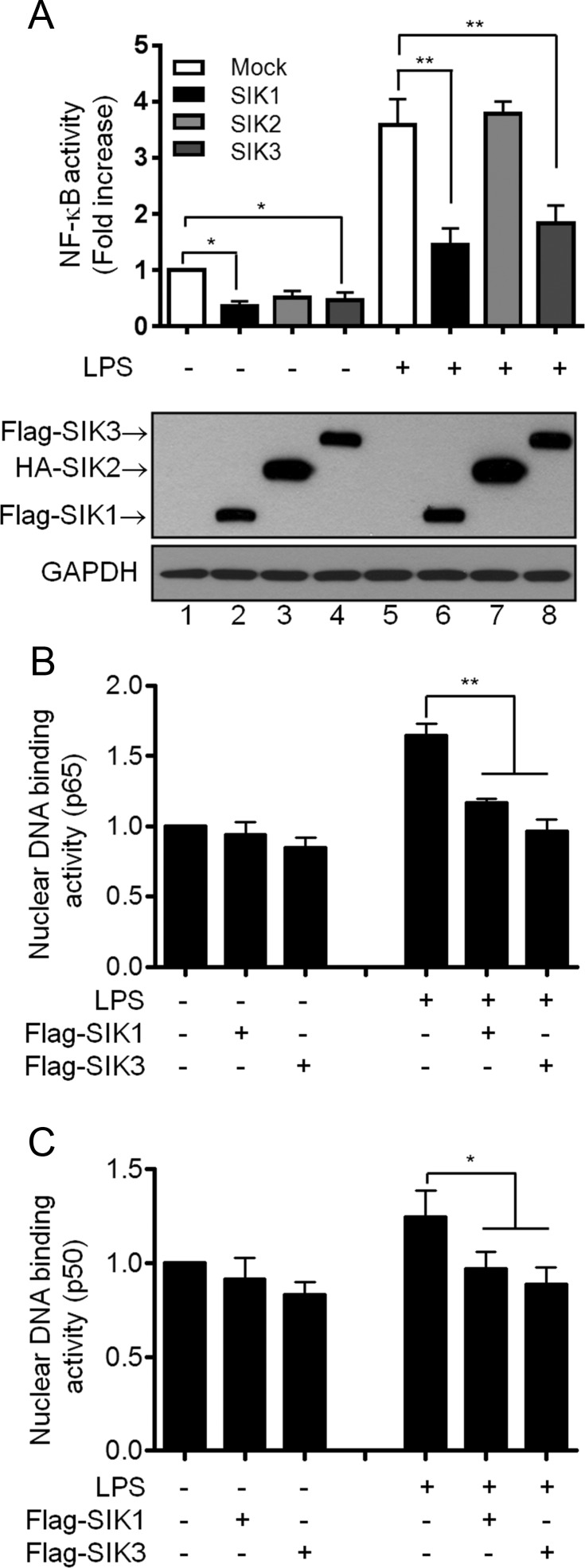
To explore whether SIK proteins are involved in TLR4-mediated signaling, Flag-tagged SIK1, HA-tagged SIK2, and Flag-tagged SIK3 proteins were overexpressed in Raw 264.7 macrophage cells (Figure 1A, Western blotting), respectively, stimulated with or without LPS, followed by NF-κB reporter assay. Overexpression of SIK1 and SIK3, but not SIK2, significantly inhibited NF-κB reporter activity in response to stimulation (Figure 1A). To verify the inhibitory effect on NF-κB activity, we further tested p65 and p50 DNA-binding activity. Consistently, p65 and p50 DNA-binding activity induced by LPS stimulation was markedly attenuated in both SIK1- and SIK3-overexpressed cells as compared with that of control cells treated with LPS (Figure 1B, p65; Figure 1C, p50), indicating that SIK1 and SIK3 may negatively regulate the TLR4-induced activation of NF-κB.
Figure 1.
SIK1 and SIK3 negatively regulate TLR4-mediated NF-κB activation. A, Raw 264.7 macrophage cells were transiently cotransfected with κ-luciferase reporter plasmid (pBIIx), Renilla plasmid, Flag-SIK1, HA-SIK2, and Flag-SIK3 as indicated. After 24 hours, the cells were untreated or treated with LPS (500 ng/mL) for 6 hours, and luciferase activities were determined by dual-luciferase assay. The values indicated represent normalized luciferase activities and are shown as the mean ± SD based on triplicate assays (n = 3). Three independent experiments produced similar results. Western blotting analyses of the same lysates from each transfection are shown in the lower panels. *, P < .05; **, P < .01. B and C, Raw 264.7 macrophage cells were transiently transfected with mock, Flag-SIK1, or Flag-SIK3 vector. After 30 hours, the cells were untreated or treated with LPS (200 ng/mL).Subunit-specific p65 (B) and p50 (C) ELISA assays were performed using 2 μg of nuclear extracts derived from each sample as indicated. Absorbance was measured at 450 nm. The values indicated represent fold increases in comparison with the mock transfectants. Data are shown as the mean ± SD based on triplicate assays (n = 3). *, P < .05; **, P < .01.
SIK1 and SIK3 attenuate expressions of proinflammatory cytokines induced by TLR4 stimulation
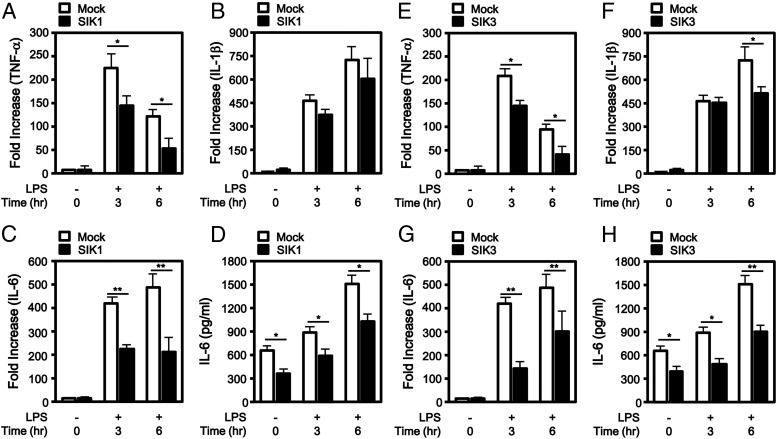
Stimulation of TLR4 activates NF-κB and induces the expression of proinflammatory cytokines such as TNF-α, IL-1β, and IL-6 (9, 17, 18). We further examined whether SIK1 and SIK3 have an effect on the expression of proinflammatory cytokines induced by TLR4 stimulation. Flag-tagged SIK1 and Flag-tagged SIK3 were expressed in Raw 264.7 cells, respectively (Figure 2A–D, SIK1; Figure 2E–H, SIK3), stimulated with LPS for different times as indicated, followed by quantitative RT-PCR assay. SIK1 overexpression significantly attenuated the gene expression of TNF-α and IL-6 but marginally attenuated IL-1β expression, induced by LPS stimulation, as compared with that of the control treated with LPS (Figure 2A, TNF-α; Figure 2B, IL-1β; Figure 2C, IL-6). Similarly, SIK3 overexpression significantly attenuated the gene expressions of IL-6 and IL-1β but marginally attenuated TNF-α expression, induced by LPS stimulation as compared with that of the control treated with LPS (Figure 2E, TNF-α; Figure 2F, IL-1β; Figure 2G, IL-6). When the level of IL-6 production was evaluated in the supernatants, marked decreases could be observed according to the expression of the SIK1 or SIK3 protein (Figure 2D, SIK1; Figure 2H, SIK3).
Figure 2.
SIK1 and SIK3 inhibit the expression of proinflammatory cytokines induced by TLR4 stimulation. A–C, Raw 264.7 macrophage cells were transiently transfected with mock and Flag-SIK1 as indicated. After 24 hours, the cells were untreated or treated with LPS (100 ng/mL) for 3 and 6 hours. Total RNAs were isolated from each sample, and quantitative RT-PCR analysis was performed with specific primers targeted to TNF-α (A), IL-1β (B), and IL-6 (C) genes. Error bars represent the mean ± SD of triplicate samples (n = 3). *, P < .05; **, P < .01. D, Raw 264.7 macrophage cells were transiently transfected with mock and Flag-SIK1 as indicated. After 24 hours, the cells were untreated or treated with LPS (100 ng/mL) for 3 and 6 hours and then analyzed for production of IL-6 in supernatants using an ELISA. Error bars indicate ± SD of triplicate samples (n = 3). *, P < .05. E–G, Raw 264.7 macrophage cells were transiently transfected with mock and Flag-SIK3 as indicated. After 24 hours, the cells were untreated or treated with LPS (100 ng/mL) for 3 and 6 hours. Total RNAs were isolated from each sample, and quantitative RT-PCR analysis was performed with specific primers targeted to TNF-α (E), IL-1β (F), and IL-6 (G) genes. Error bars represent the mean ± SD of triplicate samples (n = 3). *, P < .05; **, P < .01. H, Raw 264.7 macrophage cells were transiently transfected with mock and Flag-SIK3 as indicated. After 24 hours, the cells were untreated or treated with LPS (100 ng/mL) for 3 and 6 hours and then analyzed for production of IL-6 in supernatants using an ELISA. Error bars indicate ± SD of triplicate samples (n = 3). *, P < .05; **, P < .01.
TLR4 downstream signals are enhanced in SIK1- or SIK3-knockdown cells in response to LPS stimulation
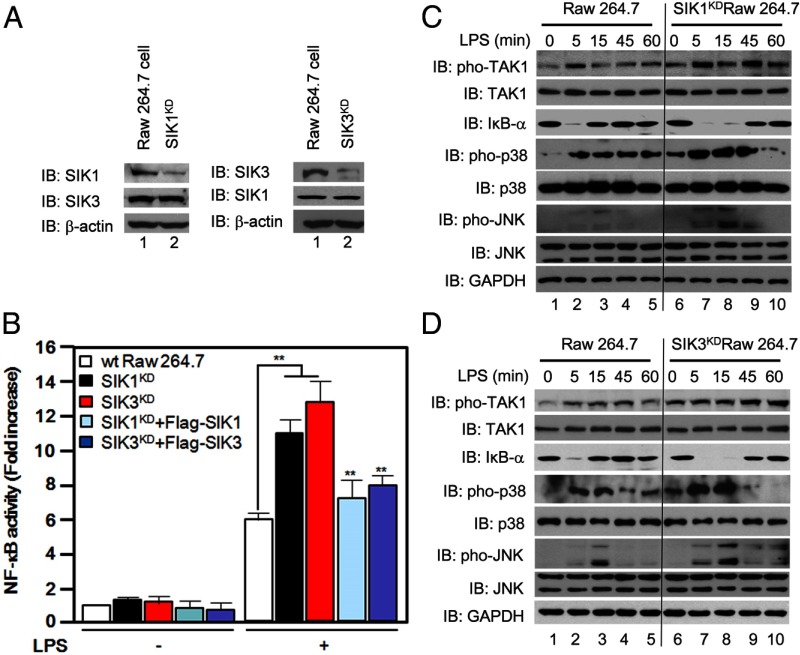
To verify the functional roles of SIK1 and SIK3 in TLR4-mediated signaling, SIK1- or SIK3-knockdown Raw 264.7 cells were generated using shRNA-targeted SIK1 and SIK3, respectively (Figure 3A), as described in Materials and Methods. SIK1- and SIK3-knockdown cells revealed enhanced NF-κB reporter activities in response to LPS stimulation as compared with those of control cells treated with LPS (Figure 3B), whereas the NF-κB reporter activities were significantly reduced by the reexpression of SIK1 or SIK3 in SIK1KD or SIK3KD cells, respectively, up to that of wild-type cell-treated LPS (Figure 3B), indicating an SIK1- or SIK3-specific effect on the NF-κB activation induced by LPS. We further examined whether SIK1 and SIK3 have an effect on downstream signaling mediated by TLR4 stimulation. Wild-type, SIK1KD, and SIK3KD Raw 264.7 cells were treated with or without LPS for different times as indicated, and then the activation of TLR4 downstream signaling molecules were analyzed by Western blotting (Figure 3C, SIK1KD cells; Figure 3D, SIK3KD cells). Upon stimulation of LPS, phosphorylations of TAK1, p38, and JNK molecules were significantly induced in wild-type Raw 264.7 cells (Figure 3, C and D, wt Raw 264. 7 cells) as compared with those of without stimulation (Figure 3, C and D, wt Raw 264. 7 cells). Interestingly, the levels of phosphorylation were markedly elevated in both SIK1KD and SIK3KD cells as compared with those of wt Raw 264.7 cells (Figure 3, C and D, SIK1KD and SIK3KD cells, respectively). Additionally, the degradation of inhibitory-κB-α in response to LPS stimulation efficiently occurred in both the SIK1KD and SIK3KD cells as compared with those of wt Raw 264.7 cells (Figure 3, C and D,). Furthermore, IL-6 production induced by TLR4 stimulation was markedly enhanced in SIK1KD cells as compared with that of wild-type Raw 264.7 cells (Supplemental Figure 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). These results strongly demonstrate that SIK1 and SIK3 are negatively involved in TLR4-mediated signaling.
Figure 3.
SIK1- and SIK3-knockdown cells enhance TLR4-mediated signaling for activation of NF-κB. A, Raw 264.7 cells were infected with lentiviral particles containing shRNA-targeted SIK1 or SIK3 according to the manufacturer's protocol. The cells were then cultured in puromycin (8 μg/mL)-containing medium for 2 weeks to select stable clones. The expression of endogenous SIK1 and SIK3 was evaluated in SIK1-knockdown or SIK3-knockdown cells with anti-SIK1 antibody and anti-SIK3 antibody. B, Wild-type, SIK1-knockdown, and SIK3-knockdown Raw 264.7 cells were transiently cotransfected with or without Flag-tagged SIK1 or Flag-tagged SIK3, respectively, together with κ-luciferase reporter plasmid (pBIIx) and Renilla plasmid. After 24 hours, the cells were untreated or treated with LPS (500 ng/mL) for 6 hours, and luciferase activities were determined by dual-luciferase assay. The values indicated represent normalized luciferase activities and are shown as the mean ± SD based on triplicate assays (n = 3). Three independent experiments produced similar results. **, P < .01. C, Wild-type and SIK1-knockdown Raw 264.7 cells were treated with or without LPS (100 ng/mL) for different times as indicated. The lysates were examined by Western blotting with antiphospho-TAK1, anti-TAK1, anti-IκB-α, antiphospho-38, anti-p38, antiphospho-JNK and anti-JNK antibodies. Immunoblotting with anti-GAPDH antibody was performed to generate a control for gel loading. IB, Immunoblotting; IκB, inhibitory κB. D, Wild-type and SIK3-knockdown Raw 264.7 cells were treated with or without LPS (100 ng/mL) for different times as indicated. The lysates were examined by Western blotting with antiphospho-TAK1, anti-TAK1, anti-IκB-α, antiphospho-38, anti-p38, antiphospho-JNK, and anti-JNK antibodies. Immunoblotting with anti-GAPDH antibody was performed to generate a control for gel loading.
SIK1 and SIK3 interact with TAB2 through the N terminus of TAB2
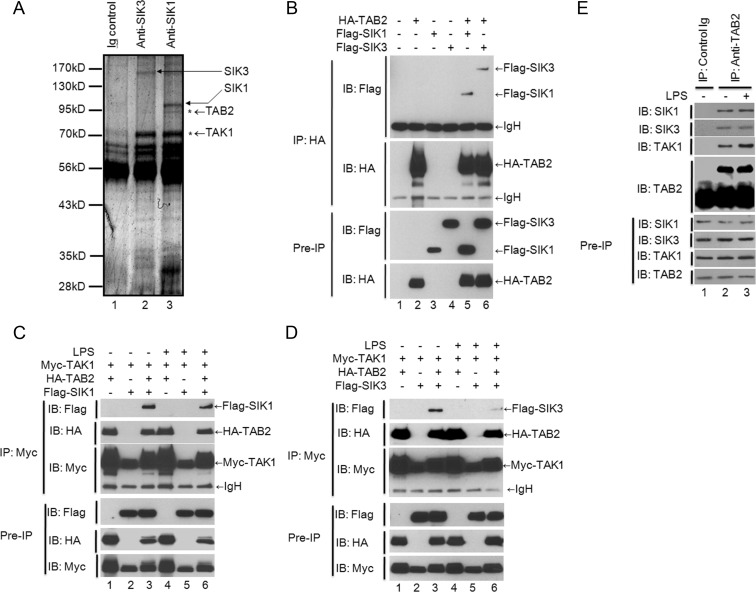
We next investigated the molecular mechanism by which SIK1 and SIK3 negatively regulate TLR4-mediated signaling. To do this, we attempted to identify interacting proteins with SIK1 or SIK3. SIK1 or SIK3 complexes were immunoprecipitated from the lysates of Raw 264.7 cells using anit-SIK1 or anti-SIK3 antibody and then analyzed by mass spectrometry. Among the interacting proteins, TAB2 and TAK1 appeared in both the SIK1 complex and SIK3 complex (Figure 4A). Because TAB2 as a TAK1-binding protein plays a pivotal role in the formation of the signaling complex TAK1-TAB2-TRAF6 in TLR4-mediated signaling (19–22), we supposed that SIK1 and SIK3 may be involved in TLR4-mediated signaling by regulating the formation of the TAK1-TAB2-TRAF6 complex (22). We first examined whether SIK1 or SIK3 interacts with TAB2 and TAK1. Flag-tagged SIK1 or SIK3 was significantly immunoprecipitated with HA-tagged TAB2 (Figure 4B, lanes 5 and 6). However, Myc-tagged TAK1 did not directly interact with Flag-tagged SIK1 or Flag-tagged SIK3, respectively (Figure 4C, lanes 2 and 5 in SIK1; Figure 4D, lanes 2 and 5 in SIK3). Interestingly, Myc-tagged TAK1 was significantly precipitated with Flag-tagged SIK1 or Flag-tagged SIK3 in the coexpression of HA-tagged TAB2 (Figure 4C, lanes 3 and 6 in SIK1; Figure 4D, lanes 3 and 6 in SIK3), indicating that SIK1 or SIK3 directly interacts with TAB2, whereas it indirectly interacts with TAK1 through SIK1- or SIK3-TAB2 interaction.
Figure 4.
SIK1 and SIK3 interact with TAB2. A, The cell lysates from Raw 264.7 cells were immunoprecipitated with anti-SIK1 or SIK3 antibodies. The precipitated proteins were separated on SDS-PAGE and identified by an electrospray ion trap mass spectrometer. B, HEK 293 cells were cotransfected with HA-TAB2, Flag-SIK1, or Flag-SIK3 as indicated. Thirty-six hours after transfection, the cells were extracted and immunoprecipitated with anti-HA antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of Flag-SIK1, Flag-SIK3, and HA-TAB2 in the pre-IP lysates was verified by Western blot analysis. C, HEK 293 cells were cotransfected with Myc-TAK1 and HA-TAB2, Myc-TAK1, and Flag-SIK1, or Myc-TAK1, HA-TAB2, and Flag-SIK1 as indicated. Thirty-six hours after transfection, the cells were untreated or treated with LPS (200 ng/mL) for 45 minutes, extracted, and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag or anti-HA antibody. The presence of Myc-TAK1, HA-TAB2, and Flag-SIK1 in the pre-IP lysates was verified by Western blot analysis. D, HEK 293 cells were cotransfected with Myc-TAK1 and HA-TAB2, Myc-TAK1 and Flag-SIK3, or Myc-TAK1, HA-TAB2, and Flag-SIK3 as indicated. At 36 hours after transfection, the cells were untreated or treated with LPS (200 ng/mL) for 45 minutes, extracted, and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag or anti-HA antibody. The presence of Myc-TAK1, HA-TAB2, and Flag-SIK3 in the pre-IP lysates was verified by Western blot analysis. E, Raw 264.7 cells (2 × 107) were untreated or treated with LPS (100 ng/mL) for 60 minutes. The cells were extracted and immunoprecipitated with normal rabbit IgG or anti-TAB2 antibodies as indicated. The interaction was detected by Western blotting with anti-SIK1, anti-SIK3, and anti-TAK1 antibodies. The presence of SIK1, SIK3, TAK1, and TAB2 in the pre-IP lysates was verified by Western blot analysis.
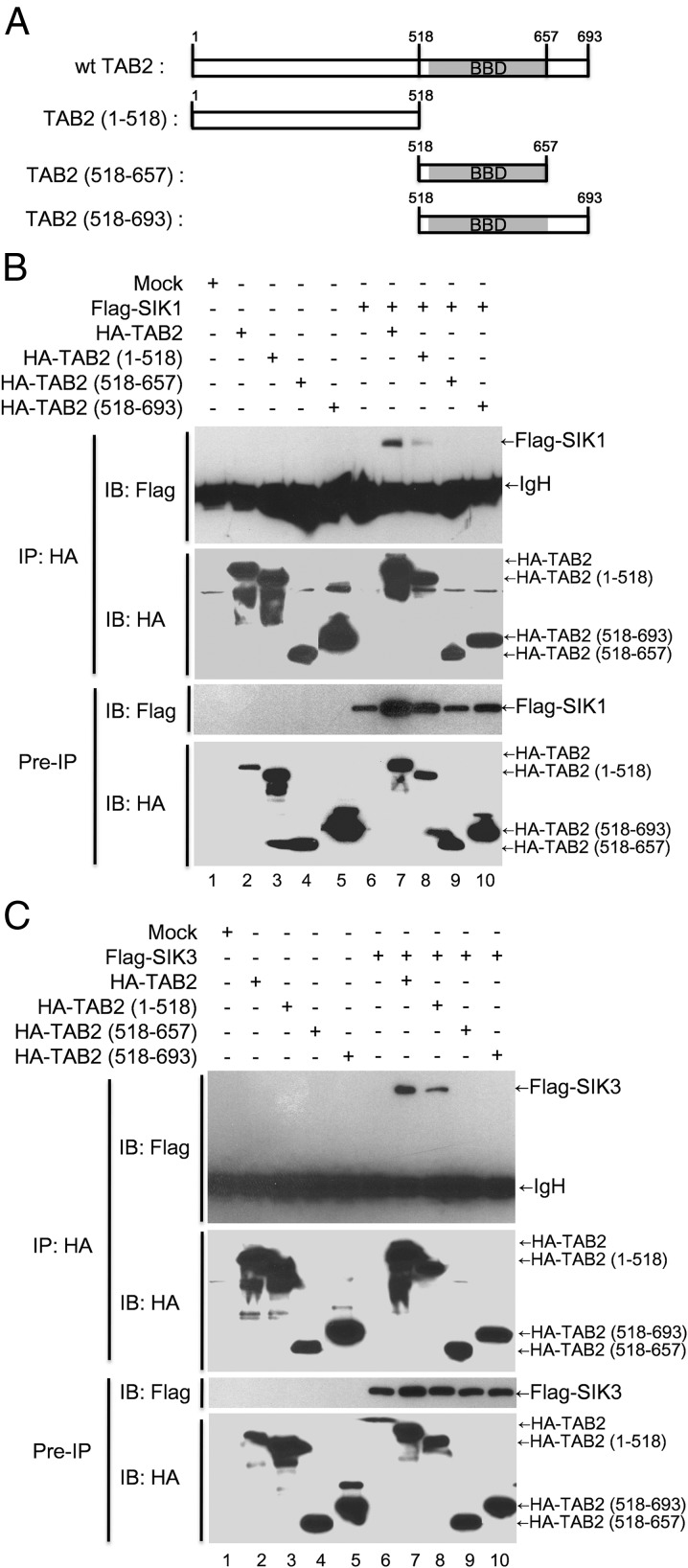
To verify the molecular complex, SIK1- or SIK3-TAB2-TAK1, the endogenous immunoprecipitation assay with anti-TAB2 antibody was performed. As shown in Figure 4E, SIK1 and SIK3 proteins were significantly coprecipitated with TAB2 protein. TAB2 has different functional domains that are capable of inducing interactions with TAK1 and TRAF6 through the coiled-coil domain and the C terminus of TAB2, respectively (23, 24). To identify the interacting domain of TAB2 and SIK1 or SIK3, we generated three different truncated mutants of TAB2 based on the functional domains (Figure 5A) and then performed a coimmunoprecipitation assay. As shown in Figure 5, B and C, Flag-tagged SIK1 or SIK3 interacted with the N terminus of TAB2, TAB2 1–518,along with wild-type TAB2 but not other truncated mutants such as TAB2 518–657 and TAB2 518–693 (Figure 5, B and C, lanes 7 and 8), indicating that SIK1 and SIK3 interact with the N terminus of TAB2, TAB2 1–518.
Figure 5.
SIK1 and SIK3 interact with N terminal 1–518 domain of TAB2. A, Three truncated TAB2 mutants as indicated were generated as described in Materials and Methods. B, HEK 293 cells were cotransfected with mock, Flag-SIK1, HA-TAB2 wild-type, HA-TAB2 1–518, HA-TAB2 518–657, and HA-TAB2 518–693 as indicated. After 24 hours, the cells were extracted and immunoprecipitated with anti-HA antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of Flag-SIK1, HA-TAB2 wild-type, HA-TAB2 1–518, HA-TAB2 518–657, and HA-TAB2 518–693 in the pre-IP lysates was verified by Western blot analysis. IB, immunoblotting. C, The HEK 293 cells were cotransfected with mock, Flag-SIK3, HA-TAB2 wild-type, HA-TAB2 1–518, HA-TAB2 518–657, and HA-TAB2 518–693 as indicated. After 24 hours, the cells were extracted and immunoprecipitated with anti-HA antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of Flag-SIK3, HA-TAB2 wild-type, HA-TAB2 1–518, HA-TAB2 518–657, and HA-TAB2 518–693 in the pre-IP lysates was verified by Western blot analysis.
SIK1 and SIK3 negatively regulate TLR4-mediated signaling by interrupting the interaction between TAB2 and TRAF6
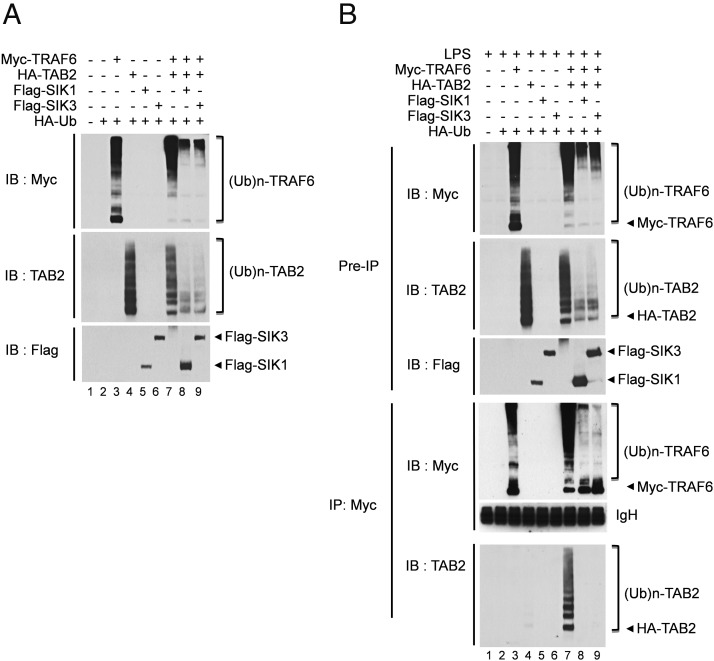
A previous report has shown that the C terminal 401–692 of TAB2 interacts with TRAF6 and that facilitates the interaction between TRAF6 and TAK1 (24). Moreover, it has been reported that TAB2 contains a zinc-finger domain that binds to polyubiquitin chains linked by K63, which is thought to be necessary for the activation of TAK1 and inhibitory-κB kinase (25) and facilitates TRAF6 ubiquitination for NF-κB activation (26). We found that SIK1 and SIK3 interact with the N terminal 1–518 of TAB2, which is partly overlapped with the interaction domain of TRAF6 (Figure 5, B and C). We supposed that the SIK1 or SIK3 protein may affect the formation of the TAB2-TRAF6 functional complex in TLR4-mediated signaling. Therefore, we first examined whether the SIK1- or SIK3-TAB2 interaction has an effect on the ubiquitination of TRAF6. Myc-tagged TRAF6 and HA-tagged TAB2 were transiently cotransfected with Flag-tagged SIK1 or Flag-tagged SIK3 along with or without the HA-Ub vector. Cotransfection of Myc-TRAF6 or HA-TAB2 with HA-Ub induced polyubiquitination of TRAF6 and TAB2 (Figure 6A, lanes 3 and 4), whereas in the presence of SIK1 or SIK3, the ubiquitination of TRAF6 and TAB2 was markedly suppressed (Figure 6A, lanes 8 and 9). Furthermore, upon TLR4 stimulation, the ubiquitination of TRAF6 and TAB2 was also attenuated [Figure 6B, lanes 8 and 9 in preimmunoprecipitation (IP) sample], and the TRAF6-TAB2 interaction was markedly abolished in the presence of SIK1 or SIK3 (Figure 6B, lanes 8 and 9 in IP sample). These results strongly suggest that SIK1 and SIK3 negatively regulate TLR4-mediated signaling through the inhibition of TAB2-TRAF6 interactions and thereby suppress TRAF6 ubiquitination.
Figure 6.
SIK1 and SIK3 inhibit the interaction between TAB2 and TRAF6 and prevent ubiquitination of TAB2 and TRAF6. A, HEK 293 cells were cotransfected with Myc-TRAF6, HA-TAB2, Flag-SIK1, Flag-SIK3, and HA-Ub vectors as indicated. After 36 hours, the cells were extracted and immunoblot analysis was performed with anti-Myc, anti-TAB2, and anti-Flag antibodies. IB, immunoblotting. B, HEK 293 cells were cotransfected with Myc-TRAF6, HA-TAB2, Flag-SIK1, Flag-SIK3, and HA-Ub vectors as indicated. After 24 hours, the cells were treated with LPS (100 ng/mL) for 45 minutes, extracted, and immunoprecipitated with anti-Myc antibody. The interaction between TRAF6 and TAB2 was detected by Western blotting with anti-TAB2 antibody. The presence of Myc-TRAF6, HA-TAB2, Flag-SIK1, and Flag-SIK3 in the pre-IP lysates was verified by Western blot analysis. Immunoblot analysis was performed with anti-Myc, anti-TAB2, and anti-Flag antibodies.
Discussion
In this study, we provide evidence that SIK1 and SIK3 expression inhibits the activation of NF-κB induced by TLR4 stimulation. The inhibitory effect was followed by decreases in expression and production of proinflammatory cytokines. Moreover, negatively regulatory effects on TLR4-mediated signaling could be confirmed in SIK1- or SIK3-knockdown cells; the knockdown cells revealed marked increases in NF-κB activity and production of proinflammatory cytokines in response to LPS stimulation, as compared with those of wild-type cells. Together, these observations provide evidence that the SIK1 and SIK3 proteins negatively regulate TLR4-mediated signaling for activation of NF-κB.
Interestingly, a mass analysis to identify the interaction proteins to be in the TLR4-mediated signal revealed that the SIK1 or SIK3 protein was significantly coprecipitated with TAB2 along with TAK1. When an immunoprecipitation assay was performed to verify the molecular interaction between SIK1 or SIK3 and TAB2 or TAK1, SIK1 and SIK3 were found to be strongly interacted to TAB2, but not TAK1, suggesting that SIK1 and SIK3 indirectly interact with TAK1 through the TAB2 protein. It has been previously demonstrated that the TRAF6 RING finger function, K63-linked ubiquitination, and the ubiquitin-binding domain of TAB2 are required for TRAF6-mediated activation of TLR4-mediated downstream signaling (19, 20, 24). A recent report has shown that IL-1 signaling leads to ubiquitination of TAB2 and TAB3 through the action of TRAF6 (25). Moreover, it has been reported that TAB2 acts as a multifunctional signaling molecule, facilitating both IL-1-dependent TRAF6 ubiquitination and the assembly of the IL-1 signaling complex (26). Based on these results, we hypothesized that the interaction between SIK1 or SIK3 and TAB2 affects the ubiquitination of TRAF6 and then affects on downstream signaling mediated by TLR4. Interestingly, coexpression of SIK1 or SIK3 along with TRAF6 and TAB2 dramatically inhibited the interaction between TRAF6 and TAB2 and resulted in the suppression of TRAF6 and TAB2 ubiquitination. These results strongly suggest that the SIK1- or SIK3-mediated inhibitory effect in TLR4-mediated signaling may be associated with inhibition of TRAF6 ubiquitination through interruption of TAB2-TRAF6 interactions.
A recent report has shown the functional effects of SIK inhibitors on macrophage function, especially in controlling the interconversion of classically activated and regulatory macrophages (16). They found that the SIK inhibitor elevates IL-10 production by inducing the dephosphorylation of CRTC3, which might be critical for the production of IL-10 via the liver kinase B (LKB)-SIK-CRTC3 signaling axis. Importantly, they suggest that the inhibition of SIKs may provide an advantage over current therapies and improve the treatment of chronic inflammation and autoimmune diseases through the kinase activity of SIKs in the function of regulatory macrophages such as M2a and M2b. Our data demonstrate that SIK1 and SIK3, but not SIK2, inhibit TLR4-mediated signaling via the interaction with TAB2 protein, leading to suppression of the ubiquitination of TRAF6, which is critical for activations of the downstream molecules including TAK1 in the TLR4-mediated signaling pathway (17–22). The inhibitory effect of SIK1 and SIK3 in the TLR4-mediated signaling was associated with the molecular intervention of the interaction between TAB2 and TRAF6, presumably in a kinase-independent manner. Increasing evidences have shown that macrophages are functionally able to adapt to pathological conditions in response to extracellular cures that allow them to play key roles throughout the inflammatory process from its onset to its resolution (13–16).
The classical macrophages, named M1, initially responded to invaded pathogen through the production of proinflammatory mediators, leading to the recruitment of other leukocytes, such as neutrophils. After clearance of infection, another type of macrophage, named M2, is developed and involved in the tissue repair and the resolution of inflammation through the production of antiinflammatory cytokines including IL-10. Moreover, a recent report has shown that prostaglandin E2 induces the production of IL-10 in the regulatory-like phenotype of macrophage via regulating the ability of SIK2 to inhibit CRTC (27). Although we could not exactly explain of how SIK proteins are implicated in this pathological condition, especially by the kinase-dependent or independent manner, we speculate that the inhibitory effects of SIK1 or SIK3 on the formation of TAK1-TAB2-TRAF6 complex that plays a key role for the production of proinflammatory mediators in response to invading pathogens may contribute to the differentiation of regulatory macrophages from recruited monocytes, leading to initially promoting tissue repair and the resolution of inflammation. And also it is not ruled out that the SIK1- or SIK3-mediated inhibition on TLR4-mediated signaling that eventually induces the production of proinflammatory cytokines in classical M1 macrophages may also facilitate the differentiation of M1 macrophages into M2 macrophages under the pathological condition. Nevertheless, the details of molecular and cellular mechanisms by which SIK proteins are involved in the different types of macrophages, especially in pathological conditions, remain to be investigated.
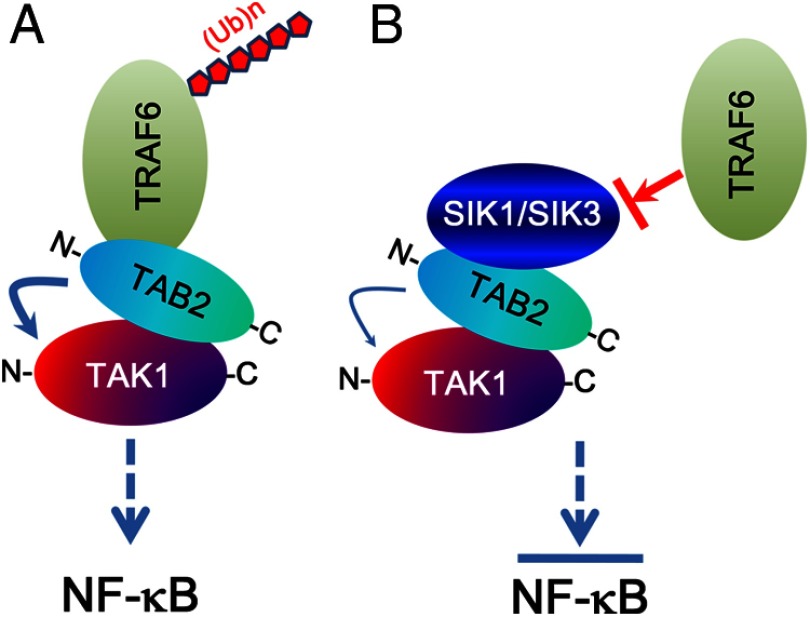
In conclusion, our results demonstrate that SIK1 and SIK3 negatively regulate the TLR4-mediated signaling pathway through interactions with the TAB2 protein (Figure 7). The SIK1- or SIK3-TAB2 interaction inhibits the interaction between TAB2 and TRAF6, and that suppresses TRAF6 ubiquitination (Figure 7B). Although the detailed pathological means of how the inhibitory effects are implicated in metabolic diseases including obesity remain unclear, we believe that our data may contribute to understanding immunometabolism interfacing immunology and metabolism in the biochemical and molecular levels and that further contribute to developing therapeutic targets for the diseases.
Figure 7.
Model of negative regulation of SIK1 and SIK3 in TLR4-mediated signaling. A, Upon the TLR4 stimulation, TAB2 interacts with C terminus of TAK1 and simultaneously interacts with TRAF6 through the N terminus of TAB2. The interaction of TAB2 to TAK1 and TRAF6 regulates TAK1 activity and facilitates TRAF6 ubiquitination, respectively, and that induces the activation of NF-κB. B, If intracellular SIK1 or SIK3 preferentially interacts with TAB2 through its N terminus, the interaction inhibits the association with TRAF6 to TAB2. Molecular inhibition between TAB2 and TRAF6 cause prevention of polyubiquitination of TRAF6 and TAB2, and that inhibits downstream signals for the activation of NF-κB.
Acknowledgments
We thank the Hyewa Forum members, namely Dr Doo Hyun Chung, Dr Jun Chang, Dr You-Me Kim, Dr Eun Sook Hwang, Dr Eui-Cheol Shin, Dr Seung-Hyo Lee, Dr Heung kyu Lee, and Dr Sang-Jun Ha for their helpful discussions. Mass analysis was supported by Research Core Facility in Sungkyunkwan University School of Medicine.
This work was supported by a grant from the Korea Healthcare Technology Research and Development Project, Ministry of Health and Welfare, Republic of Korea (Grant A111636).
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by a grant from the Korea Healthcare Technology Research and Development Project, Ministry of Health and Welfare, Republic of Korea (Grant A111636).
Footnotes
- CRTC
- cAMP response element-binding protein-regulated transcriptional coactivator
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HA
- hemagglutinin
- HEK
- human embryonic kidney
- IP
- immunoprecipitation
- JNK
- c-Jun N-terminal kinase
- LPS
- lipopolysaccharide
- NF-κB
- nuclear factor-κB
- shRNA
- short hairpin RNA
- SIK
- salt-inducible kinase
- TAB
- TAK1-binding protein
- TAK1
- TGF-β-activated kinase 1
- TLR
- Toll-like receptor
- TRAF6
- TNF receptor-associated factor 6.
References
- 1. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crozat K, Vivier E, Dalod M. Crosstalk between components of the innate immune system: promoting anti-microbial defenses and avoiding immunopathologies. Immunol Rev. 2009;227:129–149. [DOI] [PubMed] [Google Scholar]
- 3. Baker RG, Hayden MS, Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011;13:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. [DOI] [PubMed] [Google Scholar]
- 5. Yang X, Deignan JL, Qi H, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mathis D, Shoelson SE. Immunometabolism: an emerging frontier. Nat Rev Immunol. 2011;11:81–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fullerton MD, Steinberg GR, Schertzer JD. Immunometabolism of AMPK in insulin resistance and atherosclerosis. Mol Cell Endocrinol. 2013;366:224–234. [DOI] [PubMed] [Google Scholar]
- 9. Kim SY, Jeong S, Jung E, et al. AMP-activated protein kinase-α1 as an activating kinase of TGF-β-activated kinase1 has a key role in inflammatory signals. Cell Death. Dis. 2012;3:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. [DOI] [PubMed] [Google Scholar]
- 11. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. [DOI] [PubMed] [Google Scholar]
- 12. Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-α function. Nature. 1997;389:610–614. [DOI] [PubMed] [Google Scholar]
- 13. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. [DOI] [PubMed] [Google Scholar]
- 16. Clark K, MacKenzie KF, Petkevicius K, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci USA. 2012;109:16986–16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janssens S, Beyaert R. Role of Toll-like receptors in pathogen recognition. Clin Microbiol Rev. 2003;16:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. [DOI] [PubMed] [Google Scholar]
- 19. Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. [DOI] [PubMed] [Google Scholar]
- 20. Sato S, Sanjo H, Takeda K, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. [DOI] [PubMed] [Google Scholar]
- 21. Kim SY, Shim JH, Chun E, Lee KY. Reciprocal inhibition between the transforming growth factor-β-activated kinase 1 (TAK1) and apoptosis signal-regulating kinase 1 (ASK1) mitogen-activated protein kinase kinase kinases and its suppression by TAK1-binding protein 2 (TAB2), an adapter protein for TAK1. J Biol Chem. 2012;287:3381–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kishida S, Sanjo H, Akira S, Matsumoto K, Ninomiya-Tsuji J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells. 2005;10:447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. [DOI] [PubMed] [Google Scholar]
- 24. Kanayama A, Seth RB, Sun L, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. [DOI] [PubMed] [Google Scholar]
- 25. Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 2003;22:6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MAL T1 in T lymphocytes. Mol Cell. 2004;14:289–301. [DOI] [PubMed] [Google Scholar]
- 27. MacKenzie KF, Clark K, Naqvi S, et al. PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J Immunol. 2013;190:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]