Abstract
The proinflammatory cytokines IL-1β and IFN-γ decrease functional islet β-cell mass in part through the increased expression of specific genes, such as inducible nitric oxide synthase (iNOS). Dysregulated iNOS protein accumulation leads to overproduction of nitric oxide, which induces DNA damage, impairs β-cell function, and ultimately diminishes cellular viability. However, the transcriptional mechanisms underlying cytokine-mediated expression of the iNOS gene are not completely understood. Herein, we demonstrated that individual mutations within the proximal and distal nuclear factor-κB sites impaired cytokine-mediated transcriptional activation. Surprisingly, mutating IFN-γ–activated site (GAS) elements in the iNOS gene promoter, which are classically responsive to IFN-γ, modulated transcriptional sensitivity to IL-1β. Transcriptional sensitivity to IL-1β was increased by generation of a consensus GAS element and decreased correspondingly with 1 or 2 nucleotide divergences from the consensus sequence. The nuclear factor-κB subunits p65 and p50 bound to the κB response elements in an IL-1β–dependent manner. IL-1β also promoted binding of serine-phosphorylated signal transducer and activator of transcription-1 (STAT1) (Ser727) but not tyrosine-phosphorylated STAT1 (Tyr701) to GAS elements. However, phosphorylation at Tyr701 was required for IFN-γ to potentiate the IL-1β response. Furthermore, coactivator p300 and coactivator arginine methyltransferase were recruited to the iNOS gene promoter with concomitant displacement of the coactivator CREB-binding protein in cells exposed to IL-1β. Moreover, these coordinated changes in factor recruitment were associated with alterations in acetylation, methylation, and phosphorylation of histone proteins. We conclude that p65 and STAT1 cooperate to control iNOS gene transcription in response to proinflammatory cytokines by a coactivator exchange mechanism. This increase in transcription is also associated with signal-specific chromatin remodeling that leads to RNA polymerase II recruitment and phosphorylation.
Diabetes mellitus results when the function and mass of the insulin-producing islet β-cells within the pancreas are diminished. Distinct etiologies for this progression to losses in functional β-cell mass exist. These include autoimmune-mediated activation of resident macrophages and recruitment of additional leukocytes into the islets, such as that seen in type 1 diabetes (1, 2). Alternatively, accumulation of excess lipid through caloric overconsumption leads to activation of inflammatory signaling pathways that compromise tissue function and viability, which can lead to type 2 diabetes (3, 4). Islet β-cell exposure to proinflammatory cytokines, such as IL-1β, decreases insulin secretion and eventually causes decrements in pancreatic β-cell mass (1, 5). This is significant because IL-1β is associated with the onset and progression of both type 1 and type 2 diabetes (1, 6–9). Thus, both major forms of diabetes can be viewed as metabolic diseases caused by, or associated with, pathological inflammation (10, 11).
Pancreatic β-cells express the receptor activated by IL-1β (12); signaling through the IL-1β receptor pathway coordinately alters the intracellular protein milieu, which includes accumulation of the cyclooxygenase and inducible nitric oxide synthase (iNOS) proteins (13–16). iNOS catalyzes the production of micromolar amounts of nitric oxide (NO) using l-arginine as a substrate, and the massive increase in NO decreases aconitase enzyme activity and inhibits insulin secretion (17, 18). Moreover, the destructive effects of IL-1β-induced NO accumulation include reduced mitochondrial glucose oxidation (19), depletion of cellular ATP levels (20), and ultimately β-cell death (20–22). Protection from NO-mediated β-cell destruction can be achieved with selective iNOS inhibitors (13, 17, 23). In addition, inhibition of iNOS enzyme activity delays development of diabetes in nonobese diabetic mice (24, 25), a rodent model of autoimmune diabetes. The priming and potentiating effects of IFN-γ are mediated through the signal transducer and activator of transcription-1 (STAT1) in both rat islets and islets from NOD mice (26). Moreover, the amount of IL-1β required to produce iNOS and, subsequently, NO is significantly lowered in the presence of IFN-γ (27). Thus, 2 critical pathways for communicating the IL-1β and IFN-γ signals to specific gene promoters are NF-κB (15, 28, 29) and Janus kinase (JAK)-STAT (26, 27, 30, 31), respectively.
An analysis of the regulatory sequences present in rodent and human iNOS gene promoters reveals IFN-γ-activated sites (GASs) as well as binding sites for nuclear factor-κB (NF-κB) proteins (31–33). NF-κB is a dimer of proteins consisting of RelA/p65, RelB, c-Rel, p50, and p52; the prototypical heterodimer mediating increases in gene transcription is the combination of p65 and p50 (34). The GAS regulatory regions are bound by proteins of the STAT family. STAT1 was the first family member discovered and is typically activated by IFN-γ (35). Transgenic expression of a dominant-negative IFN-γ receptor (30) or genetic deletion of STAT1 (36) affords protection against cytokine-mediated β-cell damage. In both of these experimental animal models, expression of the iNOS gene was reduced, leading to diminished nitric oxide production and protection from autoimmune β-cell destruction. Whereas the signals promoting increases in the expression of the iNOS gene are appreciated, the transcriptional mechanisms responsible for cytokine-mediated activation of the iNOS gene in pancreatic β-cells are not well understood.
Therefore, in this study, we examined the molecular events underlying transcription of the iNOS gene in response to IL-1β and IFN-γ. We discovered that NF-κB and GAS sequences within the iNOS gene promoter cooperatively control transcriptional activation. Unexpectedly, we found that the GAS sequences, which are known to mediate responses to IFN-γ, also modulate the sensitivity of the iNOS gene to IL-1β. The ability of the GAS regulatory element to alter the sensitivity of the iNOS gene to IL-1β required STAT1 phosphorylated at Ser727, but not Tyr701. Furthermore, IL-1β-mediated transcriptional activation of the iNOS gene involved a coactivator exchange mechanism, whereby CREB-binding protein (CBP) is excluded from the promoter region concomitant with p300 and coactivator arginine methyltransferase (CARM1) recruitment. These molecular events are associated with chromatin remodeling, phosphorylation of RNA polymerase (RNA Pol II) at Ser5 within the core promoter and Ser2 on the coding region of the iNOS gene.
Materials and Methods
Cell culture, islet isolation, adenoviral vectors, and reagents
Selection strategy, culture, and passage of 832/13 and INS-1E rat insulinoma cells were described previously (37, 38). Islets were isolated from Wistar rats as outlined previously (39). IL-1β was from Thermo Scientific, and IFN-γ was purchased from Shenandoah Biotechnology Inc. The JAK inhibitor was from EMD Millipore. Recombinant adenoviruses expressing β-galactosidase (40), NF-κB p65 (41), IκBα superrepressor (42), and wild-type (WT) STAT1 (43) have all been described previously. The −1-kb WT rat iNOS luciferase reporter construct was a gift from Dr Hans Hohmeier (Duke University, Durham, North Carolina). We have previously described the generation of a recombinant adenovirus encoding STAT1 with the Y701F mutation (44). Primer pairs used to generate a S727A mutation in the STAT1 cDNA within the adenoviral shuttle vector pACCMV were as follows: forward (F) 5′ GAAAACCTGCTTCCCATGGCTCCAGAGGAGTTTGAC 3′ and reverse (R) 5′ GTCAAACTCCTCTGGAGCCATGGGAAGCAGGTTTTC 3′. This mutation was incorporated using a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer's recommended protocol. Recombinant adenovirus was then constructed using a protocol described previously (45).
RNA isolation, cDNA synthesis, and real-time RT-PCR
Isolation of total RNA, cDNA synthesis, and real-time RT-PCR have been described previously (44). Primers used in RT-PCR reactions were designed using Primer3Plus software and are available upon request.
Isolation of protein, immunoblot analysis, and immunofluorescence
Whole-cell lysates were prepared using M-PER (Thermo Scientific) supplemented with protease and phosphatase inhibitor cocktails (Thermo Scientific). The protein concentration of the lysate was determined using the bicinchoninic acid (BCA) assay (Thermo Scientific). SDS-PAGE, transfer to polyvinylidene difluoride membranes, membrane blocking before antibody incubation, and downstream subsequent detection using a ChemiDoc imaging system (BioRad) have been described previously (20). Antibodies were from the following sources: tubulin, PO4-Y701 STAT1, PO4-S727 STAT1, total STAT1, and p65 were all from Cell Signaling Technology; β-actin was from Sigma-Aldrich; and IκBα was from Santa Cruz Biotechnology, Inc. Fixation of cells, immunocytochemistry, and epifluorescence microscopy were fully described elsewhere (46).
Construction of iNOS promoter luciferase reporter plasmids
Mutations were generated by site-directed mutagenesis using a QuikChange Site-Directed Mutagenesis Kit, according to the manufacturer's instructions. The following PAGE-purified primer pairs were used to incorporate these mutations: proximal GASm, F 5′ GTTTGTTCCTTTTCCCCTgcTACTGTCAATATTTCAC 3′ and R 5′ GTGAAATATTGACAGTAgcAGGGGAAAAGGAACAAAC 3′; consensus GAS, F 5′ CTGTTTGTTCCTTTTCCCCgAATACTGTCAATATTTCAC 3′ and R 5′ GTGAAATATTGACAGTATTcGGGGAAAAGGAACAAACAG 3′; proximal NF-κBm, F 5′ CACACCCTACTGGGccCTCTCCCTTTGGG 3′ and R 5′ CCCAAAGGGAGAGggCCCAGTAGGGTGTG 3′; distal NF-κBm, F 5′ GGATATGCCAGGGGccTTTTCCCTCTCTCTG 3′ and R 5′ CAGAGAGAGGGAAAAggCCCCTGGCATATCC 3′. Cloning of 3.6 kb of the rat iNOS promoter was carried out using the following primer sequences: F 5′ aaaaaACGCGTTGTGGCCAATGTTCTAGTTGAC 3′ and R 5′ aaaaaCTCGAGTTACACACTCCTTAGCAGGTTG 3′. The PCR product was digested with MluI and XhoI restriction enzymes and then inserted into the pGL3 luciferase reporter plasmid multiple cloning site between the MluI (ACGCGT) and XhoI (CTCGAG) restriction sites. The following primer pair was used to mutate the distal GAS element in the −3.6 kb iNOS-luciferase plasmid: distal GASm, F 5′ GCTTTCCCATTCCCAGgcTCTACCAACTGTTGACC 3′ and R 5′ GGTCAACAGTTGGTAGAgcCTGGGAATGGGAAAGC 3′. The proximal GASm was introduced into the −3.6 kb construct using the primer described above. Mutations were confirmed by DNA sequencing at the Molecular Biology Resource Facility at the University of Tennessee (Knoxville, Tennessee).
Plasmid and small interfering (si) RNA transfection and luciferase assays
Transient transfections of luciferase reporter plasmids into 832/13 cells was achieved using TransIT-LT1 transfection reagent (Mirus Bio). The total transfected DNA was always adjusted to 100 ng/well of a 24-well plate by addition of plasmid expressing green fluorescent protein. Transfection of preannealed Silencer Select siRNA duplexes (Life Technologies) was accomplished with DharmaFECT 1 transfection reagent (Thermo Scientific). siRNA identifications were as follows: for p65, s159516 and s159517, for p50, s135617; and for STAT1, s129043 and s129044; negative control siRNA: catalog no. AM4611). DharmaFECT Duo transfection reagent (Thermo Fisher Scientific) was used for cotransfection of luciferase plasmids and siRNA duplexes. After transfection, whole-cell lysates were prepared using Passive Lysis Buffer (Promega), and luciferase assays were read according to the manufacturer's Luciferase Assay System protocol in a GloMax plate reading luminometer (Promega).
Electrophoretic mobility shift assays
Subcellular fractionation from 832/13 cells grown to confluence in 10-cm dishes was performed using an NE-PER kit from Thermo Fisher Scientific according to the manufacturer's protocol. Oligonucleotides corresponding to the distal NF-κB sequences present in the rat iNOS gene promoter were synthesized with 5′-biotin labels, purified by HPLC, and annealed before incubation with 10 μg of nuclear protein. The sequence of the sense oligonucleotide (NF-κB site underlined) is 5′ ATATGCCAGGGGGATTTTCCCTCTCTCTGT 3′. Unlabeled oligonucleotides (ie, without biotin tags) were synthesized, purified, and annealed the same as the biotin-labeled oligos and served as unlabeled (cold) competitors in the binding reaction. Protein-DNA binding reactions were set up as described for the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). These reactions were separated on 6% DNA retardation gels (Invitrogen), followed by transfer to nylon membranes. The protein-DNA complexes were cross-linked to the nylon membrane by using a UV transilluminator with 312-nm bulbs. Blocking of the membrane and subsequent downstream detection of the biotin label were as described in the LightShift Chemiluminescent EMSA Kit protocol.
Chromatin immunoprecipitation (ChIP)
832/13 cells were cultured in 10-cm dishes, using 1 dish per treatment condition. After treatment of cells, medium was aspirated, and formaldehyde in PBS was added at a final concentration of 1% and incubated at room temperature for 10 minutes. Glycine was added to a final concentration of 125 mM for 5 minutes at room temperature. Cells were washed once with cold PBS and scraped into 1-mL prechilled PBS with 1× protease inhibitors (Halt Protease Inhibitor Cocktail; Thermo Fisher Scientific). Cell pellets were collected by centrifugation at 500g for 2 minutes at 4°C. Supernatants were discarded, and pellets were reconstituted in 0.5 mL of SDS lysis buffer containing 1× protease inhibitors (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl; pH 8.0). Lysates were incubated on ice for 15 minutes, and then DNA was sheared by sonication using a Misonix Sonicator S-4000 (Thermo Fisher Scientific). The following sonication conditions generated fragments of DNA between 100 and 500 bp in length: amplitude, 4; process time, 10 minutes; pulse on time, 10 seconds; and pulse off time, 20 seconds. Sonicated fragments were centrifuged at 12,000g for 10 minutes at 4°C, and supernatants were transferred to a clean microcentrifuge tube. Preclearing was performed for 1 hour at 4°C with rotation using 5 μL of magnetic protein G beads (Thermo Scientific). Once beads were removed, 100 μL of sheared cross-linked DNA was diluted 10-fold in dilution buffer containing 1× protease inhibitors (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA,167 mM NaCl, and 17 mM Tris, pH 8.0). Then 2 μg of immunoprecipitating antibodies were added (normal rabbit or mouse IgG, p65, p50, STAT1, or Pol II; Santa Cruz Biotechnology, Inc) and incubated overnight at 4°C with rotation. Immunoprecipitation of DNA protein-antibody complexes was performed by incubation with 10 μl of protein A (for rabbit polyclonal antibodies) or G (for mouse monoclonal antibodies) Dynabeads (Invitrogen) for 1 hour at 4°C. The magnetic beads binding to the antibody-chromatin complexes were washed sequentially with 0.5 mL of the following buffers: 1× low-salt buffer, (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris; pH 8.0), 1× high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris; pH 8.0); and 1× LiCl buffer (0.25 M LiCl, 1% NP-40, 1 mM EDTA, and 10 mM Tris; pH 8.0), and 2× Tris EDTA. Once the last wash buffer was completely removed, 100 μL of 10% Chelex-100 resin in PBS was added, vortexed for 10 s, and heated to 100°C for 10 minutes. After centrifugation at 17,000g for 1 minute at 4°C, the supernatants were transferred to new tubes. Then 120 μL of RNase and DNase-free water was added, vortexed for 10 s, and centrifuged at 17,000g for 1 minute at 4°C. Supernatants were pooled. Inputs were processed as followed: 20 μL of sonicated, precleared DNA was incubated overnight at 65°C with NaCl to a final concentration of 200 mM. Input DNA was then treated with RNase A for 30 minutes at 37°C and proteinase K for 1 hour at 45°C and then purified using a QIAGEN Cleanup kit (QIAGEN); 2.5 μL of DNA was used as a template for PCR. Primer sequences for amplifying the iNOS proximal and distal NF-κB and GASs were generated using Primer3Plus software and are available upon request. Antibodies used for immunoprecipitation were as follows: acetylated histone H3 (K9, ab4441), Pol II pCTDSer5 (ab5131), and Pol II pCTDSer2 (ab5095) from Abcam; p300 (05-257), phospho-histone H3 (Ser10; 04-817) and acetylated histone H4 (K5, K8, K12, and K16; 06-866) from EMD Millipore; CBP (sc-369x), p65 (sc-372x), CARM1 (sc-5421x), p50 (sc-7178x), and STAT1 (sc-464x) from Santa Cruz Biotechnology, Inc; phospho-STAT1 (Tyr701; 9171) and phospho-STAT1 (Ser727; 9177) from Cell Signaling Technology; and rabbit (I8140) and mouse IgG (I8765) from Sigma-Aldrich.
Coimmunoprecipitation assay
832/13 cells were cultured in 10-cm dishes until confluent, using 1 dish per treatment condition. After treatment of cells as described in the figure legends, medium was aspirated, and cells were washed once with cold PBS and scraped into 1 mL of prechilled PBS with 1× protease inhibitors (Halt Protease Inhibitor Cocktail; Thermo Scientific). Cell pellets were collected by centrifugation at 500g for 2 minutes at 4°C. Supernatants were discarded, and pellets were reconstituted in 0.5 mL of immunoprecipitation buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl, pH 8.0) containing 1× protease inhibitors. Lysates were incubated on ice for 15 minutes and centrifuged at 12,000g for 10 minutes at 4°C, and supernatants were transferred to a clean microcentrifuge tube. Preclearing was performed for 1 hour at 4°C with rotation using 5 μL of magnetic protein G beads (Thermo Scientific). Once beads were removed, 200 μL of lysate was moved to a new tube, and 2 μg of immunoprecipitating antibodies was added (normal rabbit IgG; Sigma Aldrich, and p65; Santa Cruz Biotechnology, Inc) and incubated overnight at 4°C with rotation. Immunoprecipitation of antigen-antibody complexes was performed by incubation with 10 μL of protein A Dynabeads (for rabbit polyclonal antibodies; Invitrogen) or protein G Dynabeads (for mouse monoclonal antibodies) for 30 minutes at 4°C, followed by 15 minutes at room temperature. The magnetic beads binding to the antigen-antibody complexes were washed 3 times with 0.5 mL of immunoprecipitation buffer. Buffer was removed, and beads containing antigen-antibody complexes were resuspended in NuPAGE LDS Sample Buffer (Invitrogen). Precipitated proteins and input samples (10 μL of precleared cell lysate) were denatured and separated by SDS-PAGE on 4% to 12% Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride membranes (EMD Millipore). Membranes were blocked in 3% BSA for 45 minutes, followed by overnight incubation with p65, STAT1, or IκBα-specific antibodies.
Statistical analysis
A 1-way ANOVA, followed by Tukey's post hoc correction, or the Student t test, were used to identify statistical significances at the confidence levels reported within the figure legends.
Results
IFN-γ potentiates the IL-1β–mediated expression of the iNOS gene
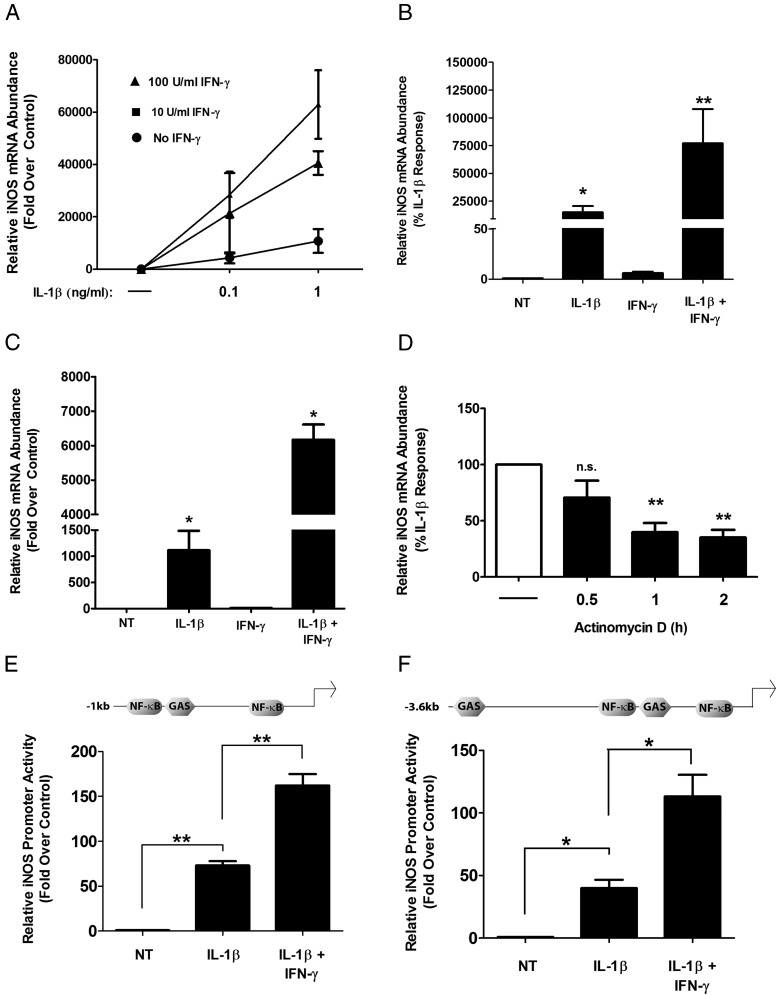
Using 832/13 rat insulinoma cells, we determined that IL-1β markedly increased iNOS mRNA accumulation and that IFN-γ potentiated the IL-1β signal in a concentration-dependent fashion (Figure 1A). We further found that INS-1E rat insulinoma cells, selected via a different strategy than that for the 832/13 cells (37, 38), also display IFN-γ-induced potentiation of the IL-1β–controlled increase in the expression of the iNOS gene (Figure 1B). The results using β-cell lines were confirmed with isolated rat islets (Figure 1C). IFN-γ alone does not stimulate iNOS mRNA to appreciable levels in either rat islets or β-cell lines (Figures 1, A–C), which is consistent with a previous report (27). We next investigated the stability of the iNOS mRNA and discovered that turnover of the iNOS transcript occurs rapidly (>50% degradation within 1 hour) after withdrawal of the IL-1β stimulus (Figure 1D). In addition, using either −1 kb or −3.6 kb (relative to the transcriptional start site), we observed that the iNOS gene promoter is strongly activated by IL-1β within 4 hour; IFN-γ potentiates this response an additional 2- to 3-fold (Figure 1, E and F). The findings reported here for rat islets and β-cell lines are congruent with similar experimental results reported for mouse, rat, and human islets (26, 27). We thus chose to carry out further analysis on transcriptional mechanisms of the iNOS gene using the 832/13 cell line.
Figure 1.
Expression of the iNOS gene is induced by proinflammatory cytokines in rat islets and β-cell lines. A, 832/13 cells were treated with 0.1 or 1 ng/mL of IL-1β alone or in combination with 0, 10, or 100 U/mL IFN-γ for 6 hours. B and C, INS-1E (B) or rat islets (C) were treated with 1 ng/mL (INS-1E) or 10 ng/mL (rat islets) IL-1β in the presence or absence of 100 U/mL IFN-γ for 6 hours as indicated on the x-axis. D, After a 6-hour stimulation with 1 ng/mL IL-1β, cells were treated with actinomycin D at time 0 (indicated by the dashed line), and total RNA was isolated after 0.5, 1, or 2 hours. A–D, Total RNA was isolated and iNOS mRNA levels were measured and normalized to those of the housekeeping gene, ribosomal S9 (RS9). **, P < .01 vs untreated (NT) (B), *, P < .05 vs NT (B and C), **, P < .01 vs no actinomycin D (D, white bar); n.s., not significant vs no actinomycin D (D, white bar). E and F, 832/13 cells were transfected with either −1 kb (E) or −3.6 kb (F) of the iNOS promoter upstream of the transcriptional start site fused to a luciferase reporter. At 24 hours posttransfection, cells were stimulated for 4 hours with 1 ng/mL IL-1β or 1 ng/mL IL-1β plus 100 U/mL IFN-γ, followed by measurement of promoter activity. **, P < .01 vs IL-1β (E); *, P < .05 vs IL-1β (F). Data are means ± SEM from 3 to 4 individual experiments.
NF-κB is required for cytokine-mediated activation of the iNOS gene
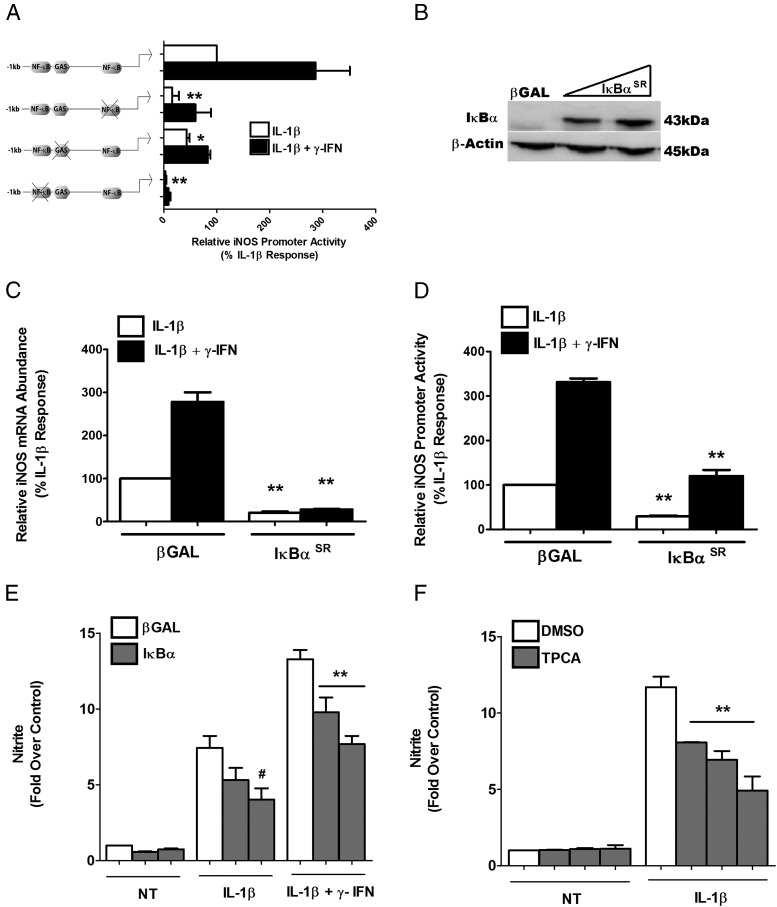
We next investigated the genomic sequences and regulatory factors required to modulate cytokine responsiveness of the iNOS gene promoter. Using specific loss-of-function mutations in the distal and proximal κB sites or in the GAS sequences, we discovered that the responses to IL-1β and IFN-γ were specific to each individual regulatory sequence. For example, specifically mutating the proximal κB element reduced the IL-1β response by 85%, although the fold potentiation by IFN-γ remained similar to that of the WT (approximately 3-fold), albeit with a reduction in the magnitude of the overall transcriptional response. In contrast, mutations in the distal κB site completely eliminated the response to both cytokines (Figure 2A). The iNOS promoter with a GAS mutation displayed a 71% decrease in the IFN-γ response, whereas the fold potentiation by IFN-γ of the IL-1β response was reduced by only 33%. The unexpected finding was that the GAS mutation, which we expected to only affect the IFN-γ response, had a substantial effect on the transcriptional activation by IL-1β (∼50% decrease) (Figure 2A).
Figure 2.
IκBα attenuates cytokine-mediated activation of the iNOS gene. A, 832/13 cells were transfected with a wild-type iNOS promoter luciferase plasmid (−1 kb) or iNOS promoter with mutations in the proximal or distal NF-κB response elements or in the GAS element. At 24 hours posttransfection, cells were stimulated with 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 4 hours. B, 832/13 cells were transduced with increasing doses of an adenovirus expressing the IκBα S32A/S36A (IκBαSR [superrepressor]), followed by collection of total protein and immunoblotting for IκBα with β-actin as the loading control. The highest concentration of virus shown in the immunoblot was used for the experiments shown in panels C and D. This blot is representative of 2 independent experiments. C, 832/13 cells were treated with recombinant adenoviruses expressing either β-galactosidase (βGAL) or IκBαSR. At 24 hours posttransduction, cells were stimulated for 6 hours with either 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ. Relative iNOS mRNA levels were quantified and normalized to RS9. **, P < .01 vs corresponding white or black bar, respectively. D, 832/13 cells were transfected with the −1 kb promoter-luciferase construct, and 4 hours later were treated with the indicated adenoviruses, cultured overnight, and then exposed to either 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for an additional 4 hours. Promoter activity was measured by luciferase assay. **, P < .01 vs the corresponding white or black bar, respectively. E, 832/13 cells were treated with recombinant adenoviruses expressing either βGAL or 5 and 10 multiplicity of infection of IκBαSR for 12 hours and then were stimulated with cytokines as indicated for an additional 12 hours. Doses used are the same as those shown in panel B above. F, Cells were treated with dimethylsulfoxide or 0.5, 1, or 2 μM TPCA and concurrently stimulated with IL-1β for 12 hours. E and F, Nitrite production was measured from the culture media using the Griess assay. #, P < .1 vs βGAL-IL-1β; **, P < .01 vs βGAL-IL-1β + IFN-γ (E); **, P < .01 vs DMSO-IL-1β (F). Data are means ± SEM from 3 to 4 individual experiments. NT, untreated.
We subsequently examined how interfering with the NF-κB signaling pathway affected the expression of the iNOS gene and promoter activity in the presence of IL-1β and IFN-γ. First, overexpressing the IκBα superrepressor, which is resistant to phosphorylation-induced degradation (42), increased IκBα protein in 832/13 cells (Figure 2B). This maneuver produced an 80% decrease in the expression of the iNOS gene in response to either IL-1β or the combination of IL-1β plus IFN-γ (Figure 2C) using the highest dose shown in Figure 2B. Next, using the iNOS promoter luciferase reporter constructs, we observed a 70% decrease in the IL-1β response and a 64% decrease in the response to IL-1β + IFN-γ (Figure 2D). Furthermore, both IL-1β and IL-1β + IFN-γ stimulated nitrite accumulation in the culture medium, which was decreased by 46% and 42%, respectively, in the presence of the highest dose of the superrepressor (Figure 2E). Because nitrite accumulation correlates with iNOS protein abundance and is an index of NO production (20, 27), our results are also consistent with the reduction in cytokine-mediated iNOS protein (data not shown). Furthermore, there were 31%, 46%, and 58% decreases in nitrite production using the compound 2-[(aminocarbonyl) amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide (TPCA) (Figure 2F), a pharmacological inhibitor of IKKβ (47). This pharmacological inhibitor experiment confirms the results seen with the IκBα superrepressor and is supportive of a major role for the NF-κB signaling pathway to regulate expression of the iNOS gene in response to cytokines.
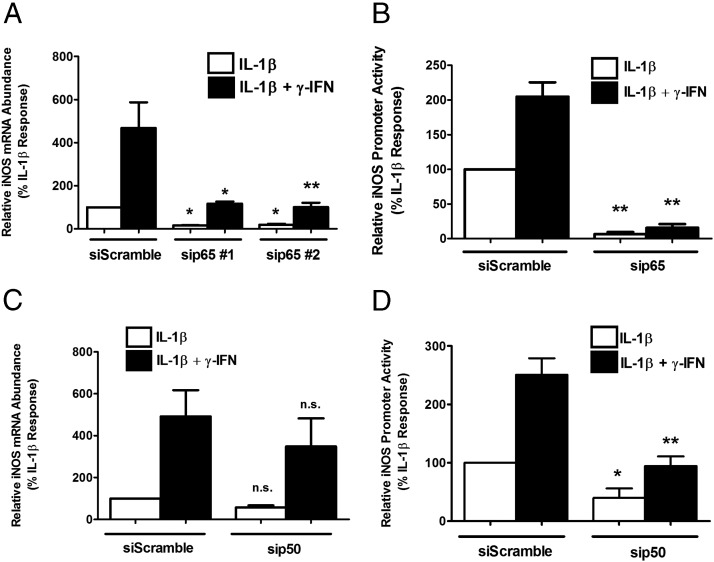
To identify whether the p65 subunit of NF-κB was involved in transcription of the iNOS gene, we used siRNA-mediated suppression of p65 (77% and 75% decreases in p65 mRNA with duplex 1 and 2, respectively) in the presence of IL-1β or IL-1β plus IFN-γ. Decreasing p65 blunted the ability of cytokines to induce iNOS mRNA accumulation by 83.9% and 80.8%, respectively, with 2 separate targeting sequences (Figure 3A). Furthermore, iNOS promoter activity was diminished by 93.4% (Figure 3B) relative to the siScramble control. We next targeted p50, one of the major heterodimer partners of p65, by also using transfected siRNA duplexes (65% decrease in p50 mRNA). IL-1β-mediated induction of iNOS mRNA and promoter activity were decreased by 43.5% (Figure 3C) and 60.2% (Figure 3D) by siRNA-directed suppression of p50, with no fold change in the IFN-γ-driven potentiation of the IL-1β response (Figure 3, C and D).
Figure 3.
Cytokine-stimulated expression of the iNOS gene requires the NF-κB subunits p65 and p50. A and C, 832/13 cells were transfected with siRNA duplexes targeting either p65 (A) or p50 (C) with a scrambled sequence as a control for siRNA transfection (siScramble). At 48 hours posttransfection, cells were exposed to either 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 6 hours. A and C, Total RNA was isolated, and iNOS transcript levels were normalized to that of RS9. *, P < .05 vs corresponding siScramble white or black bar, respectively (Α); **, P < .01 vs corresponding siScramble white or black bar, respectively (Α); n.s., not significant vs siScramble white or black bar, respectively (C). B and D, 832/13 cells were cotransfected with a −1 kb promoter-luciferase construct and siRNA duplexes targeting either p65 (B) or p50 (D). At 24 hours posttransfection, cells were stimulated with 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 4 hours. B and D, Promoter luciferase activity was measured. *, P < .05 vs corresponding siScramble white or black bar, respectively (D); **, P < .01 vs corresponding siScramble white or black bar, respectively (Β and D). Values are means ± SEM from 3 to 4 individual experiments.
IL-1β promotes p65 nuclear entry and DNA binding
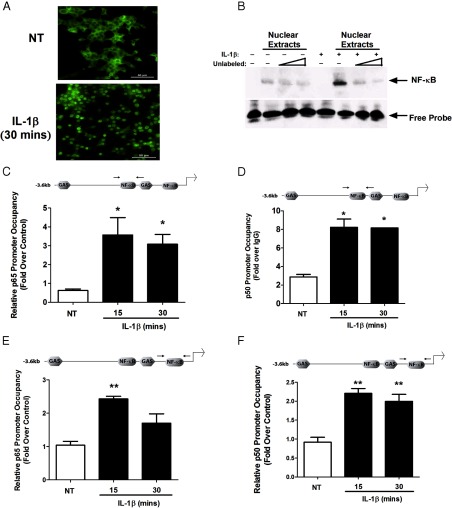
NF-κB responsive elements and the p65 transcriptional subunit are required for cytokine-mediated expression of the iNOS gene in pancreatic β-cells (Figures 2 and 3). We subsequently established that IL-1β promoted degradation of the IκBα regulatory protein (not shown), which leads to p65 nuclear accumulation detected via immunofluorescence (Figure 4A). In addition, isolation of nuclear protein from 832/13 cells exposed to IL-1β for 30 minutes reveals a complex that binds to oligonucleotides containing an iNOS κB sequence (Figure 4B). Therefore, we next examined whether p65 and its associated dimer partner p50 bound to κB sites within the endogenous iNOS gene promoter in an IL-1β-dependent manner. Using ChIP, we detected 5.83- and 5.13-fold increases in p65 occupancy at the distal κB site within 15 and 30 minutes of cellular IL-1β exposure, respectively (Figure 4C). Similar results were obtained when p50 was immunoprecipitated at the distal κB site, with an 2.8-fold increase over the IgG control within 15 minutes of IL-1β exposure (Figure 4D). The proximal κB site also displayed a significant 2.43-fold (Figure 4E) and 2.20-fold (Figure 4F) recovery with p65 and p50 antisera, respectively, after a 15-minute incubation with IL-1β.
Figure 4.
IL-1β induces recruitment of p65 and p50 to regions in the iNOS gene promoter containing κB response elements. A, 832/13 cells were untreated (NT) or exposed to 1 ng/mL IL-1β for 30 minutes. Immunofluorescence assays were used to track nuclear localization of p65. The assay was performed on 3 independent occasions, and representative images are shown (scale bar corresponds to 50 μm). B, 832/13 cells were untreated (−) or stimulated with 1 ng/mL IL-1β for 30 minutes (+). Electrophoretic mobility shift assays were performed using nuclear extracts incubated in the presence or absence of increasing concentrations of an unlabeled (“cold”) competitor oligonucleotide. The arrow indicates the specific complex bound to the NF-κB DNA sequence from the iNOS gene promoter. C–F, 832/13 cells were untreated or stimulated with 1 ng/mL IL-1β for 15, 30, or 60 minutes. ChIP assays were conducted with antibodies that immunoprecipitated p65 (C and E) or p50 (D and F). Distal (C and D) and proximal (E and F) NF-κB elements (indicated by the arrows in the schematic diagrams) in the iNOS promoter were targeted for amplification in the recovered DNA by real-time PCR. *, P < .05 vs respective NT control (C and D); **, P < .01 vs respective NT control (E and F). All data in panels C to F represent means ± SEM from 3 to 4 individual experiments, whereas the image in panel B is representative of 3 independent experiments.
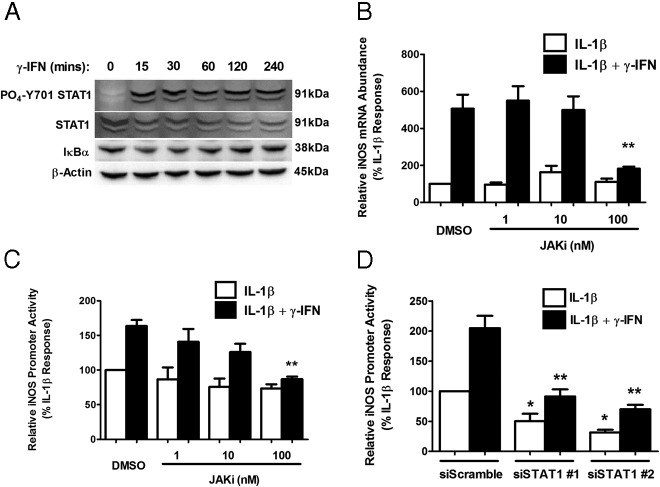
Pharmacological Inhibition of JAK1 or STAT1 knockdown attenuates the cytokine-mediated response of the iNOS gene
Because IFN-γ potentiates the IL-1β–mediated induction of the iNOS gene (Figure 1 and Ref. 27), we examined STAT1 phosphorylation in response to IFN-γ. We found that STAT1 phosphorylation at Tyr701 occurs within 15 minutes of exposure to IFN-γ and is maintained out to 4 hours (Figure 5A). In addition, pharmacological inhibition of JAK1 blocked the ability of IFN-γ to potentiate the IL-1β–mediated induction of the iNOS gene by 64% (Figure 5B). Potentiation of promoter activity by IFN-γ is also decreased (by 47%) in the presence of 100 nM JAK inhibitor (Figure 5C). To confirm the involvement of STAT1, we used siRNA duplexes targeting STAT1 mRNA and observed 55% and 66% decreases in the IFN-γ-mediated potentiation, respectively, using 2 different siRNA targeting sequences (Figure 5D, black bars). Furthermore, we note that siRNA targeting of STAT1 decreased iNOS gene promoter activity in response to IL-1β by 51% and 69%, respectively, with the 2 duplexes (Figure 5D, white bars). These data are consistent with the GAS element mutation impairing the transcriptional response to IL-1β (Figure 2A). Thus, STAT1 is required for both the IL-1β and the IFN-γ stimulation of iNOS gene transcription, which to our knowledge, has not been reported previously.
Figure 5.
Pharmacological inhibition of JAK1 attenuates the IFN-γ-mediated potentiation of IL-1β-stimulated iNOS gene expression. A, 832/13 cells were stimulated with 100 U/mL IFN-γ for 15, 30, 60, 120, and 240 minutes. Whole-cell lysates were harvested and blotted for phospho-STAT1 (Y701) and total STAT1, IκBα, and β-actin as the loading control. B, 832/13 cells were pretreated for 1 hour with the indicated doses of the JAK inhibitor (JAKi) and then were incubated for an additional 6 hours with 1 ng/mL IL-1β alone or in combination with 100 U/mL IFN-γ. iNOS mRNA abundance was quantified and normalized to that of RS9. **, P < .01 vs dimethylsulfoxide (DMSO) and IL-1β and IFN-γ. C, 832/13 cells were transfected with a −1-kb iNOS-luciferase reporter plasmid. At 24 hours posttransfection, cells were pretreated for 1 hour with the indicated doses of a JAKi and then were stimulated for 4 hours with either 1 ng/mL IL-1β or IL-1β plus 10 U/mL IFN-γ. Promoter activity was measured at the end of the 4-hour period. **, P < .01 vs DMSO and IL-1β and IFN-γ. D, 832/13 cells were transfected concomitantly with a −1-kb promoter-luciferase construct and siRNA duplexes targeting STAT1. At 24 hours posttransfection cells, were stimulated with 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 4 hours. Promoter activity was assessed by luciferase assay. *, P < .05 vs siScramble-IL-1β; **, P < .01 vs siScramble-IL-1β + IFN-γ. Data are means ± SEM from 3 individual experiments.
Sensitivity of the iNOS gene to IL-1β is modulated by response elements recognized by STAT1
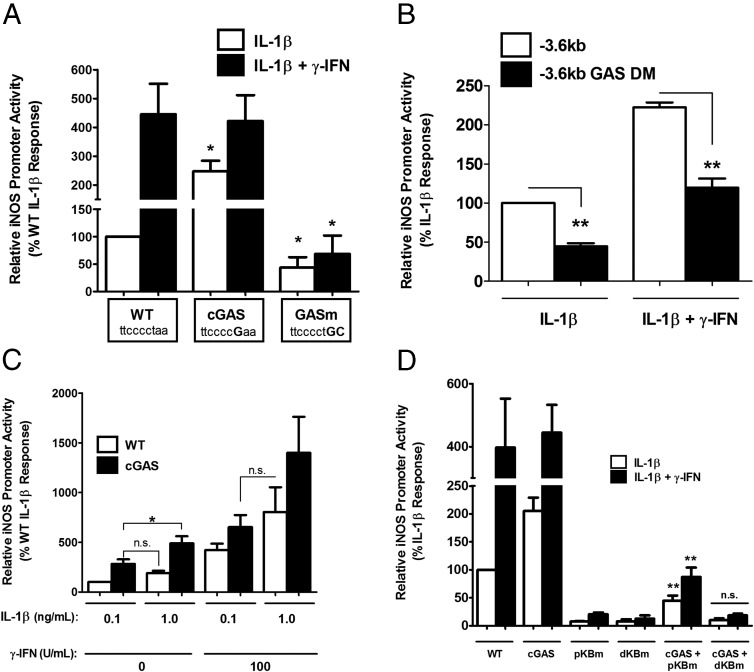
To further examine regulation of the signaling input to the iNOS gene by STAT1, we altered the GAS sequence positioned between the distal and proximal κB sites of the −1 kb gene promoter to either a perfect consensus sequence (requiring a 1-nucleotide substitution) or to a nonfunctional sequence (2-nucleotide substitution). Using this approach, we discovered that creating a consensus GAS (cGAS) element increased the response to IL-1β by 2.47-fold (Figure 6A, white bars). In contrast, the loss-of-function mutation diminished the response to IL-1β by 57% (Figure 6A, white bars) while also completely impairing the ability of IFN-γ to potentiate the IL-1β response (Figure 6A, black bars). Interestingly, converting the GAS to a perfect consensus sequence did not enhance the response to IFN-γ, despite this being a well-known STAT1 binding site (48).
Figure 6.
GAS elements within the iNOS gene promoter regulate sensitivity to IL-1β. A, 832/13 cells were transfected with a −1-kb iNOS promoter-luciferase reporter construct containing either the WT GAS, a perfect cGAS, or a GASm. At 24 hours posttransfection cells were stimulated with either 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 4 hours. *, P < .05 vs WT for intragroup comparisons. B, 832/13 cells were transfected with either a −3.6-kb iNOS promoter-luciferase construct or a −3.6-kb construct wherein both the proximal and distal GAS elements are mutated (−3.6-kb GAS double mutant [GAS DM]). After overnight culture, cells were treated with either 1 ng/mL IL-1β or IL-1β plus 100 U/mL IFN-γ for 4 hours. **, P < .01. C, 832/13 cells were transfected with either a −1-kb WT iNOS promoter-luciferase reporter or a −1-kb cGAS promoter-luciferase construct. After overnight culture, cells were treated with either 0.1 or 1 ng/mL IL-1β in the presence or absence of 100 U/mL IFN-γ for 4 hours. *, P < .05; n.s., not significant. D, 832/13 cells were transfected with the indicated −1-kb plasmids. At 24 hours posttransfection, cells were stimulated with 1 ng/mL IL-1β alone or in the presence of 100 U/mL IFN-γ. **, P < .01 vs the respective pκBm control; n.s., not significant vs dκBm. A–D, Promoter activity was assessed by luciferase assay. Data are means ± SEM from 3 individual experiments.
We also generated loss-of-function mutations in both GAS elements contained within the −3.6-kb iNOS gene promoter. Again, there was a blunting of both the response to IL-1β (56% decrease with GASm relative to WT) and of the response to IL-1β + IFN-γ by 46% (Figure 6B). To test the hypothesis that the GAS element may be increasing the sensitivity of the iNOS gene promoter to IL-1β, we compared the responses of the WT and cGAS promoters after exposure to 2 different concentrations of IL-1β: 0.1 ng/mL (submaximal) and 1 ng/mL (maximal). These concentrations of IL-1β have been validated for cellular gene expression responses in pancreatic β-cells (44, 46). We found that 0.1 ng/mL IL-1β produced a response on the cGAS promoter (Figure 6C, black bars) similar to that seen with 1 ng/mL IL-1β on the WT promoter (Figure 6C, white bars). The increase in sensitivity to IL-1β was also maintained in the presence of potentiating amounts of IFN-γ (Figure 6C, compare black bar for 0.1 ng/mL IL-1β with white bar for 1.0 ng/mL IL-1β, all in the presence of 100 U/mL IFN-γ). Thus, the GAS element in the iNOS gene promoter regulates transcriptional sensitivity to IL-1β.
In an effort to determine how the GAS element was modulating the IL-1β signal integration at the iNOS gene promoter, we next mutated either the proximal or distal κB sequences on the background of either the WT or the cGAS sequence. Using these promoter constructs, we exposed cells to either IL-1β or IL-1β + IFN-γ (Figure 6D). We discovered that mutations in either of the κB sites markedly inhibited the response to cytokines in the WT promoter, consistent with earlier results (Figure 2A). In contrast, a functional distal κB site plus cGAS element (ie, pκBm + cGAS) displayed 5.83-fold greater activity in response to IL-1β than the proximal κB mutant in the presence of WT GAS (pκBm, Figure 6D). In addition, the potentiation by IFN-γ was 4.24-fold higher in the pκBm + cGAS group compared with the pκBm group. We interpret these data to denote that the cGAS element is capable of partially rescuing the loss in cytokine responsiveness seen with mutations in the proximal κB sequences. However, mutations in the distal κB sequence cannot be offset by acquisition of a cGAS sequence, even with an intact proximal κB binding site (Figure 6D, cGAS + dκBm). Thus, the distal κB sequence and the GAS element are probably cooperating to control overall cytokine responsiveness within the iNOS gene promoter.
IL-1β promotes binding of STAT1 phosphorylated at Ser727 to the GAS regulatory element
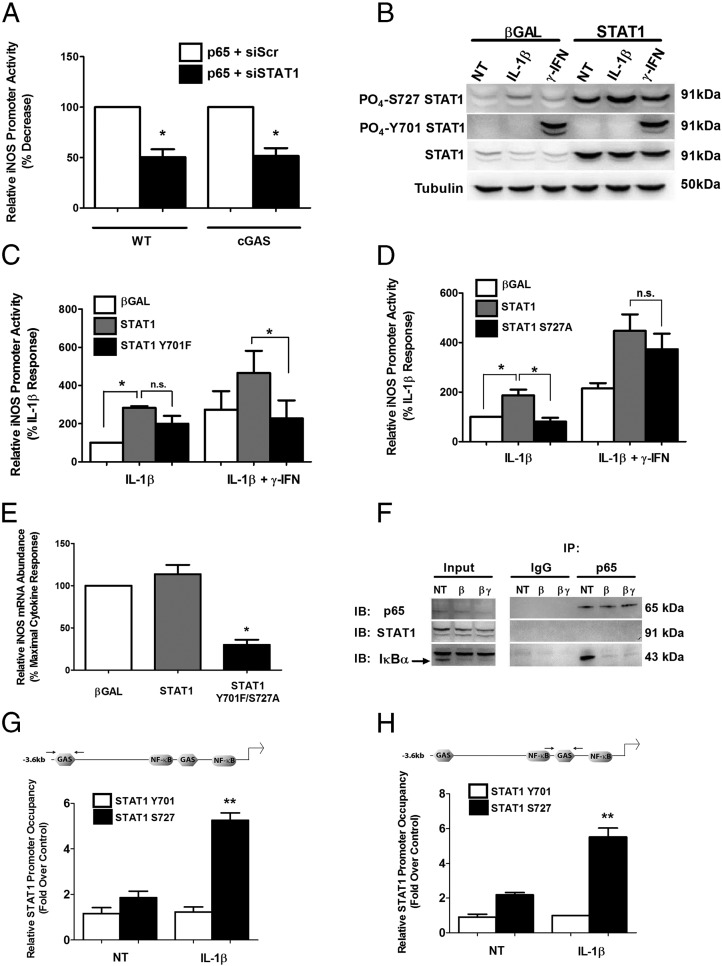
Because of the unexpected observation that the GAS element controls sensitivity of the iNOS gene to IL-1β, we subsequently examined the binding of p65 and STAT1 within the iNOS gene promoter. First, we note that transfection of p65 drives the transcription of both the WT and cGAS promoter-luciferase constructs (Figure 7A). Setting each of the promoter-luciferase responses at 100%, we observed that siRNA-mediated reductions in STAT1 (90% decrease in STAT1 mRNA) diminished the activity of each promoter by 50% (Figure 7A). Conversely, we tested the hypothesis that overexpression of STAT1 would augment expression of the iNOS gene promoter in response to either IL-1β or the combination of IL-1β + IFN-γ. In Figure 7B, we detected overexpression of the STAT1 protein and its phosphorylation in response to either IL-1β or IFN-γ. We noticed that in pancreatic β-cells, there is augmentation of phosphorylation at Ser727 within STAT1 in response to IL-1β and robust induction of phosphorylation at Tyr701 only in response to IFN-γ (Figure 7B). Therefore, we compared the impact of STAT1 overexpression with that of an adenovirus that encodes STAT1 with a Y701F mutation. We discovered that the IL-1β-mediated increase in transcriptional activity of the iNOS gene promoter is enhanced by STAT1 overexpression and that this effect is not significantly diminished in the presence of STAT1Y701F (Figure 7C). However, the inability to activate STAT1 at the Tyr701 site markedly reduced potentiation of the IL-1β response by IFN-γ (Figure 7C, black bars). In contrast, removal of the serine phosphoacceptor site at position 727 (STAT1S727A) impairs the ability of IL-1β to drive promoter activity but does not significantly affect the response to IFN-γ (Figure 7D). Further, using an adenovirus encoding a double mutation (Y701F/S727A), we observed a 71% decrease in the cytokine-stimulated expression of the iNOS gene in isolated rat islets (Figure 7E).
Figure 7.
Phosphorylation of STAT1 at Ser727 is critical for IL-1β-stimulated transcription of the iNOS gene. A, 832/13 cells were cotransfected with WT or cGAS −1-kb constructs and either an siRNA duplex containing a control sequence (siScramble) or targeting STAT1 (siSTAT1). At 24 hours posttransfection, cells were treated with adenoviruses expressing either green fluorescent protein or p65 and cultured for 12 hours. *, P < .05 vs corresponding white bar. B, 832/13 cells were treated overnight with adenoviruses expressing either β-galactosidase (βGAL) or STAT1. Cells were untreated (NT) or stimulated for 15 minutes with either 1 ng/mL IL-1β or 100 U/mL IFN-γ. Whole-cell lysates were blotted for phospho-STAT1 (S727 and Y701), total STAT1, or tubulin as the loading control. The immunoblot was repeated on 2 separate occasions. C, 832/13 cells were transfected with a −1-kb cGAS construct; 4 hours posttransfection, cells were transduced with adenoviruses expressing either βGAL, STAT1, or STAT1Y701F for overnight culture. These cells were then exposed to either 1 ng/mL IL-1β alone or IL-1β plus 100 U/mL γ-IFN for 4 hours. *, P < .05. D, 832/13 cells were transfected with cGAS −1-kb construct; 4 hours postplasmid transfection, cells were transduced with adenoviruses expressing βGAL, STAT1, or STAT1S727A overnight. After overnight culture with plasmid and virus, these cells were exposed to1 ng/mL IL-1β alone or in combination with 100 U/mL IFN-γ for 4 hours. A, C, and D, Promoter activity was assessed by luciferase assay. E, Rat islets were transduced with the indicated adenoviruses. After overnight culture, islets were treated with 10 ng/mL IL-1β and 100 U/mL IFN-γ for 6 hours. iNOS mRNA abundance was normalized to RS9. F, 832/13 cells were untreated or treated for 15 minutes with 1 ng/mL IL-1β (β) or IL-1β plus 100 U/mL γ-IFN (βγ). Lysates were immunoprecipitated (IP) with antibodies against either normal rabbit serum (IgG) or p65 and then were separated by SDS-PAGE, followed by immunoblotting (IB) with antibodies targeting p65, STAT1, or IκBα (arrow indicates IκBα as the faster migrating band). G and H, 832/13 cells were stimulated for 15 minutes with IL-1β. ChIP assays were conducted using antibodies against IgG, STAT1 Y701, and STAT1 S727. The distal and proximal GAS elements were targeted for amplification by RT-PCR using the recovered DNA as template. **, P < .01 vs STAT1 S727 untreated (black bars). Data are means ± SEM from 3 individual experiments.
We then investigated whether or not p65 and STAT1 were physically interacting in response to cellular exposure to cytokines. Using coimmunoprecipitation assays, we were unable to detect an interaction between p65 and STAT1 in the presence of either IL-1β or IL-1β + IFN-γ (Figure 7F). However, we did detect IκBα association with p65 in the basal state (absence of cytokines) and disruption of this interaction by cytokine-mediated degradation of IκBα (Figure 7F). The recovery of IκBα is consistent with a previous observation (49) and serves as a control for the robustness of the coimmunoprecipitation assay. Next, we used ChIP to determine whether occupancy of STAT1 at the GAS elements was increased in response to IL-1β. We detected an approximate 30% increase in total STAT1 occupancy at GAS elements (not shown). However, when we examined recovery using STAT1 Ser727 specific antisera, we discovered that IL-1β promoted 2.83- and a 2.52-fold increases, respectively, at the distal and proximal GAS regions (Figure 7, G and H). The Tyr701 form of STAT1 was not detected, consistent with this site not being phosphorylated in the absence of IFN-γ (Figure 7B) and not playing a significant role in activation of the promoter in response to IL-1β (Figure 7C). We interpret these data collectively to indicate that Ser727 phosphorylated STAT1 bound at the GAS element is a key contributor to IL-1β control of iNOS gene transcription.
IL-1β-mediated recruitment of the coactivator p300 and CARM1 is concurrent with removal of CBP from iNOS regulatory regions
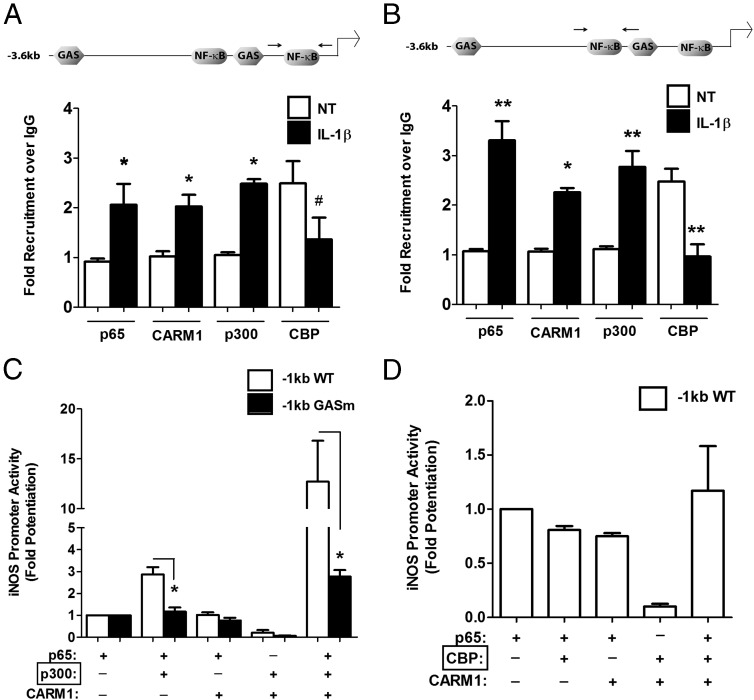
IL-1β induces nuclear entry of p65 (Figure 4A), binding of p65 and p50 at the κB sequences (Figure 4, B–F), and association of Ser727-phosphorylated STAT1 at GAS elements (Figure 7, G and H). We next examined coactivator binding at the iNOS gene promoter in response to IL-1β using ChIP assays and antibodies directed against CARM1, p300, and CBP. We discovered that IL-1β signaling induces binding of CARM1 and p300 to the iNOS gene promoter (Figure 8, A and B). Conversely, CBP was associated with the iNOS gene promoter in the basal state but was excluded concomitant with p300 and CARM1 arrival (Figure 8, A and B). To further explore this relationship between coactivators and p65, we examined whether addition of CARM1 and p300, with and without p65, was capable of enhancing iNOS promoter activity. Because loss of function mutations within the GAS element decreased sensitivity to IL-1β (Figure 6, A and B), we performed a reconstitution experiment using WT and GASm promoters. In both promoters, the ability of p65 to increase transcriptional activity over a GFP control plasmid is set at a value of 1 (Figure 8C). Coexpression of p65 and p300 increased WT iNOS promoter activity 2.86-fold over that of p65 alone (Figure 8C, white bars), whereas the GASm promoter was refractory to this combination (Figure 8C, black bars). The combination of p65 + CARM1 did not enhance activity at either of the WT or GASm promoters, whereas p65 + CARM1 + p300 induced a 12.72-fold synergistic response on the WT promoter (Figure 8C, white bars). In contrast, there was only a 2.76-fold increase on the GASm promoter, which is 78% less transcriptional activity than observed at the WT promoter (Figure 8C, black bars). Notably, we determined that CBP is not able to recapitulate the synergistic effect seen with p300, indicating specificity for the p65, p300, and CARM1 complex (Figure 8D). Collectively, these data are congruent with recruitment of p300 and CARM1 to the iNOS gene promoter via the ChIP assay in response to IL-1β (Figure 8, A and B). Moreover, the simultaneous exclusion of CBP from the same genomic region is consistent with its inability to coactivate the iNOS gene promoter (Figure 8D).
Figure 8.
IL-1β promotes exchange of coactivators to regulate transcription of the iNOS gene promoter. A and B, 832/13 cells were stimulated with 1 ng/mL IL-1β for 15 minutes. ChIP assays were performed using antibodies against IgG (control), p65, CARM1, p300, and CBP. Proximal (A) and distal (B) NF-κB elements (indicated by the arrows in the schematic diagrams) in the iNOS promoter were targeted for amplification by real-time PCR. #, P < .1 vs NT for respective antibody; *, P < .05 vs NT for respective antibody; **, P < .01 vs NT for respective antibody. C, 832/13 cells were transfected with either a −1-kb iNOS promoter-luciferase construct or a −1-kb GASm construct. In addition, cells were cotransfected with the indicated combinations of p65, p300, and CARM1 and incubated for 24 hours. D, 832/13 cells were transfected with a −1-kb iNOS promoter-luciferase construct and concurrently with the combinations of p65, CBP, and CARM1 shown. Cells were cultured for a further 24 hours posttransfection. C and D, Promoter activity was measured by luciferase assay and normalized to protein content as quantified by BCA assay. *, P < .05. Data are means ± SEM from 3 individual experiments.
IL-1β induces both chromatin remodeling and phosphorylation of RNA Pol II at serine 5 on the core promoter region and serine 2 at the coding region
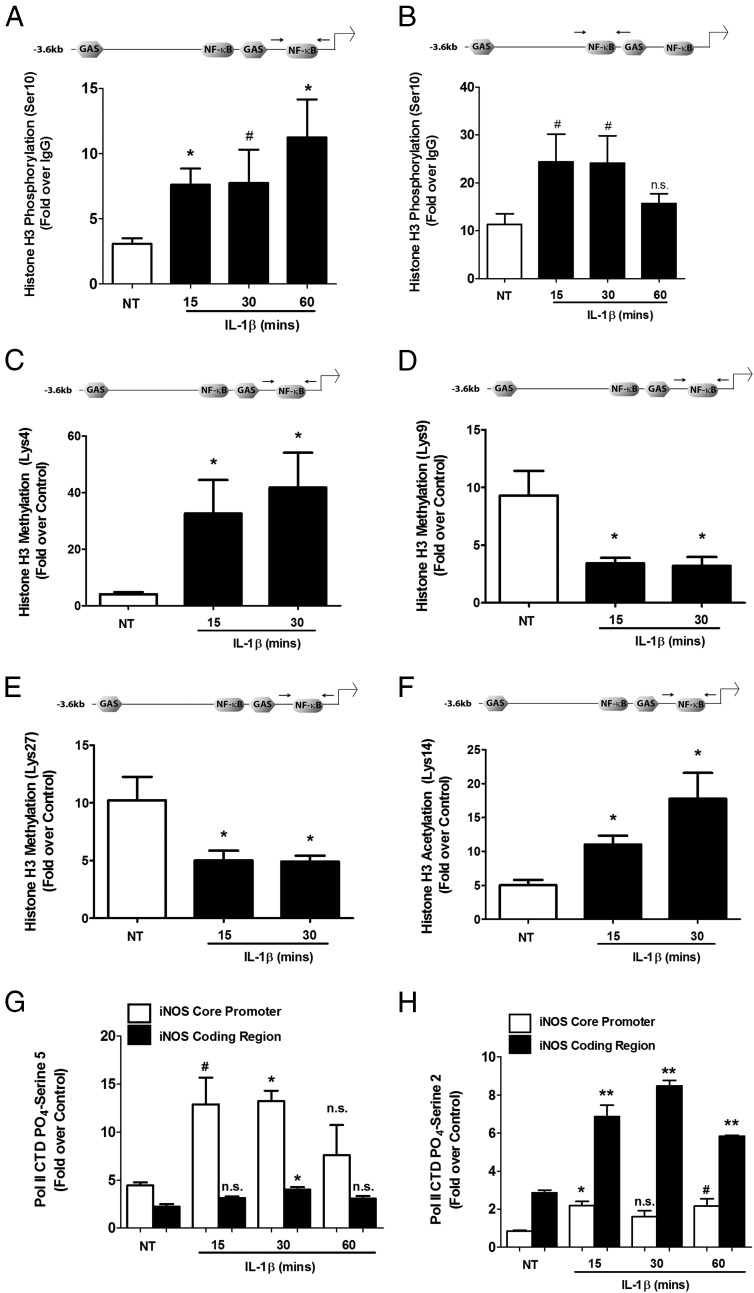
Histones are modified posttranslationally by a number of different stimuli corresponding to cellular activity (50). Because the iNOS gene is dormant in pancreatic β-cells until a stimulus is received, we investigated whether chromatin in close proximity with regulatory elements is modified posttranslationally in a stimulus-specific manner. There were 2.46, 2.50, and 3.64-fold increases in phosphorylation of histone H3 (Ser10) at the proximal κB sequence at 15, 30, and 60 minutes after the cellular exposure to IL-1β (Figure 9A). There was also a 2.43-fold enhancement in H3 (Ser10) phosphorylation at regions of the iNOS promoter containing the distal κB sequence at 15 and 30 minutes but with a return to the baseline (unstimulated) level at 60 minutes (Figure 9B). This regulatory pattern is consistent with early activation of the iNOS gene (Figure 1) and with phosphorylated histone H3 being associated with transcriptionally active genes (51). In addition, we found that H3K4 methylation was increased 8.07- and 10.35-fold after 15 and 30 minutes of exposure to IL-1β (Figure 9C). In contrast, methylation of H3K9 decreased 63.3% and 65.6% after 15 and 30 minutes of an IL-1β stimulus (Figure 9D). We also detected 51.1% and a 51.6% reductions in methylation of H3K27, respectively, with 15- and 30-minute incubations with IL-1β (Figure 9E). Alternatively, there were 2.18- and 3.52-fold enhancements in acetylation of H3K14 after 15 and 30 minutes of exposure to IL-1β (Figure 9F).
Figure 9.
Coordinated chromatin remodeling by IL-1β correlates with recruitment of phosphorylated RNA Pol II. A–H, 832/13 cells were untreated (NT) or stimulated with 1 ng/mL IL-1β for 15, 30, or 60 minutes. ChIP assays were performed with antibodies that immunoprecipitated histone H3 phosphorylated on serine 10 (A and B), histone H3 methylated on lysine 4 (C), lysine 9 (D), and lysine 27 (E), histone H3 acetylated on lysine 14 (F), and RNA Pol II phosphorylated on the CTD tail at serine 5 (G) or serine 2 (H). Proximal (A, C, D, E, and F) and distal (B) NF-κB response elements in the iNOS gene promoter (indicated by arrows in the schematic diagrams) were targeted for amplification by real-time PCR. G and H, Primers targeting either the iNOS core promoter region or coding region were used for amplification in real-time RT-PCR experiments. n.s., not significant vs respective NT control; #, P < .1 vs respective NT control; *, P < .05 vs respective NT control; **, P < .01 vs respective NT control. Data are means ± SEM from 3 to 4 individual experiments.
These chromatin modifications match the 2.93- and 3.01-fold increases in serine 5 phosphorylation of RNA Pol II observed at the iNOS core promoter region within 15 and 30 minutes after IL-1β exposure (Figure 9G). This result is significant because the phosphorylation of RNA Pol II at serine 5 indicates initiation of transcription (52). Moreover, IL-1β enhanced the serine 2 phosphorylation of the polymerase by 2.39-, 2.97-, and 2.02-fold over 15, 30, and 60 minutes, respectively, along the coding region of the iNOS gene (Figure 9H). Taken together, the results presented herein are consistent with IL-1β modifying the local chromatin regions to promote specific multiregulatory complex assembly at the iNOS gene promoter to enhance RNA Pol II recruitment and subsequent transcription.
Discussion
Regulation of the iNOS gene is an important cellular mechanism to control NO production (32). NO participates in diverse physiological effects that are important for normal tissue function; however, overproduction of this free radical signaling molecule can lead to the unwanted pathophysiological outcomes observed in a number of human diseases (53). For example, excess NO, produced by the inducible form of NOS, is detrimental to mouse, rat, and human β-cells (16, 23, 24, 30, 54). Indeed, inappropriate iNOS expression and activity have long been linked to autoimmune-mediated diabetes development (13, 55). However, the molecular mechanisms underlying regulation of the iNOS gene in response to proinflammatory cytokines in pancreatic β-cells is not well understood. Therefore, in the present study, we investigated the primary DNA binding factors, cognate DNA response elements, and epigenetic modifications required for activation of the iNOS gene by IL-1β and potentiation of this response by IFN-γ.
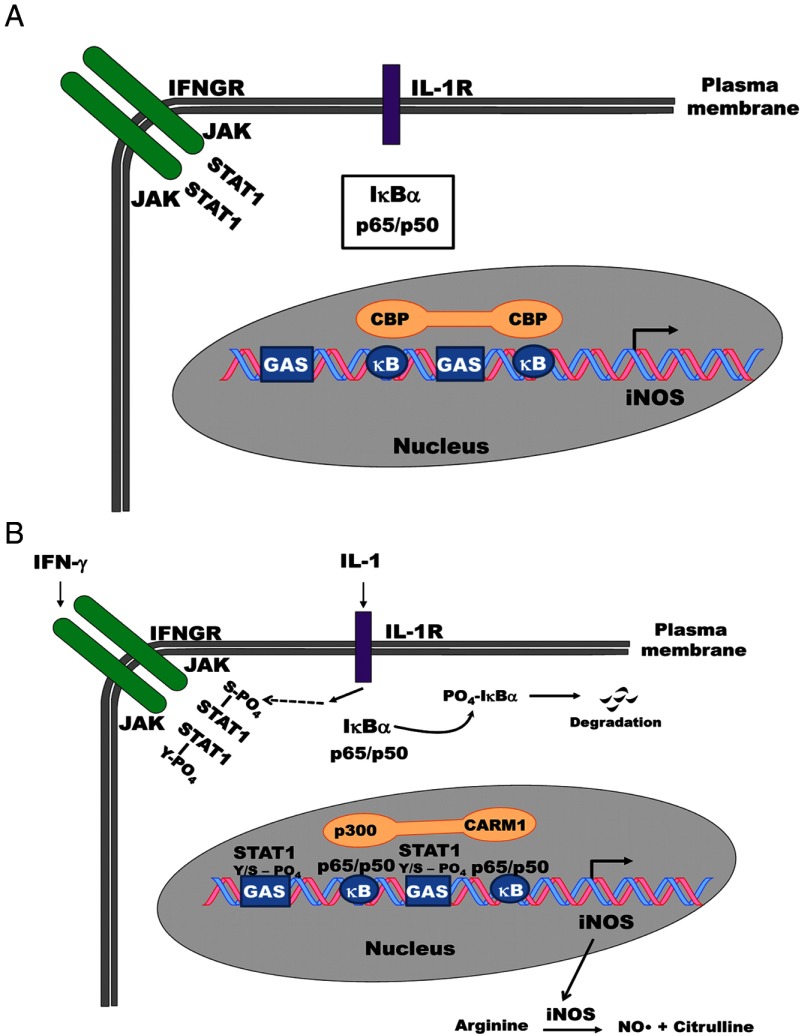
The rat iNOS gene has 2 GAS elements: a consensus distal element located in the upstream promoter region (−2397 and −3256 nucleotides away from the distal and proximal κB sequences, respectively; the numbering is relative to the transcription start site, which is +1) and a proximal GAS element that is ∼1 kb away from the transcriptional start site. The proximal GAS element differs from a consensus binding site by 1 nucleotide and is situated between the distal and proximal κB sequences. Generally, GAS elements integrate IFN-γ signals within a genomic context (56), whereas κB sequences respond to signals activating the NF-κB pathway (34). Herein we show that the NF-κB and STAT1 binding sites cooperate to regulate expression of the iNOS gene in response to IL-1β and IFN-γ (Figure 10).
Figure 10.
Cytokine-mediated regulation of the iNOS gene occurs via a coactivator switching mechanism. A, The iNOS gene is silent in pancreatic β-cells in the absence of IL-1β and IFN-γ. RelA/p65 is held in the cytoplasm by regulatory proteins, such as IκBα, whereas the coregulator molecule CBP associates with the transcriptionally inactive iNOS gene promoter in the absence of cytokines. B, Upon β-cell exposure to IL-1β, the IκBα protein is subject to phosphorylation-induced degradation; p65 enters the nucleus and binds κB elements with its heterodimer partner p50. Signaling through the IL-1 receptor (IL-1R) also increases occupancy of Ser727-phosphorylated STAT1 at GAS elements within the iNOS gene promoter. IFN-γ, through JAK-STAT signaling mechanisms, promotes Tyr701 phosphorylation of STAT1, which amplifies the cellular response to IL-1β. These signaling events induce removal of CBP from the iNOS promoter concomitant with the arrival of the coactivators p300 and CARM1.The marked increase in iNOS transcript leads to increased iNOS protein abundance. iNOS catalyzes the production of nitric oxide (NO•) and citrulline using arginine as a substrate. NO• accumulation in pancreatic β-cells is linked to losses in insulin secretion and cellular viability.
Use of site-directed mutagenesis to change the proximal GAS into a perfect consensus GAS increased the sensitivity of the iNOS gene to IL-1β but not to IFN-γ (Figure 6A). In contrast, altering the proximal GAS element by 2 nucleotides, which abolishes recognition by STAT1, decreases the ability of IL-1β to fully activate the iNOS gene promoter (Figure 6A). To our knowledge, this is the first study to report on GAS element modulation of IL-1β sensitivity at a given gene promoter in pancreatic β-cells. A key additional and unanticipated finding was that STAT1 phosphorylated at Ser727, but not at Tyr701, occupies the GAS elements in response to IL-1β (Figure 7, G and H). However, the Tyr701 site is critical for potentiation of the IL-1β response by IFN-γ (Figure 7C). These results illustrate that STAT1 can be recruited to select gene promoters without Tyr701 phosphorylation. Moreover, there is a pool of constitutively phosphorylated STAT1 at Ser727 that is capable of supporting gene transcription in pancreatic β-cells (57). In addition, IL-1β appears to enhance this serine-phosphorylated protein pool (Figure 7B) and also promotes recruitment to specific GAS-containing promoters, such as those observed in the iNOS regulatory region (Figure 7, G and H). This finding is distinct from that seen in other tissues, in which Tyr701 phosphorylation is required to recruit STAT1 into close proximity with the chromatin structure for Ser727 phosphorylation (58).
An additional intriguing possibility is that elements within a given gene promoter may, in a signal-specific manner, regulate downstream mRNA stability and translation of protein. This type of regulatory strategy is present in lower eukaryotes (59). Whether such events occur in response to cytokines or other signals in mammalian cells remains to be determined. In addition, the translation of the iNOS transcript into protein is regulated in a p38 MAPK-dependent manner in pancreatic β-cells (60). Thus, many additional regulatory mechanisms, beyond those that control gene transcription, are probably involved in the regulatory management of iNOS protein production and enzymatic activity.
Our results are in agreement with extant studies using mouse macrophages, in which the individual responses of the iNOS gene to LPS and IFN-γ require an intact GAS sequence (61). Interestingly, however, mouse macrophages do not require STAT1 phosphorylation at Ser727 to activate the iNOS gene (62). In addition, we found that the distal κB sequence was probably the site cooperating with the GAS element because its mutation could not be rescued by the generation of a cGAS within the iNOS gene promoter (Figure 6D). This finding is complementary to an earlier study in which the proximal κB site sequence alone was insufficient to support a cytokine stimulus in rat mesangial cells (63) and with additional data obtained from analysis of other genes supporting the concept that the distal κB sequence is the dominant response element controlled by NF-κB proteins (64).
We further discovered that CBP is exchanged for p300 at the iNOS gene promoter in response to IL-1β (Figure 8, A and B). CBP and p300 are important coactivator molecules that associate with more than 300 different cellular proteins and contribute to the expression of many diverse genes (65). Although these proteins have overlapping functions, they are not entirely redundant, as evidenced by their distinct genetic deletion phenotypes (66). In this study, we demonstrate for the first time that IL-1β induces recruitment of p300 to the iNOS gene promoter while simultaneously excluding CBP from the same genomic region (Figure 8, A and B). Because coactivators are often limiting factors in the expression of many genes (67), this selective coactivator exchange mechanism probably modulates specific signal integration at the iNOS gene promoter and may thus represent a novel insight into how these proteins participate in controlling inflammatory responses.
We explicitly note the inability of CBP to substitute for p300 in transcriptional activation assays (Figure 8D). We speculate that CARM1, which is also recruited to the iNOS gene promoter in response to IL-1β (Figure 8, A and B), may methylate CBP, thereby promoting removal of the latter molecule from the iNOS regulatory gene sequence regions. This type of methylation-sensitive recruitment/derecruitment strategy has been termed a “transcriptional switch” in other systems (68, 69), whereby CBP moves from CREB-associated genes to nuclear hormone receptor-regulated gene promoters after methylation by CARM1. Moreover, IKKα phosphorylates CBP at Ser1382 and Ser1386, which are phospho-acceptor sites not present within p300 (70). Thus, it is entirely possible that unique sites for posttranslational modification may also exist within p300, which could further explain its differential recruitment and responsiveness to IL-1β and other inflammation-associated signals. If so, identification of such sites could help explain the p300 association with particular genes, such as iNOS, in a manner that allows CBP to be concomitantly shuttled to other promoters in a signal-specific manner.
The requirement for p300 to regulate the iNOS gene in response to inflammatory stimuli may be a general phenomenon because it is also observed for lipopolysaccharide-mediated induction of the mouse iNOS gene in macrophage cell lines (71). However, a novel observation put forth in the present study is the loss of CBP at the iNOS gene promoter in a signal-dependent manner. We showed that IL-1β-mediated increases in expression of the rat iNOS gene in pancreatic β-cells involved recruitment of the p65 and p50 subunits of NF-κB and STAT1 phosphorylated at Ser727, as well as the coactivators p300 and CARM1. Whether p300, via acetyltransferase activity, or CARM1, via methyltransferase activity, is posttranslationally modifying p50, p65, or STAT1 in the context of the iNOS gene promoter is not known at the present time. However, a number of possibilities to explain regulation of the iNOS gene by these proteins in response to IL-1β are feasible. These include the following: (1) posttranslational modification of histone proteins by CARM1 and p300; (2) CARM1 and p300 modification of transcription factors, such as p50, p65, and/or STAT1; (3) covalent alterations to as yet uncharacterized additional accessory factors required for the expression of the iNOS gene; (4) CARM1 methylation of p300 and/or p300 acetylation of CARM1; and (5) modification of histone proteins that allow access to multiregulatory gene transcription complexes. We have demonstrated the latter and note that possibilities 1 to 4 are not mutually exclusive.
After examining a specific array of chromatin modifications associated with stimulus-induced transcription (Figure 9), we observed patterns consistent with the strong transcriptional activation of the iNOS gene by cytokines (Figure 1). We specifically note the coordinated control of acetylation, methylation, and phosphorylation of the histone H3 protein by IL-1β, indicating signal integration at key sites controlling access to gene promoter regions. For example, H3K27 methylation is often associated with transcriptional repression, whereas methylation of H3K4 typically corresponds to transcriptionally active genes (50). Moreover, H3K9 methylation is connected to gene silencing, whereas acetylation of H3K14 is frequently coupled to active transcription. These synchronized modifications probably allow for either recruitment of primary DNA binding factors, association with coactivator complexes, and/or localization of RNA Pol II with the region to be transcribed. In addition, the coordinated chromatin remodeling provides docking sites within chromatin for additional proteins, such as bromodomain proteins, which help couple posttranslational modifications to active transcription (72). Thus, the signal-induced modifications shown in this study for the iNOS gene promoter are rapid, fitting with strong activation of transcription corresponding to robust mRNA accumulation (Figure 1). Much like the DNA sequence can have allosteric effects on transcriptional regulatory proteins (73), it is quite likely that posttranslational modifications of histones also promote and facilitate extremely precise protein-protein and protein-DNA interactions.
A similar, but distinct, scenario exists for the human iNOS gene promoter. In the human iNOS gene sequence, −10 kb of the proximal promoter region, relative to the transcriptional start site, contains approximately 7 κB sequences (32). Interestingly, the human iNOS gene requires a combination of IL-1β and IFN-γ for transcriptional activation (74), whereas the rat gene will respond to IL-1β alone (Figure 1 and Ref 75). One possible explanation for this difference in species responsiveness to individual cytokines is that the human gene contains many more genetic regulatory sequences and also contains a composite κB/GAS responsive element. Composite response elements can combine regulatory inputs from multiple signals and thus integrate transcriptional responses initiated from different receptor signals (76). An additional possibility for differences in species responsiveness to cytokines is that the human gene has evolved to contain more transposable elements, which may have contributed to the alterations in spacing and positioning of regulatory information within the genome. This type of situation has been documented in stem cells (77) and in promoters of nonmammalian species (78).
It is intriguing to contemplate the possibility that the transcriptional synergy observed with the combination of p65, CARM1, and p300 (shown in Figure 8C) may be a potential therapeutic target to control inflammation. Coactivators are typically limiting reagents for transcription in a given cell; thus, anti-inflammatory compounds specifically targeting coactivator molecules in a cell through disruption and/or rerouting of vital protein components required to assemble multiregulatory complexes that control transcription may be an effective strategy. Down-regulating specific genes, such as iNOS, would probably dampen inflammation in general, because iNOS also activates additional enzymes that produce other inflammatory mediators, including cyclooxygenase-2 (79). Interestingly, the cyclooxygenase-2 gene is also regulated by the NF-κB proteins p65 and p50 in response to IL-1β in pancreatic β-cells (44) and drives PGE2 production (14, 80). Thus, further investigation into the posttranslational modifications controlling the specific protein-protein and protein-DNA interactions induced by proinflammatory signals will almost certainly reveal novel targets for the design of prospective anti-inflammatory therapies that could prevent or treat auto-inflammatory and/or autoimmune diseases.
Acknowledgments
We thank Drs Guoxun Chen, Naima Moustaid-Moussa, Christian Jobin, Hans Hohmeier, Christopher Newgard, and Jay Whelan for reagents. We also thank Emily Lazek and Dana Omari for technical assistance.
This work was supported by start-up funds provided by the University of Tennessee, Knoxville (to J.J.C.), a Summer Undergraduate Research Internship Award from the University of Tennessee Office of Research (to B.L.U.), a Professional Development Award provided by the University of Tennessee Graduate School (to J.J.C.), funds obtained through the University of Tennessee Microbiology across Campuses Educational and Research Venture (to J.J.C.), and a grant from the Physicians Medical Education Research Foundation (to J.J.C. and M.D.K.).
Current address for B.L.U.: Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, Texas 75235.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by start-up funds provided by the University of Tennessee, Knoxville (to J.J.C.), a Summer Undergraduate Research Internship Award from the University of Tennessee Office of Research (to B.L.U.), a Professional Development Award provided by the University of Tennessee Graduate School (to J.J.C.), funds obtained through the University of Tennessee Microbiology across Campuses Educational and Research Venture (to J.J.C.), and a grant from the Physicians Medical Education Research Foundation (to J.J.C. and M.D.K.).
Footnotes
- CARM1
- coactivator arginine methyltransferase
- CBP
- cAMP response element–binding protein–binding protein
- F
- forward
- GAS
- IFN-γ activated site
- iNOS
- inducible nitric oxide synthase
- NF-κB
- nuclear factor-κB
- R
- reverse
- RNA Pol II
- RNA polymerase II
- si
- small interfering
- STAT1
- signal transducer and activator of transcription-1
- TPCA-1
- 2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide
- WT
- wild-type.
References
- 1. Padgett LE, Broniowska KA, Hansen PA, Corbett JA, Tse HM. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann NY Acad Sci. 2013;1281:16–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bach JF. Insulin-dependent diabetes mellitus as a β-cell targeted disease of immunoregulation. J Autoimmun. 1995;8:439–463. [DOI] [PubMed] [Google Scholar]
- 3. Hohmeier HE, Tran VV, Chen G, Gasa R, Newgard CB. Inflammatory mechanisms in diabetes: lessons from the β-cell. Int J Obes Relat Metab Disord. 2003;27(suppl 3):S12–S16. [DOI] [PubMed] [Google Scholar]
- 4. Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:319–336. [DOI] [PubMed] [Google Scholar]
- 5. Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and β-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5:219–226. [DOI] [PubMed] [Google Scholar]
- 6. Ehses JA, Lacraz G, Giroix MH, et al. IL-1 antagonism reduces hyperglycemia and tissue inflammation in the type 2 diabetic GK rat. Proc Natl Acad Sci USA. 2009;106:13998–14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. [DOI] [PubMed] [Google Scholar]
- 8. Böni-Schnetzler M, Boller S, Debray S, et al. Free fatty acids induce a proinflammatory response in islets via the abundantly expressed interleukin-1 receptor I. Endocrinology. 2009;150:5218–5229. [DOI] [PubMed] [Google Scholar]
- 9. Donath MY, Størling J, Maedler K, Mandrup-Poulsen T. Inflammatory mediators and islet β-cell failure: a link between type 1 and type 2 diabetes. J Mol Med. 2003;81:455–470. [DOI] [PubMed] [Google Scholar]
- 10. Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11:98–107. [DOI] [PubMed] [Google Scholar]
- 11. Mandrup-Poulsen T, Pickersgill L, Donath MY. Blockade of interleukin 1 in type 1 diabetes mellitus. Nat Rev Endocrinol. 2010;6:158–166. [DOI] [PubMed] [Google Scholar]
- 12. Scarim AL, Arnush M, Hill JR, et al. Evidence for the presence of type I IL-1 receptors on β-cells of islets of Langerhans. Biochim Biophys Acta. 1997;1361:313–320. [DOI] [PubMed] [Google Scholar]
- 13. Corbett JA, Mikhael A, Shimizu J, et al. Nitric oxide production in islets from nonobese diabetic mice: aminoguanidine-sensitive and -resistant stages in the immunological diabetic process. Proc Natl Acad Sci USA. 1993;90:8992–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parazzoli S, Harmon JS, Vallerie SN, Zhang T, Zhou H, Robertson RP. Cyclooxygenase-2, not microsomal prostaglandin E synthase-1, is the mechanism for interleukin-1β-induced prostaglandin E2 production and inhibition of insulin secretion in pancreatic islets. J Biol Chem. 2012;287:32246–32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kleemann R, Rothe H, Kolb-Bachofen V, et al. Transcription and translation of inducible nitric oxide synthase in the pancreas of prediabetic BB rats. FEBS Lett. 1993;328:9–12. [DOI] [PubMed] [Google Scholar]
- 16. Corbett JA, Wang JL, Sweetland MA, Lancaster JR Jr, McDaniel ML. Interleukin 1 β induces the formation of nitric oxide by β-cells purified from rodent islets of Langerhans. Evidence for the β-cell as a source and site of action of nitric oxide. J Clin Invest. 1992;90:2384–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnush M, Heitmeier MR, Scarim AL, Marino MH, Manning PT, Corbett JA. IL-1 produced and released endogenously within human islets inhibits β cell function. J Clin Invest. 1998;102:516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scarim AL, Heitmeier MR, Corbett JA. Irreversible inhibition of metabolic function and islet destruction after a 36-hour exposure to interleukin-1β. Endocrinology 1997;138:5301–5307. [DOI] [PubMed] [Google Scholar]
- 19. Corbett JA, Wang JL, Hughes JH, et al. Nitric oxide and cyclic GMP formation induced by interleukin 1 β in islets of Langerhans. Evidence for an effector role of nitric oxide in islet dysfunction. Biochem J. 1992;287(Pt 1):229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collier JJ, Fueger PT, Hohmeier HE, Newgard CB. Pro- and antiapoptotic proteins regulate apoptosis but do not protect against cytokine-mediated cytotoxicity in rat islets and β-cell lines. Diabetes. 2006;55:1398–1406. [DOI] [PubMed] [Google Scholar]
- 21. Steer SA, Scarim AL, Chambers KT, Corbett JA. Interleukin-1 stimulates β-cell necrosis and release of the immunological adjuvant HMGB1. PLoS Med. 2006;3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collier JJ, Burke SJ, Eisenhauer ME, et al. Pancreatic β-cell death in response to pro-inflammatory cytokines is distinct from genuine apoptosis. PLoS One. 2011;6:e22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corbett JA, Sweetland MA, Wang JL, Lancaster JR Jr, McDaniel ML. Nitric oxide mediates cytokine-induced inhibition of insulin secretion by human islets of Langerhans. Proc Natl Acad Sci USA. 1993;90:1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suarez-Pinzon WL, Mabley JG, Strynadka K, Power RF, Szabo C, Rabinovitch A. An inhibitor of inducible nitric oxide synthase and scavenger of peroxynitrite prevents diabetes development in NOD mice. J Autoimmun. 2001;16:449–455. [DOI] [PubMed] [Google Scholar]
- 25. Rabinovitch A, Suarez-Pinzon WL, Sorensen O, Bleackley RC. Inducible nitric oxide synthase (iNOS) in pancreatic islets of nonobese diabetic mice: identification of iNOS-expressing cells and relationships to cytokines expressed in the islets. Endocrinology. 1996;137:2093–2099. [DOI] [PubMed] [Google Scholar]
- 26. Heitmeier MR, Scarim AL, Corbett JA. Prolonged STAT1 activation is associated with interferon-γ priming for interleukin-1-induced inducible nitric-oxide synthase expression by islets of Langerhans. J Biol Chem. 1999;274:29266–29273. [DOI] [PubMed] [Google Scholar]
- 27. Heitmeier MR, Scarim AL, Corbett JA. Interferon-γ increases the sensitivity of islets of Langerhans for inducible nitric-oxide synthase expression induced by interleukin 1. J Biol Chem. 1997;272:13697–13704. [DOI] [PubMed] [Google Scholar]
- 28. Darville MI, Eizirik DL. Regulation by cytokines of the inducible nitric oxide synthase promoter in insulin-producing cells. Diabetologia. 1998;41:1101–1108. [DOI] [PubMed] [Google Scholar]
- 29. Kwon G, Corbett JA, Rodi CP, Sullivan P, McDaniel ML. Interleukin-1 β-induced nitric oxide synthase expression by rat pancreatic β-cells: evidence for the involvement of nuclear factor κB in the signaling mechanism. Endocrinology. 1995;136:4790–4795. [DOI] [PubMed] [Google Scholar]
- 30. Thomas HE, Darwiche R, Corbett JA, Kay TW. Interleukin-1 plus γ-interferon-induced pancreatic β-cell dysfunction is mediated by β-cell nitric oxide production. Diabetes. 2002;51:311–316. [DOI] [PubMed] [Google Scholar]
- 31. Xie QW, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon γ and bacterial lipopolysaccharide. J Exp Med. 1993;177:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pautz A, Art J, Hahn S, Nowag S, Voss C, Kleinert H. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide. 2010;23:75–93. [DOI] [PubMed] [Google Scholar]
- 33. Taylor BS, de Vera ME, Ganster RW, et al. Multiple NF-κB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem. 1998;273:15148–15156. [DOI] [PubMed] [Google Scholar]
- 34. Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. [DOI] [PubMed] [Google Scholar]
- 35. Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. [DOI] [PubMed] [Google Scholar]
- 36. Kim S, Kim HS, Chung KW, et al. Essential role for signal transducer and activator of transcription-1 in pancreatic β-cell death and autoimmune type 1 diabetes of nonobese diabetic mice. Diabetes. 2007;56:2561–2568. [DOI] [PubMed] [Google Scholar]
- 37. Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. [DOI] [PubMed] [Google Scholar]
- 38. Janjic D, Maechler P, Sekine N, Bartley C, Annen AS, Wolheim CB. Free radical modulation of insulin release in INS-1 cells exposed to alloxan. Biochem Pharmacol. 1999;57:639–648. [DOI] [PubMed] [Google Scholar]
- 39. Milburn JL Jr, Hirose H, Lee YH, et al. Pancreatic β-cells in obesity. Evidence for induction of functional, morphologic, and metabolic abnormalities by increased long chain fatty acids. J Biol Chem. 1995;270:1295–1299. [DOI] [PubMed] [Google Scholar]
- 40. Herz J, Gerard RD. Adenovirus-mediated transfer of low density lipoprotein receptor gene acutely accelerates cholesterol clearance in normal mice. Proc Natl Acad Sci USA. 1993;90:2812–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prasad RC, Wang XL, Law BK, et al. Identification of genes, including the gene encoding p27Kip1, regulated by serine 276 phosphorylation of the p65 subunit of NF-κB. Cancer Lett. 2009;275:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jobin C, Panja A, Hellerbrand C, et al. Inhibition of proinflammatory molecule production by adenovirus-mediated expression of a nuclear factor κB super-repressor in human intestinal epithelial cells. J Immunol. 1998;160:410–418. [PubMed] [Google Scholar]
- 43. Chen G, Hohmeier HE, Newgard CB. Expression of the transcription factor STAT-1α in insulinoma cells protects against cytotoxic effects of multiple cytokines. J Biol Chem. 2001;276:766–772. [DOI] [PubMed] [Google Scholar]
- 44. Burke SJ, Collier JJ. The gene encoding cyclooxygenase-2 is regulated by IL-1β and prostaglandins in 832/13 rat insulinoma cells. Cell Immunol. 2011;271:379–384. [DOI] [PubMed] [Google Scholar]
- 45. Becker TC, Noel RJ, Coats WS, et al. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43(Pt A):161–189. [DOI] [PubMed] [Google Scholar]
- 46. Burke SJ, Goff MR, Updegraff BL, et al. Regulation of the CCL2 gene in pancreatic β-cells by IL-1β and glucocorticoids: role of MKP-1. PLoS One. 2012;7:e46986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Podolin PL, Callahan JF, Bolognese BJ, et al. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of IκB kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell proliferation. J Pharmacol Exp Ther. 2005;312:373–381. [DOI] [PubMed] [Google Scholar]
- 48. Pearse RN, Feinman R, Shuai K, Darnell JE Jr, Ravetch JV. Interferon γ-induced transcription of the high-affinity Fc receptor for IgG requires assembly of a complex that includes the 91-kDa subunit of transcription factor ISGF3. Proc Natl Acad Sci USA. 1993;90:4314–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. [DOI] [PubMed] [Google Scholar]
- 50. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. [DOI] [PubMed] [Google Scholar]
- 51. Nowak SJ, Corces VG. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 2000;14:3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. [DOI] [PubMed] [Google Scholar]
- 53. Hill BG, Dranka BP, Bailey SM, Lancaster JR Jr, Darley-Usmar VM. What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem. 2010;285:19699–19704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eizirik DL, Leijerstam F. The inducible form of nitric oxide synthase (iNOS) in insulin-producing cells. Diabete Metab. 1994;20:116–122. [PubMed] [Google Scholar]
- 55. Corbett JA, McDaniel ML. Does nitric oxide mediate autoimmune destruction of β-cells? Possible therapeutic interventions in IDDM. Diabetes. 1992;41:897–903. [DOI] [PubMed] [Google Scholar]
- 56. Stark GR, Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burke SJ, Goff MR, Lu D, Proud D, Karlstad MD, Collier JJ. Synergistic expression of the CXCL10 gene in response to IL-1β and IFN-γ involves NF-κB, phosphorylation of STAT1 at Tyr701, and acetylation of histones H3 and H4. J Immunol. 2013;191:323–336. [DOI] [PubMed] [Google Scholar]
- 58. Sadzak I, Schiff M, Gattermeier I, et al. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci USA. 2008;105:8944–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. [DOI] [PubMed] [Google Scholar]
- 60. Nishiki Y, Adewola A, Hatanaka M, Templin AT, Maier B, Mirmira RG. Translational control of inducible nitric oxide synthase by p38 MAPK in islet β-cells. Mol Endocrinol. 2013;27:336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-γ-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-γ and lipopolysaccharide. J Biol Chem. 1997;272:1226–1230. [DOI] [PubMed] [Google Scholar]
- 62. Farlik M, Reutterer B, Schindler C, et al. Nonconventional initiation complex assembly by STAT and NF-κB transcription factors regulates nitric oxide synthase expression. Immunity. 2010;33:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Beck KF, Sterzel RB. Cloning and sequencing of the proximal promoter of the rat iNOS gene: activation of NFκB is not sufficient for transcription of the iNOS gene in rat mesangial cells. FEBS Lett. 1996;394:263–267. [DOI] [PubMed] [Google Scholar]
- 64. Leung TH, Hoffmann A, Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453–464. [DOI] [PubMed] [Google Scholar]
- 65. Kasper LH, Fukuyama T, Biesen MA, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. [DOI] [PubMed] [Google Scholar]
- 67. Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. [DOI] [PubMed] [Google Scholar]
- 68. Xu W, Chen H, Du K, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. [DOI] [PubMed] [Google Scholar]
- 69. Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol Cell. 2007;26:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guo H, Mi Z, Kuo PC. Characterization of short range DNA looping in endotoxin-mediated transcription of the murine inducible nitric-oxide synthase (iNOS) gene. J Biol Chem. 2008;283:25209–25217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72. LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. [DOI] [PubMed] [Google Scholar]
- 74. de Vera ME, Shapiro RA, et al. Transcriptional regulation of human inducible nitric oxide synthase (NOS2) gene by cytokines: initial analysis of the human NOS2 promoter. Proc Natl Acad Sci USA. 1996;93:1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Corbett JA, Kwon G, Turk J, McDaniel ML. IL-1 β induces the coexpression of both nitric oxide synthase and cyclooxygenase by islets of Langerhans: activation of cyclooxygenase by nitric oxide. Biochemistry. 1993;32:13767–13770. [DOI] [PubMed] [Google Scholar]
- 76. Miner JN, Yamamoto KR. Regulatory crosstalk at composite response elements. Trends Biochem Sci. 1991;16:423–426. [DOI] [PubMed] [Google Scholar]
- 77. Kunarso G, Chia NY, Jeyakani J, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. [DOI] [PubMed] [Google Scholar]
- 78. Lerman DN, Feder ME. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol Biol Evol. 2005;22:776–783. [DOI] [PubMed] [Google Scholar]
- 79. Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. [DOI] [PubMed] [Google Scholar]
- 80. Heitmeier MR, Kelly CB, Ensor NJ, et al. Role of cyclooxygenase-2 in cytokine-induced β-cell dysfunction and damage by isolated rat and human islets. J Biol Chem. 2004;279:53145–53151. [DOI] [PubMed] [Google Scholar]