Abstract
High-throughput sequencing technology has shown tremendous promise for microbial community composition and diversity. Illumina MiSeq platform was exploited to study the microbial community associated with the different stages of the life-cycle of ovine Haemonchus contortus field strains using two distinct amplification primer sets (targeting V3–V4, and V5–V7). Scanning electron microscope and polymerase chain reaction coupled with Illumina MiSeq platform were employed to confirm the absence of any parasite surface contamination by intact bacteria or their DNA products. Results showed 48 (V3–V4 tags) and 28 (V5–V7 tags) bacterial genera comprised the microbial flora of H. contortus microbiome. The dominant bacterial genera belonged to Escherichia-Shigella, Pseudomonas and Ochrobactrum, which were shared in all the stages of the parasite life-cycle using V3–V4 and V5–V7 amplicons. Moreover, the parasite microbiome could reflect the external micro-organisms (i.e. micro- and macro-habitats). There is abundant room for further progress in comparing microbiome of different helminths, which has, and will continue to offer considerable potential for better understanding a wide-variety of devastating animal and human diseases.
Introduction
Among the gastrointestinal parasites that cause losses to the farming industry, for example, Haemonchus, Ostertagia, Trichostrongylus, Nematodirus and Cooperia, the barber’s pole worm Haemonchus contortus is the predominant, blood- sucking, highly pathogenic, and economically important nematode that infects small ruminants. It has been demonstrated that larvae provoked tiny haemorrhages as early as 3-day post-infection (dpi). Emergence of the larvae into the abomasal lumen commenced between 7- and 11-dpi and all worms had moulted to the 4th stage by 4-dpi. The early 4th stage (L4) has a provisional buccal capsule, which facilitated larvae to attach to the abomasal mucosa and suck blood for the first time1. All mammals harbor a wide diversity of microbes that live in harmony with their host and colonize the mucosal surfaces, including digestive tract, respiratory surfaces and reproductive tract2. Our knowledge of the symbiosis between bacteria and eukaryotes has grown rapidly in recent years. It has conclusively been shown that the α-proteobacterium Wolbachia pipientis causes reproductive manipulation of the arthropod and filarial nematode hosts in various ways, including feminization, parthenogenesis, male killing and sperm-egg incompatibility3,4. Apart from the well-studied nematode-associated Wolbachia, there appear to be few reports of studies designed to identify the bacteria associated with Ascaris suum 5,6 and Trichuri muris 7. More recent attention has focused on the alteration of nematode entomopathogenicity by Rhabditis regina microbiome8. The hypervariable regions of 16S ribosomal RNA (rRNA) provide species-specific signature sequences. The Polymerase Chain Reaction (PCR)-amplified bacterial 16S rRNA gene sequencing has been emerged as the backbone of microbial community studies over recent decades owing to major breakthroughs in nucleic acids sequencing technology (the development of Next-Generation Sequencing technology) and molecular techniques9. DNA sequencing of 16S rRNA genes or gene fragments has played a key role in large ongoing microbial community studies, such as the National Institutes of Health (NIH) funded human microbiome project10, human and animal microbiome studies during parasitic infection11,12 to index the bacterial communities in a sample without the bias or cultivation effort. Currently, this is the most reliable approach to explore the diversity of microbial community and to determine its composition. We employed 16S rRNA gene amplification and Illumina MiSeq platform to portray, for the first time to our knowledge, the differences in composition and relative abundance of diverse H. contortous microbiome. This study provides new insights into the microbial community naturally associated with field strains of H. contortus life-cycle stages, with the long-term novel approach of manipulating them to control ruminant gastrointestinal parasites.
Materials and Methods
Ethics approval and consent to participate
Animal experiments were conducted in accordance with the guidelines of Beijing the Municipality on the Review of Welfare and Ethics of Laboratory Animals approved by the Beijing Municipality Administration Office of Laboratory Animals (BAOLA), and under the protocol (CAU-AEC-2010–0603) approved by the China Agricultural University Animal Ethics Committee. All experimental procedures were also approved by the Institutional Animal Care and Committee of China Agricultural University (The certificate of Beijing Laboratory Animal employee, ID: 15883).
Animals and parasites
Field microscopic investigation on H. contortus in 107 sheep in an animal farm in Jin Zhan, Cun, Chaoyang, Beijing, China to determine whether any general characteristics of the sheep were associated with infections were performed to select H. contortus-infected animals for the current study. Parasitological examination of rectal fecal samples from the sheep was analyzed with the modified McMaster technique13. In addition, FAMACHA© scores were performed for detecting anemic sheep on the farm by matching the color of the ocular conjunctival mucous membranes of each animal to the FAMACHA© eye color chart, with the scale ranging from 1 to 5, where a score of 1 is valued not anemic and 5 is valued highly anemic14. Body condition scoring (BCS) was assessed by observing and palpating the loin area (behind the ribs and in front of the pelvis), and assigning a score of 1–10 according to the procedure used by15 with 1 being emaciated and 10 being extremely fat. Blood was collected for haemato-biochemical to reconfirm the presence of H. contortous infection. The aforementioned parasitological tests, FAMACHA© scores and body condition scoring and haemato-biochemical analyses were reassessed after two weeks to ensure the consistency of the results.
A total of three out of fourteen H. contortus-infected sheep; two-month-old sheep (Ovis aries) that were randomly selected and brought from the local farmer in Jin Zhan village, Chaoyang, Beijing, China. Upon arrival in China Agricultural University facilities, all sheep were tagged (each sheep was assigned an arbitrary number for sample identification purposes). Feces from the infected sheep were collected and cultured at room temperature for 7 days to allow the third stage larvae (the infectious stage) to develop from the eggs, and larvae were harvested using the Baermann technique16. The remaining feces together with the abomasal contents were collected from H. contortus-infected sheep immediately after death, and eggs were extracted employing sucrose step-density gradient centrifugation (unpublished data). The collected eggs were washed on a 35 µm sieve with distilled water to remove the sugar under sterile conditions. The mesh sieve is present at the top of ceramic funnel and the flask with one side-arm attached to rubber tube connected with another double-side arm flask. The latter is connected with vacuum pump, which has an advantage over the ordinary gravitational force. The adult worms were recovered from the abomasal mucosa using forceps and dissecting microscope. Before the DNA was extracted H. contortus adult worm, exsheathed L3 and eggs were washed with phosphate-buffered saline (PBS, pH 7.4) and incubated in an antibiotic solution (1 mg/ml ampicillin/1 mg/ml gentamicin) for 2 h to kill external bacteria. Then, the parasites were washed twice with 2% sodium hypochlorite for 20 s each followed by five times with sterile PBS.
Ruling out bacterial contamination associated with the parasite surfaces
Several tests, such as scanning electron microscope (SEM) and polymerase chain reaction (PCR) were conducted to rule out any abomasal microbial contamination associated with the surface of the distinct life-cycle stages of H. contortus.
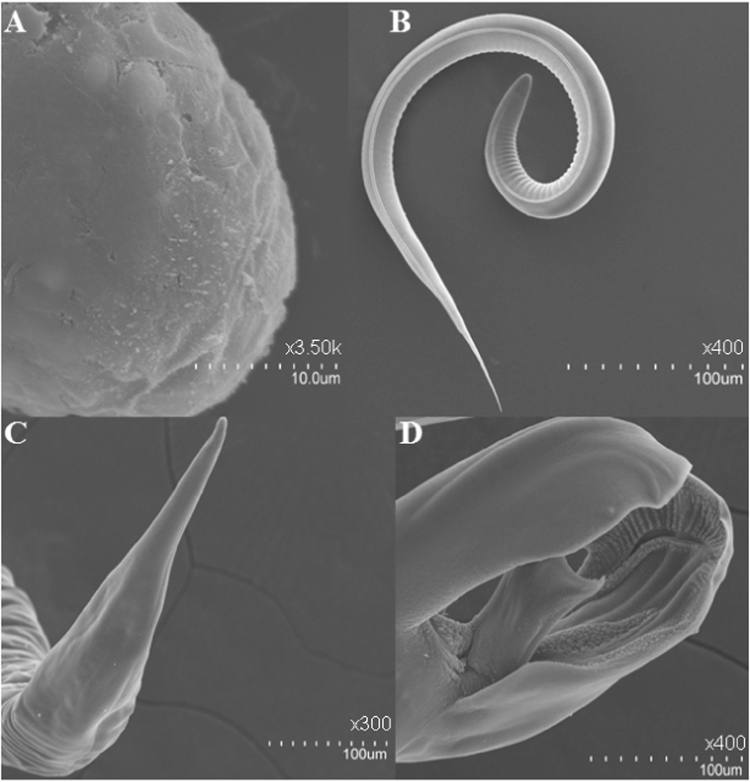
Parasites were prepared for SEM as previously reported by17 and Hitachi S-3400N SEM was employed to assure the absence of any surface bacteria on the selected life-cycle stages of H. contortus (Fig. 1). So in addition to SEM, PCR further confirmed the absence of bacterial contamination from the abomasal microbial community. The approach to distinguishing between unwashed and washed individual female worms, surface washes, which were washed following the procedure above, involved PCR and next-generation sequencing. Parasites were stored at −20 °C for downstream applications. The procedures launched are described in detail in the Materials and Methods section below.
Figure 1.
Transmission electron microscope (TEM) images of the different life -cycle stages of H. contortus. (A) H. contortus egg. (B) H. contortus larva. (C) H. contortus adult female worm (posterior end). (D) H. contortus adult male worm (posterior end).
DNA extraction, amplification and processing of samples for high-throughput sequencing
Microbial DNA extraction
H. contortus genomic DNA together with microbial genomic DNA from samples was extracted using the E.Z.N.A.® Bacterial DNA Kit (Omega Bio-tek, Norcross, GA, US) according to manufacturer’s instruction. DNA products were assessed by electrophoresis in 1% agarose gel run at 100 V for 30 min, and the sizes of the products (23 kb) were validated by comparison with a molecular size standard. The quantity and quality of the DNA were evaluated by measuring the absorbance employing an ultraviolet spectrophotometer (DU1 640, Beckman Instruments, Inc., CA, USA) at 260 and 280 nm. All microbial genomic DNA was stored at −20 °C before further analysis.
Amplicon generation and high-throughput sequencing
The 16S ribosomal RNA (rRNA) gene contains of nine hypervariable regions enclosed by regions of more conserved sequence. As shown in Fig. 2, regions of approximately 468 bp and 394 bp encircling the V3-V4 and V5-V7 hypervariable regions respectively within the 16S rRNA gene was subjected to high-throughput sequencing to maximize the effective length of sequencing reads18.
Figure 2.
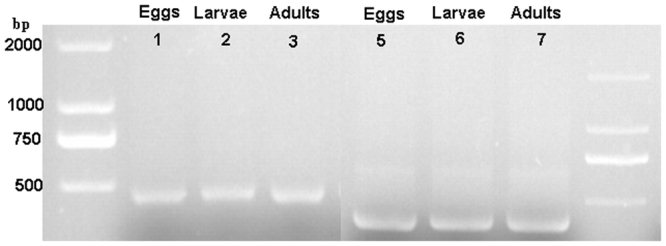
GoldViewTM Nucleic Acid Stained 1.5% Agarose gel of V3-V4 region of 16S RNA gene sequence (1, 2 and 3) and V5-V7 region of 16S RNA gene (4, 5 and 6) amplified product of eggs, L3 larvae and adults of H. contortus. Lane 1 and 8: Direct-load™ Star Marker Plus (D2000 Plus); GenStar Biosolutions Co. Ltd, Beijing, China.
Sequencing was done on the Illumina MiSeq® platform at Beijing Allwegene Technology Co., Ltd, China. The V3-V4 and V5-V7 regions of the bacterial 16S rRNA gene sequences were amplified employing a set of commonly used primers for the analysis of H. contortus-associated bacterial communities using Illumina MiSeq platform (Table 1).
Table 1.
Summary of primers used in the current study.
| Primer pairs | Primer sequence (5′−3′) | Hypervariable region of the 16S rRNA gene sequence targeted by primer pairs. | References |
|---|---|---|---|
| 338F | 5′-ACTCCTACGGGAGGCAGCAG-3′ | V3-V4 region | Wu, X. et al.18 |
| 806R | 5′-GGACTACHVGGGTWTCTAAT-3′ | ||
| 799F | AACMGGATTAGATACCCKG | V5-V6-V7 region | Beckers et al.32 |
| 1193R | ACGTCATCCCCACCTTCC |
Briefly, each 50 μL of PCR reaction contains 10 ng of abomasal or ruminal microbial genomic DNA as the template, 0.25 μL Pyrobest™ DNA Polymerase (Takara Biotechnology, Dalian, CO., LTD) and 1 μL of 10 μM of each primer. PCR reactions were carried out using the following protocol: (1) an initial denaturation step performed at 95 °C for 5 min followed by 30 cycles of denaturation (95 °C, 30 s), annealing (56 °C, 30 s) and extension (72 °C, 40 s), and a final elongation of 10 min at 72 °C. The PCR products were separated by 1% agarose gel electrophoresis, and the 468 bp and 394 bp fragments were purified by using E.Z.N.A.® Gel Extraction kit (Omega Bio-tek, GA, US). Sequencing libraries were generated using NEBNext® Ultra DNA Library Prep Kit for Illumina® (Illumina, USA) following manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Then, the library was sequenced on an Illumina MiSeq platform (Beijing Allwegene Technology Co., Ltd, China) and 300 bp paired-end reads were generated.
Statistical and bioinformatics analysis
After trimming the adaptor and primer sequences from Illumina reads, the raw sequences were assembled for each sample according to the unique barcode using Qiime (V1.7.0, http://qiime.org/scripts/split_libraries_fastq.html).
Paired-end reads from the original DNA fragments were merged using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/).
Quality filtering was performed under specific filtering conditions to obtain the high-quality clean tags according to the pipeline tool quantitative insights into microbial ecology Qiime (V1.7.0, http://qiime.org/scripts/split_libraries_fastq.html).
The clean tags were compared against the reference database (Gold database, http://drive5.com/uchime/uchime_download.html) employing UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) to detect and remove chimeric sequences. Paired-end reads were assigned to each sample according to the unique barcodes. Sequence analysis was performed by UPARSE software package (Uparse v7.0.1001, http://drive5.com/uparse/) using the UPARSE-OTU and UPARSE-OTU ref algorithms. Alpha (within samples) and beta (among samples) diversities were analyzed employing in-house Perl scripts. Sequences with ≥97% identities were assigned to the same OTUs. Representative sequences for each OTU were picked up to be used in the ribosomal database project (RDP) classifier to annotate the taxonomic information for each representative sequence. In order to estimate alpha diversity, we rarified the operational taxonomic units (OTUs) and calculated five metrics: Phylogenetic diversity (PD) Whole tree, Good’s coverage, Chao1, Shannon and Observed species. Rarefaction curves were created based on this five metrics.
The Kruskal-Wallis test19 was used to compare diversity metrics, such as genus evenness, Simpson and Shannon-Wiener species diversity indices. The level of significance was determined at P < 0.05.
The Principal Coordinate Analysis (PCoA) was conducted to explore the differences in the bacterial community structures and was displayed with the WGCNA, stat and ggplot2 packages in the R software (Version 2.15.3). Weighted and un-weighted unifrac distances were calculated employing QIIME20. Graphical representation of the relative abundance of bacterial diversity from phylum to genus was visualized using a bar plot with horizontal bars.
Results
Parasitological tests, FAMACHA© scores and body condition scoring
For coprological examinations, the rectal fecal samples were collected for fecal egg count examination employing the modified McMaster technique. H. contortus were found in 14 (13.08%) of the 107 sheep studied. Additionally, FAMACHA© scores and body condition scoring for all 14 infected animals were assessed. Our results showed that FAMACHA© scores of all 14 infected animals fell between 2 and 3 out of 5 and no sheep decreased below 6 or increased above 8 of all 14 infected sheep for body condition scores. Blood samples were collected for haemato-biochemical analyses to confirm the presence of H. contortus infection. Interestingly, these results further support the coprologic examinations (unpublished data). The aforementioned parasitological tests, FAMACHA© scores, body condition scoring and haemato-biochemical analyses were reassessed after two weeks; the results are consistent with the previously obtained data.
Morphological forms of the ovine H. contortus life-cycle stages
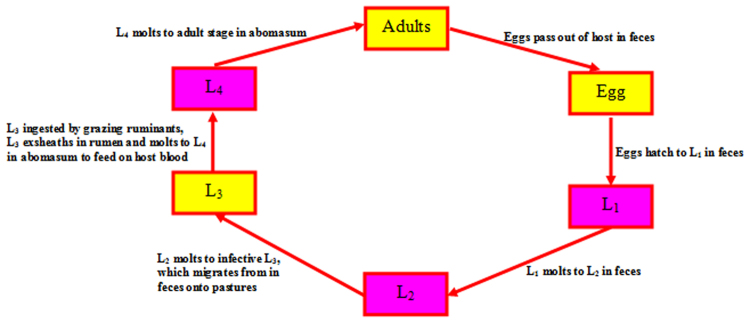
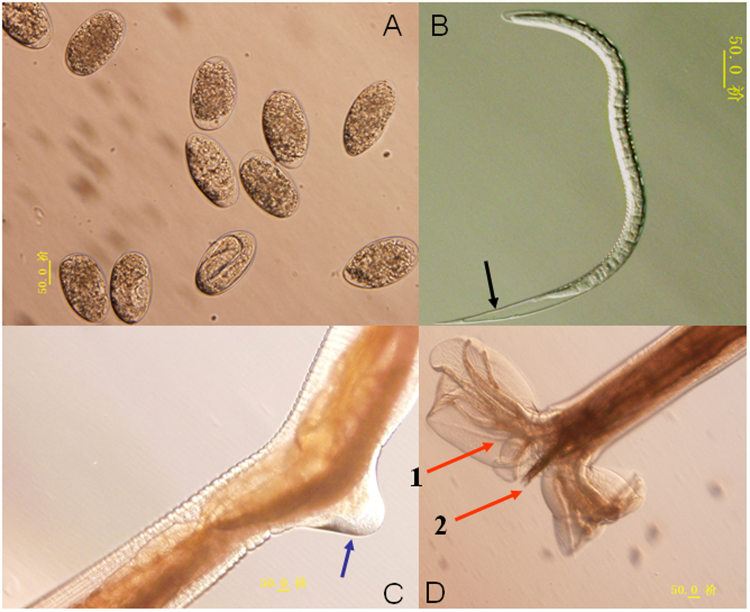
The life-cycle of H. contortus consists of six stages: the egg, four larval stages (L1, L2, L3, L4) and the adult. Each stage in the life cycle is separated by a molt except for the egg - L1. The embryo develops to a 1st-stage larva within the egg, hatches and undergoes four molts before developing into the sexually mature adult. H. contortus passes through diverse environments during completion of its life-cycle. The adults and 4th-stage larvae are parasitic and adapted to the environment within the host, while the other stages are free-living (Fig. 3). Three distinct stages of the life-cycle of H. contortus are selected for the characterization of their microbiota, which are: eggs, L3 larval stage and adults (Fig. 4).
Figure 3.
The basic life-cycle pattern of H. contortus.
Figure 4.
H. contortus life-cycle stages. (A) H. contortus egg. (B) H. contortus L3 larva. Slender larva, tail of sheath medium length (>50 µm <100 µm), tapering to point and often kinked. (C) H. contortus adult female vulval flap. The blue arrow is pointing to the vulval flap. (D) H. contortus adult male bursa. The red arrows are pointing to the capulatory bursa (1) spicules used to hold open the female worms genital opening (2).
Investigation of parasite surfaces for abomasal microbial contamination
Combining the evidence from the two distinct approaches, including SEM and next-generation sequencing of 16S rRNA gene (V3-V4 region), the obvious result is there is no contamination (i.e. intact bacteria and their DNA products) originating from the abomasal microbial community and consequently; the results of the Haemonchus microbiome are sufficient to remove an important obstacle about the abomasal bacterial contamination (Figure S1 and Table S1).
Larval-stage of H. contortus displaying internalized transgenic Escherichia coli
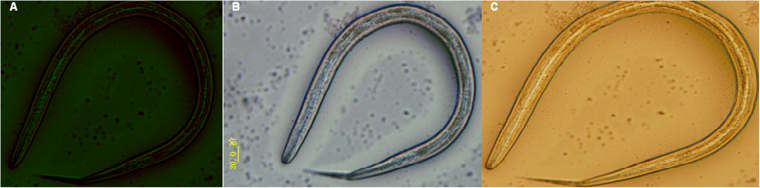
The expression cassette plasmid, H4-DHFR-EYFP-ACT (Fig. 5), was constructed from the recombinant plasmid pd2EYFP-N1+ formerly described by21. The mic1-EYFP and RFP were replaced with H4-DHFR. Therefore, the tandem dihydrofolate reductase thymidylate synthase- enhanced yellow fluorescent protein (DHFR-EYFP) coding region is flanked by the Histone 4 (H4) promoter from E. tenella and 3′region of actin. With respect to plasmid transformation, the procedure was conducted according to the manufacturer’s recommendations using BL21 (DE3) chemically competent cell (Beijing TransGen Biotech Co., Ltd.). A fluorescent microscopy image (Olympus, Tokyo, Japan) using the filter set at 488-nm excitation, 508-nm emission22 of the free-living larval-stage that incubated for 72 hours with yellow fluorescent protein (YFP)-expressing E. coli is presented in Fig. 6.
Figure 5.

Schematic representation of YFP expressing plasmid: Expression cassette is shown as colored boxes.
Figure 6.
H. contortus showing internalized yellow fluorescent protein (YFP)-expressing Escherichia coli. Fluorescent (A), merged (B) and Bright (C) images were taken.
Diversity index analysis of microbial community
Samples were evaluated for alpha diversity (microbial diversity within samples) and beta diversity (community diversity between samples) analysis.
Alpha rarefaction curves
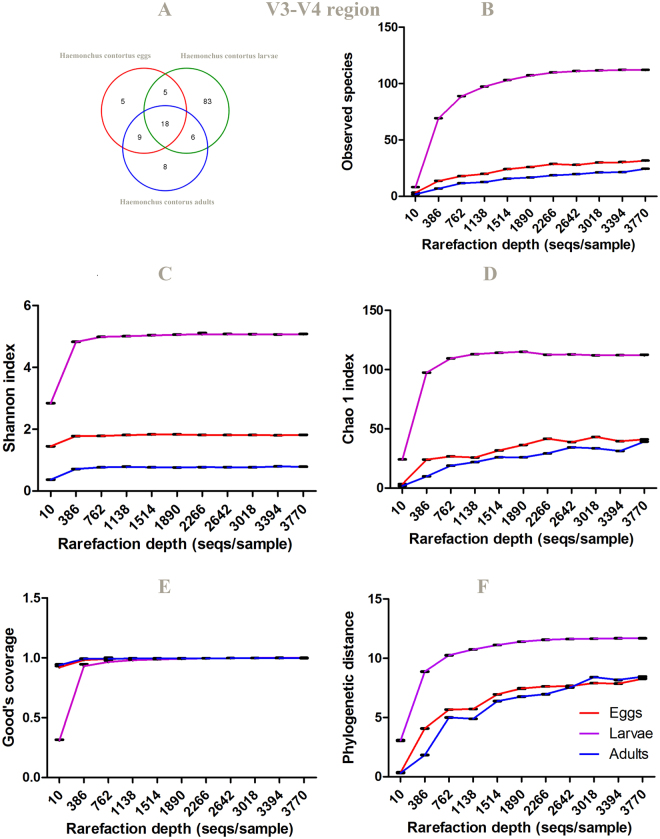
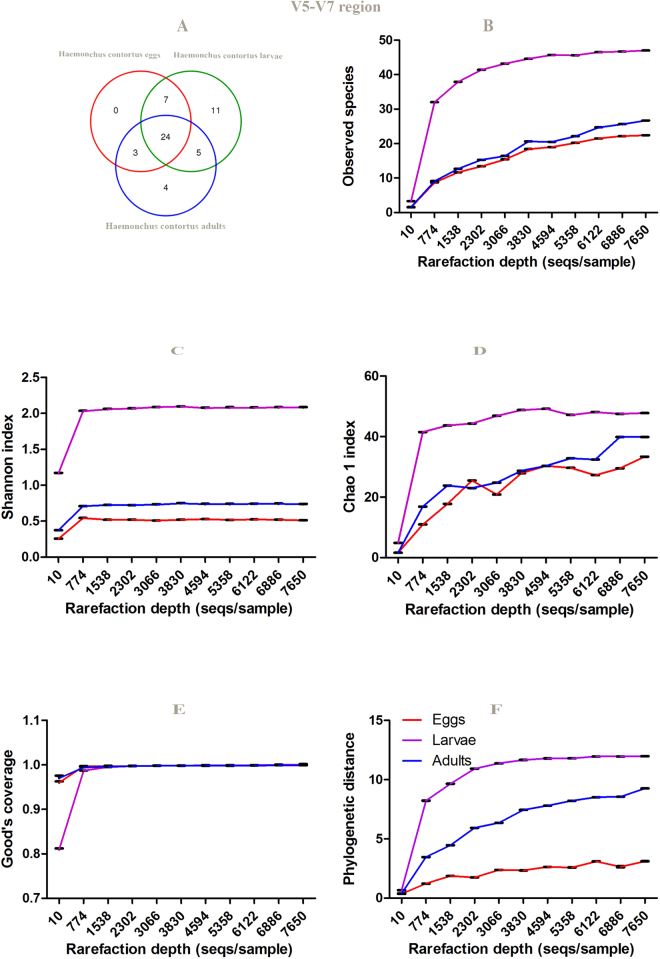
It can be seen from the data in Table 2, Figs 7A–F and 8A–F, Tables S2 and S3 that V5–V6 amplicons showed reduced observed OTUs, regardless of H. contortus life- cycle stage, compared to V3-V4 amplicons, which could be due to greater conservation in this region of the 16S RNA gene sequence. The alpha diversity indexes represent species richness/number of OTUs per H. contortus life-cyle stages using various metrics. Alpha rarefaction curves of H. contortus life-cycle stages were computed exploiting the alpha_rarefaction.py in Qiime. Alpha diversity metrics, such as Phylogenetic diversity (PD) whole tree, Good’s coverage, Chao1, Shannon and Observed species were plotted using QIIME tools. They illustrated the number of observed species at a random pool of sequences in different depths (number of sequences). The rarefaction curves for all stages of the parasite reached the near plateau phase depicting satisfying sampling depth. The estimators, such as Shannon, Chao1, Good’s coverage and Observed species showed that the third larval-stage (L3) of H. contortus could significantly harbor a more abundant and diversified microbial community compared to those of the adult- and egg- stages, respectively. Furthermore, common microbial diversity indices, such as Simpson’s index, Shannon-Wiener species diversity index (H), richness (Table 2), were evaluated.
Table 2.
Overview of alpha diversity metrics (interquartile range (IQR) and median) for the different life-cycle stage microbiome. P-values were calculated using Kruskal-Wallis test. The level of significance was determined at P < 0.05.
| 16S rRNA gene sequence region | Diversity index | H. contortus egg-stage microbiomea | H. contortus larval-stage Microbiomea | H. contortus adult -stage microbiomea | H. contortus eggs vs H. contortus larvae | H. contortus eggs vs H. contortus adults | H. contortus larvae vs H. contortus adults | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Median, IQR | Median, IQR | Median, IQR | Median, IQR | Median, IQR | Chi-Square | Asymp. Sig. | Chi-Square | Asymp. Sig. | ||
| V3-V4 region | OTUs number of each sample (total species) | 37.05, 0.45 | 111.8, 0.8 | 41.04, 0.64 | 3.8571 | 0.0495 | 3.8571 | 0.0495 | 3.8571 | 0.0495 |
| Shannon-Wiener species diversity index | 1.22827, 0.000004 | 2.3507, 0 | 0.531, 0 | 4.5 | 0.0339 | 4.5 | 0.0339 | 5 | 0.0253 | |
| Genus evenness (equitability) | 0.340028, 0.00115 | 4.716712, 0.007137 | 0.143, 0.1302 | 3.8571 | 0.0495 | 0.4412 | 0.5066 | 3.9706 | 0.0463 | |
| Simpson’s index | 0.589393, 0.000393 | 0.825784, 0 | 0.771, 0 | 4.5 | 0.0339 | 4.5 | 0.0339 | 5 | 0.0253 | |
| V5-V7 region | OTUs number of each sample (total species) | 34, 0.2 | 47.2, 0.6 | 36.1, 0.5 | 3.9706 | 0.0463 | 3.8571 | 0.0495 | 3.9706 | 0.0463 |
| Shannon-Wiener species diversity index | 0.327, 0 | 1.25429, 0 | 0.48614, 0 | 5 | 0.0253 | 5 | 0.0253 | 5 | 0.0253 | |
| Genus evenness (equitability) | 0.0927, 0.0001 | 0.32542, 0.001084 | 0.013617, 0.121984 | 3.9706 | 0.0463 | 0.4412 | 0.5066 | 3.8571 | 0.0495 | |
| Simpson’s index | 0.121, 0 | 0.514703, 0 | 0.771735, 0.190294 | 5 | 0.0253 | 4.5 | 0.0339 | 4.5 | 0.0339 | |
aData represent median and interquartile range (IQR) values from pooled populations (i.e. from each sheep) of eggs/larvae/adults (male and female) come from three different sheep (natural infection).
Figure 7.
Venn graph (A) and alpha diversity analysis (B–F) of H. contortus eggs, larvae and adults based on 16S rRNA gene sequence (V3-V4 region) of parasite microbiome and 97% identity. Rarefraction curve for (B). Observed species (C). Shannon, (D). Chao1, (E). good’s coverage and (F). PD whole tree. Data represent average values from pooled populations of eggs/larvae/adults (male and female) come from three different sheep.
Figure 8.
Venn graph (A) and alpha diversity analysis (B–F) of H. contortus eggs, larvae and adults based on 16S rRNA gene sequence (V5-V7 region) of parasite microbiome and 97% identity. Rarefraction curve for (B). observed species (C). Shannon, (D). Chao1, (E). good’s coverage and (F). PD whole tree. Data represent average values from pooled populations (i.e from each sheep) of eggs/larvae/adults (male and female) come from three different sheep.
Our results suggested that H. contortus larval-stage microbiome had been more taxon-rich compared to H. contortus egg and adult-stage microbiomes, respectively using two different amplification primer sets (targeting V3–V4, and V5–V7) and Illumina MiSeq. Moreover, H. contortus egg-stage microbiome compared to H. contortus larval-stage microbiome, and the later compared to H. contortus adult-stage microbiome displayed significant differences employing genus evenness, Simpson and Shannon-Wiener species diversity indices at the genus-level. Similarly, there was a consequential difference between H. contortus egg-stage microbiome in comparison with H. contortus adult-stage microbiome at the genus-level (targeting V3–V4, and V5–V7) regarding Simpson and Shannon-Wiener species diversity indices; however, genus evenness did not show any significant differences (Table 2).
Beta diversity
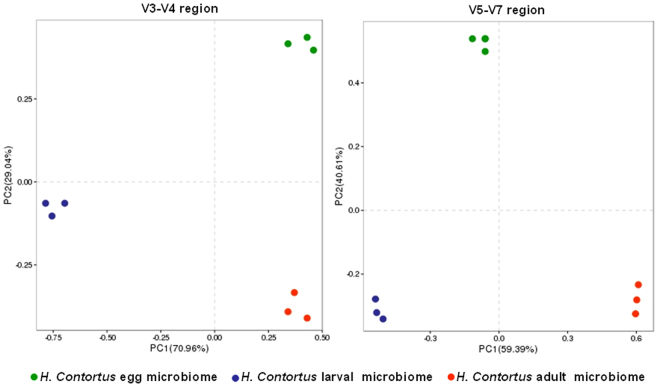
Computing differences between microbial communities were calculated exploiting the default beta diversity metrics of weighted and unweighted UniFrac23 (beta_diversity_through_plots.py). Principal Coordinate Analysis (PCoA) was plotted using the result of UniFrac distance matrices to determine the similarity between groups of samples/time-points. The three-dimensional PCoA plots were visualized employing the Emperor tool24. The beta diversity analysis showed three very distinct clusters separating the different life-cycle stages of H. Contortus (Fig. 9).
Figure 9.
Principal coordinate analysis (PCoA) of H. contortus microbiome using primer pairs targeting V3–V4 (A) and V4–V5 (B) regions. The percentage variation explained with first two principal components (PC1 and PC2). Data represent average values from pooled populations (i.e. from each sheep) of eggs/larvae/adults (male and female) come from three different sheep.
The microbial community composition of the ovine abomasum and rumen in the context of H. contortus infection at the genus level
Two-month-old sheep (Ovis aries) that were raised under confined living conditions to avoid worm exposure were moved from a local farmer in Jin Zhan village, Chaoyang, Beijing, China with an average liveweight of 25.02 ± 2.2. Twelve parasite-naive sheep were divided into four treatments, uninfected-control groups and 7- and 50- day post-infection (dpi) Haemonchus- infected groups, in triplicate. 6 sheep were inoculated with 5,000 of H. contortus infective larvae laboratory strain and followed for 7 and 50 days. The animals were fed a fixed-formula diet containing forage and concentrate in a ratio of 1 to 1. The abomasal and ruminal microbial communities (V3-V4 amplicon) of the early (7-day post-infection) and late (50-day post-infection) H. contortus -infected sheep at the genus level are presented in Tables S4 and S5.
The analysis of microbial community structure and dominant taxa associated with distinct life-cycle stages of H. contortus
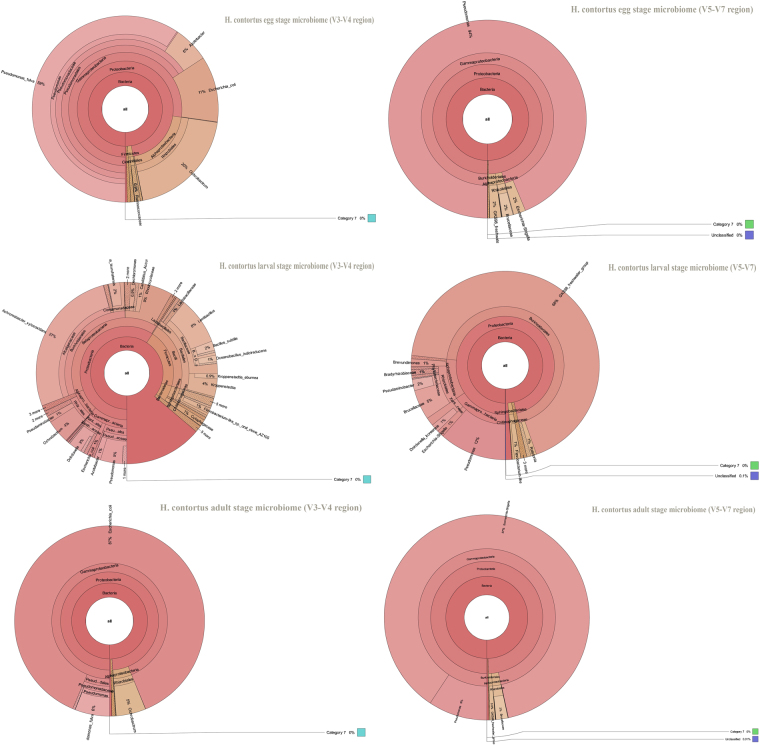
To explore the bacterial community composition associated with the different life -cycle stages of H. contortus, we carried out high-throughput Illumina paired end sequencing. For a better survey, taxonomic abundances of all taxa can be visualized in a Krona plot25 that is based on the relative abundance of the microbial community associated with the different life-cycle stages of H. contortus (Fig. 10). Additionally, Kruskal-Wallis test of all microbial taxa are presented in Tables S6 and S7.
Figure 10.
Krona plot of H. contortus egg-, larval-, adult- stage microbiome based on 16S rRNA gene sequence V3-V4 region (A) and V5-V7 region (B). Data represent average values from pooled populations (i.e. from each sheep) of eggs/larvae/adults (male and female) come from three different sheep.
Four phyla for egg-stage microbiome (Proteobacteria 97.39%, Firmicutes 1.64%, Actinobacteria 0.323% and Bacteroidetes 0.31%) and four phyla for adult stage microbiome (Proteobacteria 99.02%, Firmicutes 0.37%, Tenericutes 0.131%, and Actinobacteria0. 0873%), eight for larval-stage microbiome (Proteobacteria 59.24%, Firmicutes 20.29%, Bacteroidetes 5.4%, Planctomycetes 0.29%, Acidobacteria 0.3972%, 0.2913%, Elusimicrobia 0.1059%, Candidate_division_WS3 0.1589%) using V3-V4 amplicons was identified. For all stages, the parasite microbiome was highly dominated (i.e. had a relative abundance greater than 1.0%) by microbes of the following bacterial phyla, Proteobacteria 97.39% and Firmicutes 1.64% for egg-stage microbiome, Proteobacteria 99.02% for adult-stage microbiome and Proteobacteria 59.24%, Firmicutes 20.29% and Bacteroidetes 5.4% for larval-stage microbiome. Targeting the hypervariable V5-V7 region, four phyla for egg-, larval- and adult- stage microbiome (Bacteroidetes, Proteobacteria, Gemmatimonadetes, Actinobacteria and Firmicutes) were characterized with different relative abundances. The predominant phyla observed in the three stages of H. contortus life-cycle were Proteobacteria for egg- and adult- stage microbiome with relative abundances 99.66% and 99.49% respectively, Proteobacteria 96.29% and Bacteroidetes 1.53% for larval-stage microbiome. Intriguingly, experimental sequencing of the V5-V7 region revealed phylum Gemmatimonadetes with relative abundances 0.46%, 0.019% and 0.015% for the larval-, egg- and adult- stage microbiome respectively, emphasizing the necessity for experimental sequencing of this region.
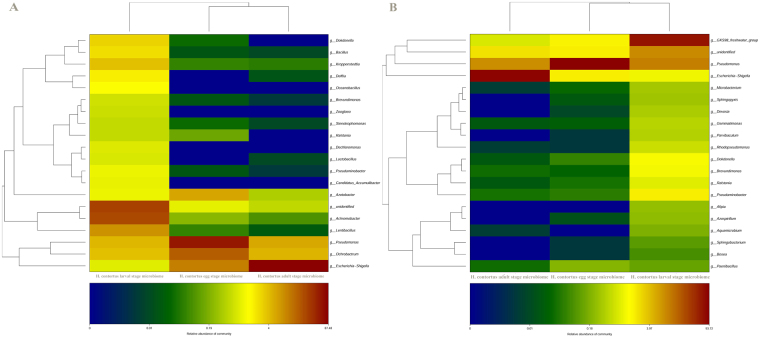
The results of the microbial community composition associated with the different life -cycle stages of ovine H. contortus (V3-V4/V5-V7 amplicons) at the genus level are set out in Tables S8 and S9. Furthermore, we assessed the relative distribution of bacterial taxa at the level of genus for each stage of H. contortus life-cycle using V3-V4 tags. Pseudomonas (59.58%), Ochrobactrum (19.64%), Escherichia-Shigella (11.44%) and Azotobacter (6.193%) were abundant in the egg-stage microbiome. The dominant genera of larval-stage microbiome are Achromobacter (26.67%), Lentibacillus (8.45%), Pseudomonas (4.69%), Ochrobactrum (4.212%), Kroppenstedtia (4.105%), Dokdonella (2.97), Bacillus (2.52%), Delftia (2.04%), Oceanobacillus (1.51%), Azotobacter (1.17%), Pseudaminobacter (1.17%) and Candidatus_Accumulibacter (1.09%). Additionally, the three most-abundant genera identified in the adult-stage microbiome are Escherichia-Shigella (87.5%), Pseudomonas (5.91%) and Ochrobactrum (5.06%). The representative members of genera, Pseudomonas and Ochrobactrum were prevalently shared in all the stages of the parasite life-cycle. Hierarchical clustering heatmap (Fig. 11A) depicts the relative percentage of each bacterial genus (variables clustering on the Y-axis) within each life -cycle stage (X-axis clustering). As can be seen from the heat map analysis below, the larval-stage microbiome using V3-V4 region did not cluster together indicating that the bacterial diversity of the larval-stage is varied and distinct is distinct from the other life- cycle stages. For V5-V7 tags, the dominant bacterial genera in the egg-stage belonged to Pseudomonas 93.7%, Escherichia-Shigella 2% and GKS98 freshwater group 1.84%, whereas in the adult-stage, the predominant genera belonged to GKS98 freshwater group 67.895%, Pseudomonas 11.5%, Pseudaminobacter 1.95%, Dokdonella 1.37%, Brevundimonas 1.28% and Escherichia-Shigella 1.2%. Furthermore, the adult-stage bacterial population was found to be prevalently composed of two genera: Escherichia-Shigella 87.32% and Pseudomonas 9.305%. Additionally, Escherichia-Shigella and Pseudomonas were predominantly shared by all stages. Hierarchically clustered heatmap analysis based on the bacterial community profiles at genus level disclosed that egg-stage microbiome and larval-stage microbiome clustered together (Fig. 11B).
Figure 11.
Double hierarchical dendrogram of the top 50 ranked H. contortus microbiota showing the relative abundance of the bacterial community at the genus level for different life-cycle stages of H. contortus using primer pairs targetingV3–V4 (A) and V4–V5 (B) regions. The relative values for bacterial species are depicted by color intensity, with legend indicated at the bottom of the figure. Data represent average values from pooled populations (i.e. from each sheep) of eggs/larvae/adults (male and female) come from three different sheep.
The analysis of microbial community structure associated with individual adult female worms of H. contortus
The average individual-level data (single female adult worm) from the same population of sheep were consistent with the group-level data (clusters of male and female worms) collected during the winter 2017 and summer 2016 seasons, respectively with minor differences regarding the less common bacteria (Table S10).
Discussion
The bacterial profiles of complex microbial communities associated with the different life-cycle stages of H. contortus field strains were explored with the aid of Illumina MiSeq sequencing technology. Furthermore, in this study the authors have advocated a set of measures that resulted in the absence of the cross-contamination by unwanted abomasal microbial community.
Bacteria are presumably transmitted vertically (from parents), horizontally (from the environment or unrelated animals) or in some cases a mixture of horizontally and vertically. As the barber’s pole worm, H. contortus is exposed to the microbiota of the rumen and abomasum; the nematode gut microbiota may reflect the external micro-organisms (i.e. from microhabitat [abomasum and rumen] and macrohabitat [pasture and manure]), which continually pass through its alimentary canal as evidenced by haemonchine larvae with internalized transgenic Escherichia coli. These results match those observed in earlier studies. Relatedly, significant changes in the abundance and composition of abomasal and ruminal microbiota were observed throughout H. contortus infections under the same diet regimen (forage (grass silage): concentrate (barley, maize, wheat, wheat bran and bruised soya) 1:1 ad libitum. The luminal abomasum microbiota is considerably dominated by microbes of the following bacterial phyla, Bacteroidetes 65.25%, Firmicutes 24.96%, Fibrobacteres 2.79%, Lentisphaerae 2.18%, Proteobacteria 1.41%, Spirochaetae 1.07% and Tenericutes 1.05% for control uninfected group, Bacteroidetes 71.23%, Firmicutes 18.43%, Proteobacteria 5.6% and Lentisphaerae 1.23% for 7-dpi group, Bacteroidetes 57.7%, Firmicutes 35.42%, Proteobacteria 2.86%, Fibrobacteres 1.14% for 50-dpi group. However, the highly dominant ruminal OTUs belonged to Bacteroidetes 79.82%, Firmicutes 14.33%, Proteobacteria 1.43% and Spirochaetae 1.3% for control uninfected group, Bacteroidetes 61.26%, Firmicutes 31.25%, Proteobacteria 2.23%, Lentisphaerae 1.36% and Spirochaetae 1.28% for 7-dpi group, Bacteroidetes 76.16%, Firmicutes 5.99%, Proteobacteria 15.75% and Spirochaetae 1.44% for 50- dpi group.
Similarly, for all stages, the parasite microbiome is highly dominated by microbes of the following bacterial phyla, Proteobacteria and Firmicutes for egg-stage microbiome, Proteobacteria for adult-stage microbiome and Proteobacteria, Firmicutes and Bacteroidetes for larval-stage microbiome using V3-V4 tags. However, the predominant phyla observed in the three stages of the H. contortus life-cycle were Proteobacteria for egg- and adult- stage microbiome and Proteobacteria 96.29% and Bacteroidetes 1.53% for larval stage microbiome employing V5-V7 tags. Furthermore, the free-living stages of H. contortus, such as L1 (rhabditiform), and L2 (rhabditiform) and L3 filiariform infective larvae on pasture may be a vector or reservoir, of environmental microbial community26. These results corroborate the ideas of27, who suggested that the representative members from these phyla are universal and are not limited to the peculiar environment. Among the aforementioned genera (see result section), Pseudomonas, Ochrobactrum and Escherichia-Shigella were predominantly shared by all the life-cycle stages of H. contortus (eggs, larvae and adults) using V3–V4 and V5–V7 amplicons.
Ochrobactrum spp., which are observed as emerging opportunistic pathogens, have been isolated from soil, hospital water sources, the rhizosphere of potato (Solanum tuberosum), rice (Oryza sativa), and wheat (Triticum sativum). The expression of AmpC beta-lactamases immensely complicates the therapy of infections caused by Ochrobactrum spp.28. Similarly, in 2009, Lacharme-Lora, et al.26 reported the potential role of helminths as pathogen vectors. Intriguingly, α-Proteobacteria group has the ability to avoid the host innate recognition29,30. Additionally, it has been demonstrated that O. anthropi infection provokes a mix of Th1 and Th2 type responses in the mice30. It can therefore be assumed that the H. contortus microbiome play a role in shaping the ovine immune response. Of special concern is the supporting evidence that the repression of Beta-2-microglobulin (beta-2-M), MHC class I alpha chain and MHC class II, DR alpha at day 7 post-infection and MHC class I alpha chain and MHC class II Alpha chain A alpha chain expression at day 50 post-infection in the abomasal tissue of sheep experimentally infected with H. contortus employing Illumina RNA-Seq technology (unpublished data).
There is a fierce competition for obtaining iron among microorganisms inside the ovine body to keep bacterial invaders under control. Iron is an indispensable element for many reduction–oxidation (redox) reactions and the virulence of pathogenic bacteria31. The predominant microbial genera, especially, Pseudomonas and Escherichia have developed an innovative way to confront with a variety of iron limitations by colonization of all the life-cycle stages of the blood-sucking parasite, H. contortus (eggs, larvae and adults). The probable association between the predominant microbial genera, such as Pseudomonas and Escherichia, and ovine hosts were further observed by data from H. contortus in naturally infected caprine hosts (unpublished data). The next few years will likely witness an immense number of studies uncovering the exact nature of this microbiome-H. contortus interaction, and finally reveal the specific molecular mechanisms underlying the nematode microbiome as an achilles’ heel of the gastrointestinal parasites. There is abundant room for further progress in comparing microbiome of different helminths, which has, and will continue to offer an important parallel goal for the removal of a wide-variety of devastating animal and human diseases.
Electronic supplementary material
Acknowledgements
This research was supported by the National Key Basic Research Program (973 program of China (Grant No. 2015CB150300). Additionally, the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceived and designed the experiments: S.E. and X.S. Performed the experiments: S.E. Analyzed the data: S.E. Contributed reagents/materials/analysis tools: S.E. and X.S. Wrote the paper: S.E. and X.S. All authors read and approved the final version of the manuscript.
Competing Interests
The authors declare no competing financial interests.
Footnotes
An erratum to this article is available at https://doi.org/10.1038/s41598-017-07623-9.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00171-2
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has not been fixed in the paper.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rahman WA, Collins GH. The establishment and development of Haemonchus contortus in goats. Veterinary parasitology. 1990;35:189–193. doi: 10.1016/0304-4017(90)90053-E. [DOI] [PubMed] [Google Scholar]
- 2.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (New York, N.Y.) 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annual review of microbiology. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Slatko BE, Luck AN, Dobson SL, Foster JM. Wolbachia endosymbionts and human disease control. Molecular and biochemical parasitology. 2014;195:88–95. doi: 10.1016/j.molbiopara.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Shahkolahi AM, Donahue MJ. Bacterial flora, a possible source of serotonin in the intestine of adult female Ascaris suum. The Journal of parasitology. 1993;79:17–22. doi: 10.2307/3283271. [DOI] [PubMed] [Google Scholar]
- 6.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. Journal of bacteriology. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes KS, et al. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science (New York, N.Y.) 2010;328:1391–1394. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez-Cortes JG, et al. Microbiota from Rhabditis regina may alter nematode entomopathogenicity. Parasitology research. 2016;115:4153–4165. doi: 10.1007/s00436-016-5190-3. [DOI] [PubMed] [Google Scholar]
- 9.Tremblay J, et al. Primer and platform effects on 16S rRNA tag sequencing. Frontiers in microbiology. 2015;6:771. doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A framework for human microbiome research. Nature486, 215-221, doi:10.1038/nature11209 (2012). [DOI] [PMC free article] [PubMed]
- 11.Cantacessi C, et al. Impact of experimental hookworm infection on the human gut microbiota. The Journal of infectious diseases. 2014;210:1431–1434. doi: 10.1093/infdis/jiu256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li RW, et al. The effect of helminth infection on the microbial composition and structure of the caprine abomasal microbiome. Scientific reports. 2016;6:20606. doi: 10.1038/srep20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zajac, A., Conboy, G. A. (Chichester, West Sussex, UK: Wiley-Blackwell, 2012).
- 14.Kaplan RM, et al. Validation of the FAMACHA eye color chart for detecting clinical anemia in sheep and goats on farms in the southern United States. Veterinary parasitology. 2004;123:105–120. doi: 10.1016/j.vetpar.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Khan, K., Meyer, H. H. and Thompson, J. M. In Proceedings Western Section American Society of Animal Science (1992).
- 16.Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PloS one. 2013;8:e53608. doi: 10.1371/journal.pone.0053608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenback JD. A comparison of techniques useful for preparing nematodes for scanning electron microscopy. Journal of nematology. 1986;18:479–487. [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, et al. Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3-V4 region of the 16S rRNA gene. Applied microbiology and biotechnology. 2016;100:3577–3586. doi: 10.1007/s00253-015-7257-y. [DOI] [PubMed] [Google Scholar]
- 19.Qi J, Asl HF, Bjorkegren J, Michoel T. kruX: matrix-based non-parametric eQTL discovery. BMC bioinformatics. 2014;15:11. doi: 10.1186/1471-2105-15-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, et al. Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PloS one. 2012;7:e35470. doi: 10.1371/journal.pone.0035470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin G, Liu X, Zou J, Huang X, Suo X. Co-expression of reporter genes in the widespread pathogen Eimeria tenella using a double-cassette expression vector strategy. International journal for parasitology. 2011;41:813–816. doi: 10.1016/j.ijpara.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Hao L, Liu X, Zhou X, Li J, Suo X. Transient transfection of Eimeria tenella using yellow or red fluorescent protein as a marker. Molecular and biochemical parasitology. 2007;153:213–215. doi: 10.1016/j.molbiopara.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacharme-Lora L, Salisbury V, Humphrey TJ, Stafford K, Perkins SE. Bacteria isolated from parasitic nematodes–a potential novel vector of pathogens? Environmental health: a global access science source. 2009;8(Suppl 1):S17. doi: 10.1186/1476-069X-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravindraja C, Viszwapriya D, Karutha Pandian S. Ultradeep 16S rRNA sequencing analysis of geographically similar but diverse unexplored marine samples reveal varied bacterial community composition. PloS one. 2013;8:e76724. doi: 10.1371/journal.pone.0076724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoma B, et al. Identification and antimicrobial susceptibilities of Ochrobactrum spp. International journal of medical microbiology: IJMM. 2009;299:209–220. doi: 10.1016/j.ijmm.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Barquero-Calvo E, et al. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PloS one. 2007;2:e631. doi: 10.1371/journal.pone.0000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barquero-Calvo E, et al. The differential interaction of Brucella and ochrobactrum with innate immunity reveals traits related to the evolution of stealthy pathogens. PloS one. 2009;4:e5893. doi: 10.1371/journal.pone.0005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.EP, S. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6(8), doi:10.1371/journal.ppat.1000949 (2010). [DOI] [PMC free article] [PubMed]
- 32.Beckers B, et al. Performance of 16s rDNA Primer Pairs in the Study of Rhizosphere and Endosphere Bacterial Microbiomes in Metabarcoding Studies. Frontiers in microbiology. 2016;7:650. doi: 10.3389/fmicb.2016.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.