Abstract
The first version of a glycoprotein hormone receptor (GPHR) information resource was designed to link functional with structural GPHR information, in order to support sequence-structure-function analysis of the LH, FSH, and TSH receptors (http://ssfa-gphr.de). However, structural information on a binding- and signaling-sensitive extracellular fragment (∼100 residues), the hinge region, had been lacking. A new FSHR crystal structure of the hormone-bound extracellular domain has recently been solved. The structure comprises the leucine-rich repeat domain and most parts of the hinge region. We have not only integrated the new FSHR/FSH structure and the derived homology models of TSHR/TSH, LHCGR/CG, and LHCGR/LH into our web-based information resource, but have additionally provided novel tools to analyze the advanced structural features, with the common characteristics and distinctions between GPHRs, in a more precise manner. The hinge region with its second hormone-binding site allows us to assign functional data to the new structural features between hormone and receptor, such as binding details of a sulfated tyrosine (conserved throughout the GPHRs) extending into a pocket of the hormone. We have also implemented a protein interface analysis tool that enables the identification and visualization of extracellular contact points between interaction partners. This provides a starting point for comparing the binding patterns of GPHRs. Together with the mutagenesis data stored in the database, this will help to decipher the essential residues for ligand recognition and the molecular mechanisms of signal transduction, extending from the extracellular hormone-binding site toward the intracellular G protein-binding sites.
The family of glycoprotein hormone receptors (GPHRs), a subgroup of the family A G protein-coupled receptors (GPCRs), consists of the TSH receptor (TSHR), the FSH receptor (FSHR), and the lutropin/choriogonadotropin receptor (LHCGR).
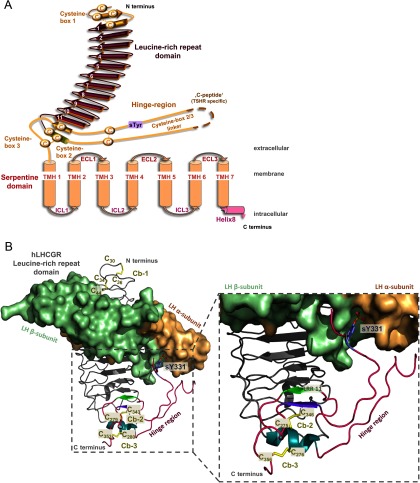
The general structural topology of the homologous GPHRs is identical with that of other GPCRs and is characterized by an N-terminal extracellular region (N-ECR) and an intracellular C-terminal section, with 7-transmembrane helices (TMHs) connected by 3 intracellular loops and 3 extracellular loops. The TMHs and loops constitute the serpentine domain or 7-transmembrane domain (7-TMD), which spans the membrane from the extra- to the intracellular site. A special structural feature of all GPHRs, in contrast to other family A GPCRs, is a large N-ECR (Figure 1A), which is made up of more than 320 amino acids (reviewed in Reference 1).
Figure 1.
Schematic Illustration of GPHR Topology and a Homology Model of hLHCGR N-ECR with Bound Lutropin. A, GPHRs possess a 7-TMH topology (serpentine domain consists of 7-TMHs connected by 3 extracellular loops [ECLs] and 3 intercellular loops [ICLs]), as do all GPCRs. In contrast to most family A GPCRs, they have a very large N-ECR. This N-ECR is composed of the LRRD and the hinge region. The LRRD is made up of 11 full repeats. Cb-1 comprises a cysteine from the N terminus, 2 cysteines from an antiparallel β-strand upstream and one from the first regular repeat. The hinge region, now structurally defined by the new FSHR crystal structure, starts after a short helix-element of the 11th repeat and contains the first 2 cysteines of Cb-2, which are linked via disulfide bridges to Cb-3. After the helix, an unstructured region harbors the cysteine box 2/3 linker and the cleavable peptide (C-peptide), in the case of TSHR. The sulfotyrosine is known from mutagenesis studies and contributes to hormone binding in all 3 GPHR subtypes. The C-terminal section of the hinge region contains a β-strand that is located parallel to the last repeat of the LRRD and is connected via a disulfide bridge. B, Homology model of the N-ECR of hLHCGR in complex with hLH. The gray ribbon presentation visualizes the Cα atom backbone of the LRRD with 9 repeats, according to previous information concerning the LRRD dimension. The new structural information for the LRRD (33) is colored magenta and includes an extension of the LRRD by repeat 11 (β-strand [light green] and a short helix [light blue]), a coiled region with the sulfated tyrosine 331 (blue sticks) and finally a β-strand orientated parallel to the last strand of the LRRD. The 2 cysteines of the short helix are bridged via disulfide bonds between the β-strand and the C terminus of the hinge region. The hLH is visualized as a surface (α-subunit in pale orange, β-subunit in pale green).
The major binding region for the hormones TSH, LH, chorionic gonadotropin (CG), and FSH (2) has been identified by experimental studies on the LRRDs (3–7). A complementary pattern of amino acid side-chain properties is responsible for specific hormone recognition (8). Furthermore, the hinge region (comprising about 90–130 residues) has also been shown to be important for hormone binding and signal induction (examples in References 9–15). On the basis of site-directed mutagenesis, it has been concluded that the extracellular hinge region of GPHRs is necessary for stabilization of a signaling-competent basal receptor conformation (12, 15–23), and it has been proposed that the hinge region plays an important role in the regulation and amplification of receptor activity (11, 16, 21, 24).
The first crystal structure of the FSHR leucine-rich repeat domain (LRRD) complex with the hormone FSH (PDB [Protein Database] entry code: 1XWD) was published in 2005 (26). Moreover, the structural features of the FSHR LRRD were subsequently confirmed by crystal structure complexes between the TSHR LRRD and an activating (27) (PDB entry code: 3G04) or inactivating (28) autoantibody (PDB entry code: 2XWT), respectively.
However, the detailed molecular mechanism of receptor activation and the regulation of signaling activity at the extracellular site are not fully understood. One major reason for this has been the lack of structural information for the hinge region containing the second hormone-binding site, which would be a prerequisite to clarify the exact arrangement of receptor components to each other or to describe the entire hormone-binding process. This missing structural section was also absent in the freely available GPHR information resource (web-address: http//:ssfa-gphr.de). This database and linked web applications were designed to collect relevant GPHR information, with the aim of supporting sequence-structure-function analysis (SSFA) of this GPCR subfamily (29). The SSFA-GPHR information resource allows a focused analysis of semiquantitative mutant data from GPHR subtypes and diverse experimental approaches (30, 31). A second and complementary GPHR information resource with mutant data and several online applications has been provided by others (Reference 32; http://gris.ulb.ac.be/).
A new GPHR crystal structure (hFSHR in complex with hFSH) that is a milestone in GPHR research has recently been published (33). This structure presents structural information on almost the entire extracellular region, including the LRRD and most parts of the hinge region that had not yet been solved. Binding site analysis revealed that both the LRRD and the hinge region interact with the hormone. In consequence, combination of this novel structural information with available functional information allows us to answer important questions and to interpret experimental findings on GPHRs. For example, a section of the hinge region interacts directly with the hormone, and structural details of this second binding site can now be elucidated (33).
This advanced information is also relevant to the homologous TSH and LH receptor and demands a revision and upgrade of our GPHR research resource. Therefore, we have now implemented not only the new extracellular human FSHR structure, but have also provided the derived new homologous structural models for GPHR subtypes (hTSHR, hLHCGR, rLHCGR, rFSHR) and linked functional information with these new structural insights. This enables more detailed extraction of data for these receptors and analyses of differences or similarities among the GPHRs. This upgrade includes, for the first time, not only the extracellular receptor region, but also the bound hormones. To ensure optimal exploitation of the complete hormone-binding interface at the extracellular region, a new Interface Analysis Tool has been added that offers visualization and analysis options to illuminate the contacts and interactions between the hormone and receptor complex.
Materials and Methods
Homology modeling of different GPHRs bound with hormone subtypes
Structural homology models for the extracellular domains of the TSHR and LHCGR were built on the basis of the newly available FSHR crystal structure (33). The procedure of hTSHR modeling has been recently published (34), and this model (TSHR extracellular region with bound bTSH) was also used for the database. In brief, the incomplete LRRD of hTSHR with 9 repeats was extracted from the LRRD-antibody complex (27) (PDB entry code: 3G04) and extended by adding amino acids from the superimposed full FSHR N-ECR crystal structure with 11 repeats (33). The 11-repeat is followed by a helical element comprising cysteine box 2 (Cb-2).
Consecutively to Cb-2, the new FSHR N-ECR crystal structure includes the N-terminal fragment of the hinge region (hFSHR positions, Trp291–Ser295; hTSHR positions, Gln289–Ser304). This portion was also added to the hTSHR LRRD model, and side chains of TSHR were substituted. Disulfide bridges between cysteines of Cb-2 and Cb-3 were built as suggested by the FSHR crystal structure. Furthermore, the corresponding region of the hFSHR crystal structure was used as a structural template (hFSHR positions, Thr331–Ile359) in the design of the hTSHR C-terminal hinge region fragment (hTSHR positions, Ser383-Ile411).
The sulfated hFSHR Tyr335 was already known as mandatory for hormone binding and signaling and interacts tightly with amino acids of the hormone subunits between the β-subunit loop 2 and the α-loop 1 (33). Therefore, if it is assumed that this sulfated tyrosine in TSHR also interacts with the same hormone-binding site as that observed for the FSHR-FSH complex in the TSHR model Tyr385 (termed Tys385), it then substitutes for the sulfated FSHR Tyr335.
The hormone bTSH was modeled on the basis of hFSH, so that the general mode of orientation between the hormone and receptor was maintained as suggested by the human FSHR-FSH complex.
The same procedure was repeated for the complex structures of human and rat LHCGR. For hLHCGR, both interaction models, hLHCGR/hLH and hLHCGR/hCG, are provided. For the complex with bound hCG, hFSH in the template structure of the hFSH-hFSHR complex was substituted with the crystallized structure of hCG (35). In the hLH model, this hormone structure was used as a template. The CG interaction model is currently only preliminary, due to the lack of any structural information about its C-terminal tail (24 amino acids), which might influence the binding mode of hCG (36). In analogy, the rLHCGR model was created. In rodents, only LH is responsible for rLHCGR activation (37); therefore, the respective interaction model, rLHCGR/CG, is not provided.
The rFSHR model was designed by substituting subtype-specific residues, while keeping the hFSHR/hFSH at conserved positions.
Glycosylation was not considered in any structures or structural models. Side chains of the homology models were subjected to conjugate gradient minimizations (until they converged at a termination gradient of 0.05 kcal/[mol*Å]). The AMBER F99 force field was used. Finally, the models were minimized without constraints. Structural modifications and homology modeling procedures were performed with Sybyl X2.0 (Tripos, Inc, St Louis, Missouri).
General modifications of the database according to the new information from the FSHR crystal structure
The new FSHR structure revealed new structural definitions of specific receptor sections, eg, the length of the LRRD or the hinge region (33). We, therefore, refined our previous annotations in the database for specific sections and amino acids, in accordance with the new set of structural descriptions, and also revised our structural annotations in the alignments, numbering scheme, and snake-plot designer.
In the analysis of the hormone receptor-binding interface and for localization of functional information in proximity to the binding area, and in contrast to former database versions, we added the whole complex and the derived homology models. The 3-dimensional interactive windows to visualize functional information of the hormone-receptor complex were adapted and optimized. For example, in the 3D-Structural Search, one can easily switch between solely the N-ECR, the complex of N-ECR and hormone, or only the 7-transmembrane domain.
Furthermore, in the previous version of our web application, amino acids in the hinge region with functional insights (mutations) were not linked to any structural information, even though the hinge region is known to be important for signaling and ligand interaction. We have now added available structural information for the hinge regions of GPHR subtypes, in order to visualize specifically wild-type amino acids with known functional data from mutagenesis studies.
New tool for the interface analysis of hormone-receptor complex
The new FSHR structure and homologous models for TSHR and LHCGR provide additional insights into hormone recognition, as well as binding and receptor activation. For the identification and analysis of essential amino acids, a new tool was implemented to analyze opposing binding patterns by selectively visualizing the contact surface, either for the hormone or for the receptor, or by highlighting interaction partners.
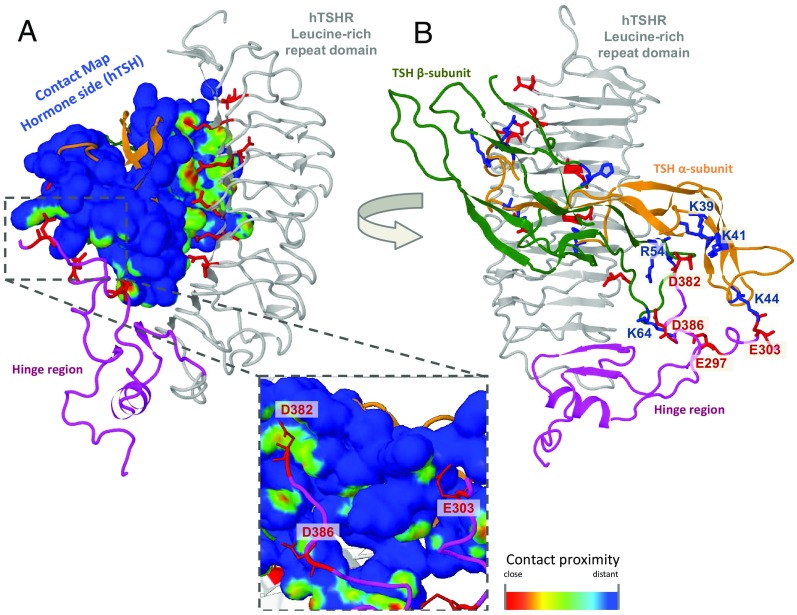
For displaying intermolecular interactions via a surface, a new Jmol (38) function for contact maps is applied (Figure 2A). The color of the surface is based on the contact proximity of the van der Waals surface between potentially interacting partners. Red spots are the closest contacts and usually correspond to hydrogen bonds. On the other hand, yellow and green spots highlight more distant side chains and often correspond to hydrophobic interactions. The user can visualize contacting residues with distinct properties on opposing sides of the complex, eg, charge-charge interactions − positive vs negative charges, with the tool to highlight them separately (Figure 2B).
Figure 2.
Protein-Protein Interface Analysis Tool. The display shows the homology model of hTSHR (LRRD in gray; hinge region in magenta) in complex with the hormone bTSH (α-subunit in orange, β-subunit in green) as visualized at the SSFA-GPHR website. The surface of the hormone is colored according to the receptor-hormone contact proximity of the van der Waals radius between the interaction partners. Red spots are the closest contacts and usually correspond to hydrogen bonds. Yellow and green spots highlight more distant side chains and often correspond to hydrophobic interactions. As an example, in panel A, all negatively charged N-ECR residues in close contact to the hormone are visualized together with the contact surface displayed on bTSH. The inset shows a close-up of particular negatively charged side chains of the hinge region interacting with the hormone. In panel B the interaction partners (positively charged side chains) on the interface are highlighted (labeled manually for representation).
Results and Discussion
We now report an updated and revised version of a Glycoprotein-Hormone Receptor Database (29), which has been improved significantly by implementation of new structural information. This mainly comprises missing sections (90–130 residues) of the N-ECR, namely the extracellular hinge region that was recently solved for the FSHR by x-ray crystallography in a complex with FSH (33). In addition, extended insights into the structure of the LRRD were observed, namely by enlarging this domain (already predicted in Reference 11). Finally, the second hormone-binding site at the hinge region is deciphered in detail around a sulfated tyrosine and can be analyzed in complex with the hormone for all GPHR subtypes.
In addition to the participation of Cb-1 in the N-terminal fold of the LRRD, the new structure revealed that the LRRD continues through specific amino acids of Cb-2 (Figure 1). Thus, the LRRD is constituted by 11 LRRs, instead of 9 as assumed previously. The short helical structure at repeat 11 contains the 2 consecutive cysteines that are bridged to the last 2 extracellular cysteines of Cb-3. These are located on one side in a short β-strand parallel to the LRRD β-strand 11 and on the other side are adjacent to TMH-1 (Figure 1). In conclusion, the previous prediction of tight spatial proximity between Cb-2 and Cb-3 (10) is confirmed, as well as the suggested cysteine bridges (17, 36, 39). Regarding the short helix linking the extended LRRD and the hinge region toward the transmembrane domain via 2 disulfide bridges, it is conceivable that this may act as a pivot and center, where the conformational effects of 2 hormone-binding sites (LRRD and hinge) converge toward the transmembrane domain (25).
Moreover, a specific feature appearing at the second hormone-binding site of the new crystal structure is a sulfated tyrosine at the C terminus of the hinge region that binds in a pocket located between the α- and β-subunit of the hormone. All GPHRs exhibit such a sulfotyrosine (FSHR Tyr335, LHR Tyr331, TSHR Tyr385). Several functional studies had already confirmed the general importance of this sulfated residue for hormone binding and signal transduction at GPHRs (36, 40). Tyrosylprotein sulfotransferases (such as tyrosylprotein sulfotransferase 2) are enzymes that catalyze the posttranslational modification of specific tyrosines by adding an SO3 group. This addition enhances the negative electrostatic potential of the side chain and therefore increases the number of possible hydrogen bonds formed. Their relevance for TSHR might also be indicated in vivo by the absence of tyrosylprotein sulfotransferase 2 in knockout mice, which leads to hypothyroidism (41).
Interestingly, distinct sulfated tyrosines at extracellular N-terminal domains of several chemokine-receptors have also been shown to interact with the corresponding endogenous ligands: CXCR4, CCR2B, CCR5, CXCR3, CX3CR1, C5a receptor, and CCR8 (eg, Refs. 42–44). As observed in the FSHR crystal structure, the crystal structure of the N-terminal tail of the CXCR4 (PDB entry code 2K03) contains a sulfated tyrosine, with strong hydrogen bond interactions between arginine and asparagine side chains in the chemokine (45). Finally, tyrosine sulfation is here of pathophysiologic relevance too, as indicated by interrelations with HIV infection mechanisms via chemokine receptors (46).
Moreover, several examples of important functions for sulfotyrosines are also known in non-GPCR proteins: 1) direct contacts in selectins, 2) proteolytic processing (eg, gastrin processing), and 3) proteolytic activation of extracellular proteins (eg, factor V and VIII activation) (47).
The more extensive implications for ligand/protein recognition reflect the general importance of this issue and are relevant to the understanding of the hormone (ligand)-receptor interaction in more detail. In the new version of our database, we have therefore implemented a visualization tool of interactions between the receptor and hormone. Particular amino acids with specific biophysical properties can be depicted and highlighted (Figure 2); on the other hand, surface rendering combined with distance mapping can be used for contact analyses. In particular, this might guide comparative studies between the GPHRs in the now almost complete extracellular complexes. Additionally, it might also be helpful in explaining the diverse functional properties of the GPHRs (48, 49).
Finally, this current report and work provide a progressive step toward a comprehensive understanding of GPHRs, including important aspects such as signal transduction and structural organization.
Acknowledgments
This work was supported by research grants KL2334/2–2 and KR1273/4–1 from the Deutsche Forschungsgemeinschaft.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by research grants KL2334/2–2 and KR1273/4–1 from the Deutsche Forschungsgemeinschaft.
Footnotes
- Cb-1
- -2, and -3
- cysteine box-1
- -2, and -3
- CG
- chorionic gonadotropin
- FSHR
- FSH receptor
- GPCR
- G protein-coupled receptor
- GPHR
- glycoprotein hormone receptor
- LHCGR
- lutropin/CG receptor
- LRRD
- leucine-rich repeat domain
- N-ECR
- N-terminal extracellular region
- SSFA
- sequence-structure-function analysis
- TMH
- transmembrane helix.
References
- 1. Vassart G, Pardo L, Costagliola S. A molecular dissection of the glycoprotein hormone receptors. Trends Biochem Sci. 2004;29(3):119–126. [DOI] [PubMed] [Google Scholar]
- 2. Hearn MT, Gomme PT. Molecular architecture and biorecognition processes of the cystine knot protein superfamily. I. The glycoprotein hormones. J Mol Recognit. 2000;13(5):223–278. [DOI] [PubMed] [Google Scholar]
- 3. Braun T, Schofield PR, Sprengel R. Amino-terminal leucine-rich repeats in gonadotropin receptors determine hormone selectivity. EMBO J. 1991;10(7):1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagayama Y, Rapoport B. Role of the carboxyl-terminal half of the extracellular domain of the human thyrotropin receptor in signal transduction. Endocrinology. 1992;131(2):548–552. [DOI] [PubMed] [Google Scholar]
- 5. Nagayama Y, Wadsworth HL, Chazenbalk GD, Russo D, Seto P, Rapoport B. Thyrotropin-luteinizing hormone/chorionic gonadotropin receptor extracellular domain chimeras as probes for thyrotropin receptor function. Proc Natl Acad Sci USA. 1991;88(3):902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aizen J, Kowalsman N, Kobayashi M, et al. . Experimental and computational study of inter- and intraspecies specificity of gonadotropins for various gonadotropin receptors. Mol Cell Endocrinol. 2012;364(1–2):89–100. [DOI] [PubMed] [Google Scholar]
- 7. Angelova K, de Jonge H, Granneman JC, Puett D, Bogerd J. Functional differences of invariant and highly conserved residues in the extracellular domain of the glycoprotein hormone receptors. J Biol Chem. 2010;285(45):34813–34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caltabiano G, Campillo M, De Leener A, et al. . The specificity of binding of glycoprotein hormones to their receptors. Cell Mol Life Sci. 2008;65(16):2484–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonomi M, Busnelli M, Persani L, Vassart G, Costagliola S. Structural differences in the hinge region of the glycoprotein hormone receptors: evidence from the sulfated tyrosine residues. Mol Endocrinol. 2006;20(12):3351–3363. [DOI] [PubMed] [Google Scholar]
- 10. Kleinau G, Jäschke H, Neumann S, Lättig J, Paschke R, Krause G. Identification of a novel epitope in the thyroid-stimulating hormone receptor ectodomain acting as intramolecular signaling interface. J Biol Chem. 2004;279(49):51590–51600. [DOI] [PubMed] [Google Scholar]
- 11. Kleinau G, Mueller S, Jaeschke H, et al. . Defining structural and functional dimensions of the extracellular thyrotropin receptor region. J Biol Chem. 2011;286(25):22622–22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mizutori Y, Chen CR, McLachlan SM, Rapoport B. The thyrotropin receptor hinge region is not simply a scaffold for the leucine-rich domain but contributes to ligand binding and signal transduction. Mol Endocrinol. 2008;22(5):1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mueller S, Kleinau G, Jaeschke H, Paschke R, Krause G. Extended hormone binding site of the human thyroid stimulating hormone receptor: distinctive acidic residues in the hinge region are involved in bovine thyroid stimulating hormone binding and receptor activation. J Biol Chem. 2008;283(26):18048–18055. [DOI] [PubMed] [Google Scholar]
- 14. Mueller S, Kleinau G, Szkudlinski MW, Jaeschke H, Krause G, Paschke R. The superagonistic activity of bovine thyroid-stimulating hormone (TSH) and the human TR1401 TSH analog is determined by specific amino acids in the hinge region of the human TSH receptor. J Biol Chem. 2009;284(24):16317–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nurwakagari P, Breit A, Hess C, Salman-Livny H, Ben-Menahem D, Gudermann T. A conformational contribution of the luteinizing hormone-receptor ectodomain to receptor activation. J Mol Endocrinol. 2007;38(1–2):259–275. [DOI] [PubMed] [Google Scholar]
- 16. Agrawal G, Dighe RR. Critical involvement of the hinge region of the follicle-stimulating hormone receptor in the activation of the receptor. J Biol Chem. 2009;284(5):2636–2647. [DOI] [PubMed] [Google Scholar]
- 17. Ho SC, Van Sande J, Lefort A, Vassart G, Costagliola S. Effects of mutations involving the highly conserved S281HCC motif in the extracellular domain of the thyrotropin (TSH) receptor on TSH binding and constitutive activity. Endocrinology. 2001;142(7):2760–2767. [DOI] [PubMed] [Google Scholar]
- 18. Jaquette J, Segaloff DL. Constitutive activation of the LH receptor is associated with an alteration in the conformation of the ectodomain. Mol Cell Endocrinol. 2002;194(1–2):211–215. [DOI] [PubMed] [Google Scholar]
- 19. Nakabayashi K, Kudo M, Hsueh AJ, Maruo T. Activation of the luteinizing hormone receptor in the extracellular domain. Mol Cell Endocrinol. 2003;202(1–2):139–144. [DOI] [PubMed] [Google Scholar]
- 20. Sangkuhl K, Schulz A, Schultz G, Schöneberg T. Structural requirements for mutational lutropin/choriogonadotropin receptor activation. J Biol Chem. 2002;277(49):47748–47755. [DOI] [PubMed] [Google Scholar]
- 21. Vlaeminck-Guillem V, Ho SC, Rodien P, Vassart G, Costagliola S. Activation of the cAMP pathway by the TSH receptor involves switching of the ectodomain from a tethered inverse agonist to an agonist. Mol Endocrinol. 2002;16(4):736–746. [DOI] [PubMed] [Google Scholar]
- 22. Zhang M, Tong KP, Fremont V, et al. . The extracellular domain suppresses constitutive activity of the transmembrane domain of the human TSH receptor: implications for hormone-receptor interaction and antagonist design. Endocrinology. 2000;141(9):3514–3517. [DOI] [PubMed] [Google Scholar]
- 23. Zhang ML, Sugawa H, Kosugi S, Mori T. Constitutive activation of the thyrotropin receptor by deletion of a portion of the extracellular domain. Biochem Biophys Res Commun. 1995;211(1):205–210. [DOI] [PubMed] [Google Scholar]
- 24. Hamidi S, Chen CR, Mizutori-Sasai Y, McLachlan SM, Rapoport B. Relationship between thyrotropin receptor hinge region proteolytic posttranslational modification and receptor physiological function. Mol Endocrinol. 2011;25(1):184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krause G, Kreuchwig A, Kleinau G. Extended and structurally supported insights into extracellular hormone binding, signal transduction and organization of the thyrotropin receptor. PloS One. 2012;7(12):e52920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan QR, Hendrickson WA. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433(7023):269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanders P, Young S, Sanders J, et al. . Crystal structure of the TSH receptor (TSHR) bound to a blocking-type TSHR autoantibody. J Mol Endocrinol. 2011;46(2):81–99. [DOI] [PubMed] [Google Scholar]
- 28. Sanders J, Chirgadze DY, Sanders P, et al. . Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody. Thyroid. 2007;17(5):395–410. [DOI] [PubMed] [Google Scholar]
- 29. Kleinau G, Brehm M, Wiedemann U, Labudde D, Leser U, Krause G. Implications for molecular mechanisms of glycoprotein hormone receptors using a new sequence-structure-function analysis resource. Mol Endocrinol. 2007;21(2):574–580. [DOI] [PubMed] [Google Scholar]
- 30. Kleinau G, Kreuchwig A, Worth CL, Krause G. An interactive web-tool for molecular analyses links naturally occurring mutation data with three-dimensional structures of the rhodopsin-like glycoprotein hormone receptors. Hum Mutat. 2010;31(6):E1519–E1525. [DOI] [PubMed] [Google Scholar]
- 31. Kreuchwig A, Kleinau G, Kreuchwig F, Worth CL, Krause G. Research resource: Update and extension of a glycoprotein hormone receptors web application. Mol Endocrinol. 2011;25(4):707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Durme J, Horn F, Costagliola S, Vriend G, Vassart G. GRIS: glycoprotein-hormone receptor information system. Mol Endocrinol. 2006;20(9):2247–2255. [DOI] [PubMed] [Google Scholar]
- 33. Jiang X, Liu H, Chen X, et al. . Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci USA. 2012;109(31):12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krause G, Kreuchwig A, Kleinau G. Extended and structurally supported insights into extracellular hormone binding, signal transduction and organization of the thyrotropin receptor. PLoS One. 2012;7(12):e52920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lapthorn AJ, Harris DC, Littlejohn A, et al. . Crystal structure of human chorionic gonadotropin. Nature. 1994;369(6480):455–461. [DOI] [PubMed] [Google Scholar]
- 36. Bruysters M, Verhoef-Post M, Themmen AP. Asp330 and Tyr331 in the C-terminal cysteine-rich region of the luteinizing hormone receptor are key residues in hormone-induced receptor activation. J Biol Chem. 2008;283(38):25821–25828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merz WE. Biosynthesis of human chorionic gonadotropin: a review. Eur J Endocrinol. 1996;135(3):269–284. [DOI] [PubMed] [Google Scholar]
- 38. Jmol: an open-source Java viewer for chemical structures in 3D. [computer program].http://www.jmol.org/
- 39. Ho SC, Goh SS, Su Q, Khoo DH. Cysteine 390 mutation of the TSH receptor modulates its ectodomain as an inverse agonist on the serpentine domain with decrease in basal constitutive activity. Mol Cell Endocrinol. 2005;245(1–2):158–168. [DOI] [PubMed] [Google Scholar]
- 40. Costagliola S, Panneels V, Bonomi M, et al. . Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. EMBO J. 2002;21(4):504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westmuckett AD, Hoffhines AJ, Borghei A, Moore KL. Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2 deficiency. Gen Comp Endocrinol. 2008;156(1):145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gutiérrez J, Kremer L, Zaballos A, Goya I, Martínez AC, Márquez G. Analysis of post-translational CCR8 modifications and their influence on receptor activity. J Biol Chem. 2004;279(15):14726–14733. [DOI] [PubMed] [Google Scholar]
- 43. Preobrazhensky AA, Dragan S, Kawano T, et al. . Monocyte chemotactic protein-1 receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved extracellular N-terminal region. J Immunol. 2000;165(9):5295–5303. [DOI] [PubMed] [Google Scholar]
- 44. Farzan M, Mirzabekov T, Kolchinsky P, et al. . Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–676. [DOI] [PubMed] [Google Scholar]
- 45. Veldkamp CT, Seibert C, Peterson FC, et al. . Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1(37):ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Y, Hoffhines AJ, Moore KL, Leary JA. Determination of the sites of tyrosine O-sulfation in peptides and proteins. Nat Methods. 2007;4(7):583–588. [DOI] [PubMed] [Google Scholar]
- 47. Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem. 2003;278(27):24243–24246. [DOI] [PubMed] [Google Scholar]
- 48. Casarini L, Lispi M, Longobardi S, et al. . LH and hCG action on the same receptor results in quantitatively and qualitatively different intracellular signalling. PloS one. 2012;7(10):e46682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galet C, Ascoli M. The differential binding affinities of the luteinizing hormone (LH)/choriogonadotropin receptor for LH and choriogonadotropin are dictated by different extracellular domain residues. Mol Endocrinol. 2005;19(5):1263–1276. [DOI] [PubMed] [Google Scholar]