Supplemental Digital Content is Available in the Text.
Key Words: HIV, drug resistance, drug resistance mutations, non-B subtype, Africa, first-line failure
Abstract
Objective:
To determine drug resistance mutation (DRM) patterns in a large cohort of patients failing nonnucleoside reverse transcriptase inhibitor (NNRTI)-based first-line antiretroviral therapy regimens in programs without routine viral load (VL) monitoring and to examine intersubtype differences in DRMs.
Design:
Sequences from 787 adults/adolescents who failed an NNRTI-based first-line regimen in 13 clinics in Uganda, Kenya, Zimbabwe, and Malawi were analyzed. Multivariable logistic regression was used to determine the association between specific DRMs and Stanford intermediate-/high-level resistance and factors including REGA subtype, first-line antiretroviral therapy drugs, CD4, and VL at failure.
Results:
The median first-line treatment duration was 4 years (interquartile range 30–43 months); 42% of participants had VL ≥100,000 copies/mL and 63% participants had CD4 <100 cells/mm3. Viral subtype distribution was A1 (40%; Uganda and Kenya), C (31%; Zimbabwe and Malawi), and D (25%; Uganda and Kenya), and recombinant/unclassified (5%). In general, DRMs were more common in subtype-C than in subtype-A and/or subtype-D (nucleoside reverse transcriptase inhibitor mutations K65R and Q151M; NNRTI mutations E138A, V106M, Y181C, K101E, and H221Y). The presence of tenofovir resistance was similar between subtypes [P (adjusted) = 0.32], but resistance to zidovudine, abacavir, etravirine, or rilpivirine was more common in subtype-C than in subtype-D/subtype-A [P (adjusted) < 0.02].
Conclusions:
Non-B subtypes differ in DRMs at first-line failure, which impacts on residual nucleoside reverse transcriptase inhibitor and NNRTI susceptibility. In particular, higher rates of etravirine and rilpivirine resistance in subtype-C may limit their potential utility in salvage regimens.
INTRODUCTION
Large databases of HIV drug resistance mutations (DRMs), invaluable for individual patient clinical decision-making, are largely populated with data from subtype-B viruses, dominant in Europe and North America. The prevalence and pattern of DRMs differ between B and non-B subtypes. Studies comparing different non-B subtypes have typically had small numbers of participants failing first-line therapy, and thus limited power to detect subtype differences, given confounding associated with setting-specific factors, such as first-line regimens and monitoring approaches.1
More than 15 million people receive antiretroviral therapy (ART) in sub-Saharan Africa, mostly delivered using the WHO public health approach that recommends using 2 nucleoside reverse transcriptase inhibitor (NRTI) drugs combined with a nonnucleoside reverse transcriptase inhibitor (NNRTI) drug for first-line treatment and with a protease inhibitor (PI) for second-line treatment.2,3 Regular viral load (VL) monitoring is not widely available, and treatment failure is consequently detected late, at which time there is extensive resistance.3 Current WHO guidelines recommend selecting the NRTI drugs for second-line therapy using an algorithm based on first-line NRTI drug exposure.3 An understanding of subtype differences in mutational patterns and residual drug susceptibility might allow more refined NRTI selection predictions in areas where a single subtype predominates and might also clarify the role of second-generation NNRTIs in third-line therapy.
We assessed resistance in a large cohort of sub-Saharan African adults/adolescents with first-line treatment failure to determine the impact of viral subtype on mutational patterns and corresponding drug susceptibility.
METHODS
This study included participants failing NNRTI-based first-line therapy enrolled in the EARNEST trial, a large trial testing several PI-based treatment regimens for second-line therapy, and was performed at 13 sites in 4 sub-Saharan African countries (1 trial site excluded from this study, see below).4 In the 3–5 years preceding this trial, these sites were delivering ART using the public health approach with standardized first-line ART regimens (stavudine- and nevirapine-based regimens predominating in earlier years, with increasing use of tenofovir- and efavirenz-based regimens subsequently). At most sites, first-line failure was detected using clinical monitoring, sometimes supplemented with intermittent CD4 monitoring. In the year before enrollment, some sites were performing targeted VL testing in suspected treatment failures, but none had implemented regular routine VL testing.
Trial eligibility required participants to have been taking an NNRTI-based first-line regimen for >12 months and to be currently adherent (no more than 3 ART doses missed in the month before screening). Failure of first-line ART was defined by clinical, immunological, or virological criteria (modified from WHO 2010 guidelines and confirmed by VL >400 copies/mL).4 A baseline plasma sample was obtained before the switch to second-line ART, stored locally, and shipped to a central repository at the Joint Clinical Research Centre, Kampala, Uganda, within 12 months of collection.
All 37 participants recruited at 1 site in Zambia (3% of the total sample size), and 41 (3%) participants from the other 13 sites had no baseline samples stored and were excluded. Three participants not taking standard 2NRTI + NNRTI at failure (2 previously NNRTI exposed, 1 ineligible4) were also excluded. Samples from all remaining participants randomized to PI/NRTI (N = 398) and PI/raltegravir (N = 393) arms were selected, and 15 samples from those randomized to PI monotherapy who had received only tenofovir and lamivudine/emtricitabine (with NNRTI) in their first-line regimen were added to increase numbers receiving the current WHO-recommended first-line regimen (total 806 baseline samples assayed). Patient demographics and medical and treatment history (including antiretroviral drugs) were obtained from case records and patient self-report. VL and CD4 counts were performed at trial screening (<6 weeks before baseline) using standard methods at local sites.
HIV genotyping was performed using an in-house sequencing method encompassing codons 1–300 of reverse transcriptase at a WHO-designated laboratory (Joint Clinical Research Centre, Kampala, Uganda).5 In brief, RNA was extracted using the Qiagen RNA extraction kit and reverse transcribed followed by a nested PCR. The cleaned PCR product was cycle sequenced using the ABI 3730xl. Sequences were edited using the SeqScape version 2.7 and subsequently on Recall as recommended by WHO.6 Amino acid sequences were compared with consensus subtype-B, DRMs were defined using the International AIDS Society—USA list,7 and susceptibility was predicted using the Stanford algorithm version 7.0 using the full sequence data available.8 Subtype was determined by the REGA algorithm version 3.0.9 Sequences of viral isolates in this study were submitted to GenBank.
Statistical Analysis
Multivariable logistic regression was used to determine associations between specific DRMs and 2 main factors of interest: subtype and ART exposure at first-line failure (stavudine, tenofovir, zidovudine, or other NRTI for NRTI DRMs; efavirenz or nevirapine for NNRTI DRMs). As well as including these 2 factors, models adjusted for the following potential confounders: ART drug exposure before the regimen they failed on, time on first-line ART, CD4 and log10 VL at failure, and presence/absence of clinical failure. Exact logistic regression (continuous factors dichotomized at approximate midpoints) was used where logistic regression failed because of small numbers. Similar approaches were used to determine associations between Stanford intermediate-/high-level resistance to key drugs and the factors above. Participants with recombinant viruses or where no subtype could be determined were excluded from all models. Models did not adjust for country because national programs determined the specific NRTIs used in first line (Table 1); analyses assumed that any relationship between country and DRMs or susceptibility could only be realized through ART received and the other factors above.
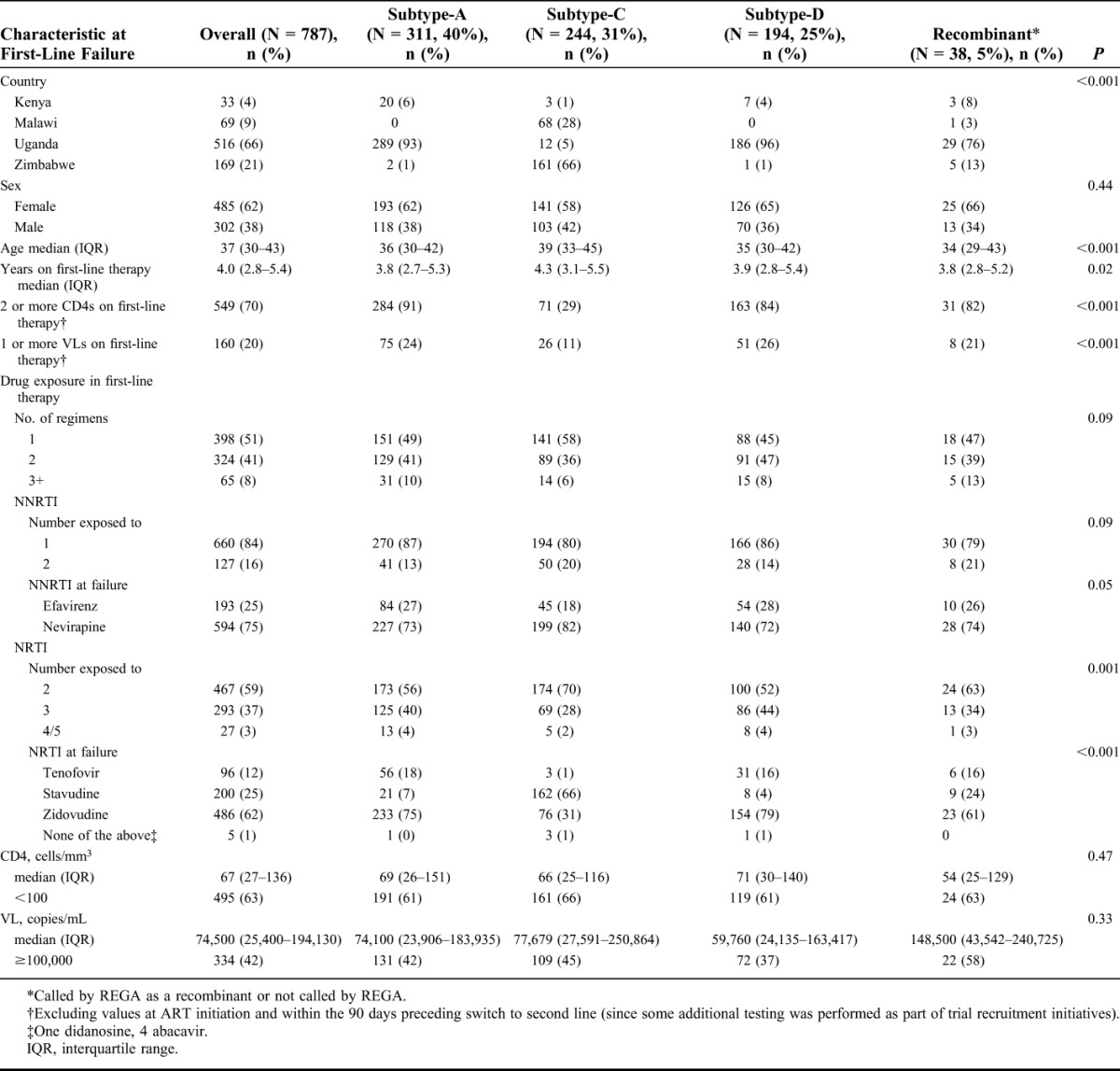
TABLE 1.
Characteristics of Study Population at First-Line Failure
All statistical tests presented are 2-sided, without adjustment for multiple comparisons. All analyses were performed in Stata version 13.1.
RESULTS
Of the 806 samples taken between 12 April 2010 and 29 April 2011 and assayed, sequences were obtained in 787 (98%) samples. Forty-two percent of the participants had VL ≥100,000 copies/mL, and 63% participants had CD4 <100 cells/mm3 (Table 1). The predominant viral subtypes were A1 (40%; Uganda/Kenya), C (31%; Zimbabwe/Malawi), and D (25%; Uganda/Kenya) with 5% recombinants/not predicted by REGA. Subtypes differed significantly in duration of first-line ART and drugs prescribed: more stavudine/nevirapine use and longer duration on first-line ART with subtype-C and more tenofovir use and substitutions in first-line NRTIs (zidovudine/stavudine to tenofovir) with subtype-A/subtype-D infection.
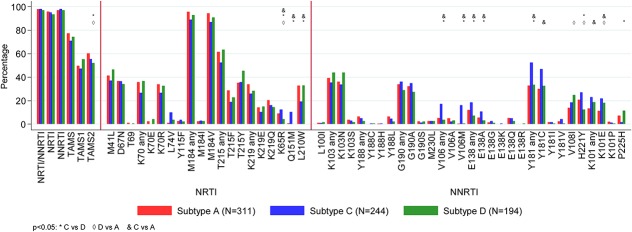
The overall prevalence of major DRMs by subtype is shown in Figure 1 and Supplemental Digital Content, Tables 1a/b, http://links.lww.com/QAI/A970. One or more major NRTI or NNRTI DRMs were present in 769 (98%) participants; 3 (0.4%) participants had only NRTI DRMs, 21 (3%) participants had only NNRTI DRMs, and 745 (95%) participants had both. Only 18 (2%) participants had no NRTI or NNRTI major DRMs.
FIGURE 1.
Prevalence of major International AIDS Society (IAS)-USA DRMs by HIV-1 subtype. Prevalence is percentage of successful sequences.
Overall DRM prevalence (after adjustment) was broadly similar between subtypes, but some significant differences were seen (global P < 0.05). For NRTI DRMs (Fig. 1, Supplemental Digital Content, Table 1a, http://links.lww.com/QAI/A970), type-2 thymidine analog DRMs were more common in A and C than in D, K65R and Q151M were more common in C than in both A and D (K65R was more common in A than in D), whereas L210W was less common in C than in both A and D. In particular, Q151M was seen in 10% of the subtype-C vs <1% of subtype-A/subtype-D. For NNRTI DRMs (Fig. 1, Supplemental Digital Content, Table 1b, http://links.lww.com/QAI/A970), (1) E138A, V106M, and Y181C were more common in C than in both A and D, (2) K101E was significantly more common in C and D than in A, (3) H221Y was more common in A and C than in D (and more common in C than in D), (4) V108I was more common in D than in A, and (5) P225H was less common in C than in D (the only DRM significantly less common in C). In particular, V106M occurred in 16% subtype-C compared with 1% A/D, whereas P225H occurred in 2% C vs 7% A and 11% D.
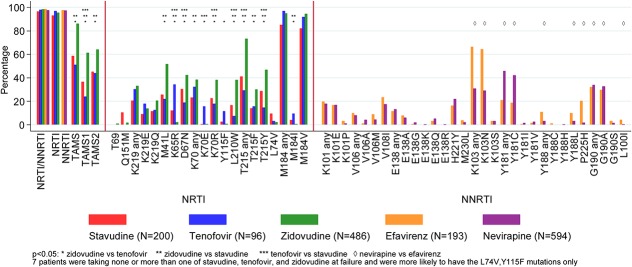
Mutation prevalence by drug exposure is shown in Figure 2 and Supplemental Digital Content, Tables 2a/b, http://links.lww.com/QAI/A970. After adjustment, participants on zidovudine at failure were more likely to have T215F, T215Y, M41L, K70R, D67N, L210W, type-1 thymidine analog, type-2 thymidine analog, and any thymidine analogue mutations (TAMs); those on tenofovir to have K65R, K70E, Y115F, and M184I; those on efavirenz to have K103N, P225H, Y188L, and L100I; and those on nevirapine to have Y181C and G190A. Intermediate-/high-level resistance to tenofovir and lamivudine was predicted in 57% and 95%, respectively (Fig. 3), and to etravirine and rilpivirine in 55% and 65%, respectively. The proportion with resistance to all 3 NRTIs (zidovudine, abacavir, and tenofovir) that might be used with lamivudine/emtricitabine in a second-line regimen was 50% (Table 2), and to all NNRTIs (including second generation), it was 55%. Much of the individual drug resistance was due to cross-resistance rather than previous direct exposure to that drug (Fig. 4).
FIGURE 2.
Prevalence of major IAS-USA DRMs by drug exposure at first-line failure. Prevalence is percentage of successful sequences.
FIGURE 3.
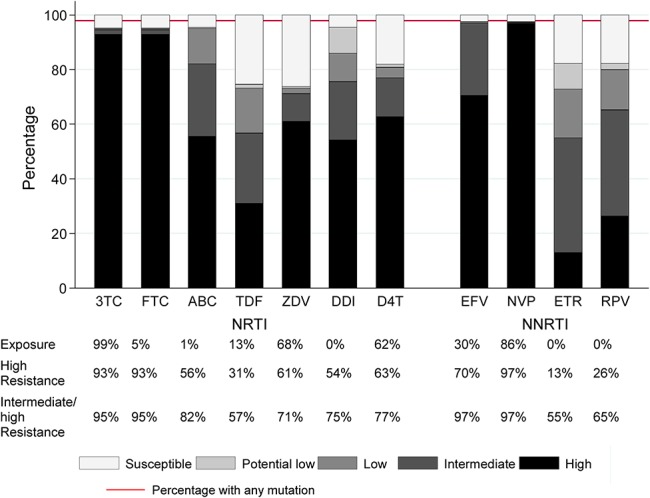
Overall resistance to NRTI and NNRTI drugs. 3TC, lamivudine; ABC, abacavir; D4T, stavudine; DDI, didanosine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NVP, nevirapine; RPV, rilpivirine; TDF, tenofovir; ZDV, zidovudine. Prevalence is percentage of successful sequences.
TABLE 2.
Resistance to Potential Future NRTI and NNRTI Drug Options
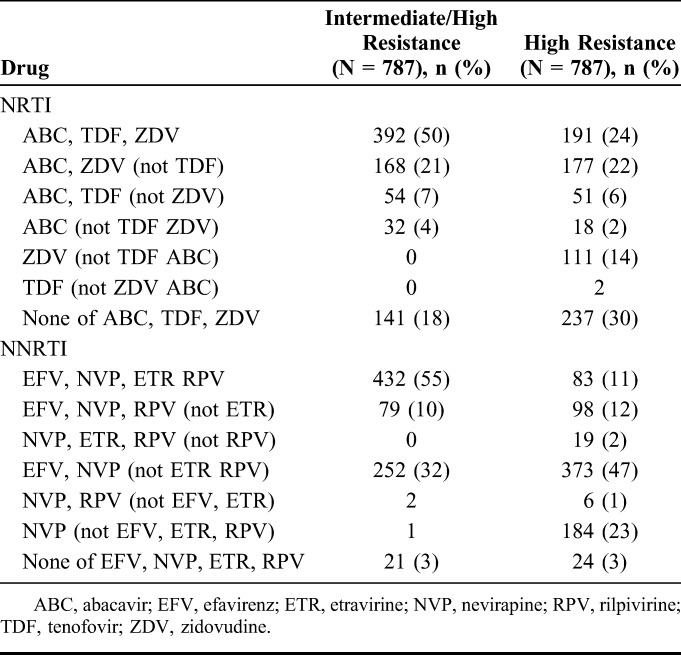
FIGURE 4.
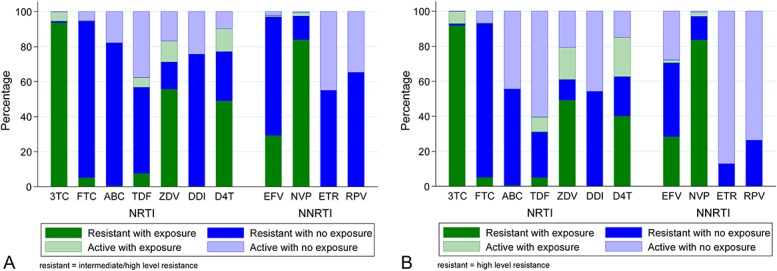
A, Intermediate-/high-level resistance according to exposure or cross-resistance. B, High-level resistance according to exposure or cross-resistance. 3TC, lamivudine; ABC, abacavir; D4T, stavudine; DDI, didanosine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NVP, nevirapine; RPV, rilpivirine; TDF, tenofovir; ZDV, zidovudine. Prevalence is percentage of successful sequences.
After adjustment, subtype-C was associated with greater abacavir and zidovudine resistance compared with subtype-A and subtype-D (P < 0.01), with prevalence of intermediate-/high-level resistance to abacavir and zidovudine of 95% and 86%, respectively, in subtype-C compared with 84% and 72%, respectively, in subtype-A, and 81% and 70%, respectively, in subtype-D. There was no significant impact of subtype on tenofovir resistance (global P = 0.32) (Table 3). Subtype-C was associated with greater etravirine and rilpivirine resistance than subtype-A and subtype-D (P < 0.003) with prevalence of intermediate-/high-level resistance to etravirine and rilpivirine of 69% and 80%, respectively, in subtype-C compared with 49% and 63%, respectively, in subtype-A, and 55% and 61%, respectively, in subtype-D (percentages adjusted to average over other model factors).
TABLE 3.
Factors Predicting Intermediate-/High-Level Resistance to Drugs for Potential Use in a Second-Line Regimen
Those on tenofovir at failure had less zidovudine resistance compared with those on zidovudine/stavudine (P < 0.01), whereas those on zidovudine appeared to have more tenofovir resistance than those on tenofovir (P = 0.06); there was no difference in abacavir resistance in those receiving zidovudine vs tenofovir (P = 0.26). Those with first-line nevirapine exposure had more etravirine and rilpivirine resistance (P < 0.01). After adjustment, higher VL at failure was strongly associated with greater tenofovir, zidovudine, and abacavir resistance (P < 0.01), and more weakly with greater etravirine resistance (P = 0.04). Lower CD4 count at failure was independently associated with higher risk of resistance to all drugs (P < 0.01).
DISCUSSION
We found DRMs conferring resistance to 1 or more NRTI/NNRTI drugs in 98% of participants failing NNRTI-based first-line ART (resistance to both classes in 95%), similar to smaller studies in Malawi,10 Nigeria,11 and the A5230 study.12 Previous studies have shown that patients monitored without VL accumulate DRMs at a rate of 1 new TAM approximately every 15 months (low-income setting) to 4.3 years (high-income setting), and 1 new NNRTI DRM every 1.6 years (high-income setting) with continued drug exposure after virological failure.13–15 The high VL/low CD4 in this cohort suggests that participants had protracted virological failure before second-line switch, consistent with the fact that they were managed in programs using immunological and clinical monitoring (without regular VL testing) to detect treatment failure. The very high level of accumulated DRMs in this cohort is therefore not surprising. Conversely, although regular VL monitoring detects failure earlier and may allow the chance for resuppression (one-third after 3 months' adherence support),16 resistance often occurs simultaneously with VL rebound; so, regular VL testing may not always prevent its development. In a research cohort taking WHO-recommended NNRTI-based first-line regimens with VL tested 1 year after starting ART, 70% of those with detectable VL had 1 or more DRMs, with 53% and 60% having 184V and NNRTI DRMs, respectively.17 Similarly, a large South African program in which VL was tested 6-monthly found 86% of patients had at least 1 DRM at second-line switch, likely reflecting delays in acting on VL results and illustrating programmatic barriers to reducing resistance development.18
This study, the largest to date of resistance in patients failing NNRTI-based first-line therapy in sub-Saharan Africa, makes a substantial contribution to the existing literature on differences in mutational patterns in non-B subtypes. In general, resistance-conferring DRMs were more common in subtype-C than in either or both of subtypes-A and subtype-D. K65R was more common in subtype-C than in A or D, supporting previous observations in subtype-C compared with subtype-AE and subtype-B in a global study of patients failing stavudine-containing regimens,19 and in comparison with non-C subtypes in an African cohort study (although this effect was not significant after adjustment for drugs).17 Differential codon usage has been hypothesized to underlie this difference and could also contribute to other variations in resistance at failure. Q151M, a rare mutation that confers cross-class NRTI resistance, was present in 11% of subtype-C, but negligible levels in other subtypes (not previously described). Subtype differences in L210W have previously been noted (higher in A than other subtypes in Nigeria).20
We found more intersubtype differences in NNRTI DRMs, with many being substantially more common in subtype-C than in 1 or both other subtypes. The (almost) exclusive occurrence of V106M in subtype-C has been noted previously.17 This mutation confers resistance to efavirenz and nevirapine and was found in 31% of those in a small cohort failing first-line therapy in South Africa.21 Another substitution at this position, V106I (not seen in our study), is significantly more common in subtype-G.20 Substitutions at position 138 (found in 13% overall) confer resistance to rilpivirine and etravirine and are therefore particularly concerning, given that these drugs may be considered for use in third-line therapy.7 A Kenyan study reported substitutions at this position in 14% of those failing nevirapine or efavirenz-based first-line therapy, mainly subtype-A22; and a study in Nigeria, mainly subtype-G and circulating recombinant form (CRF)-02_AG, found these substitution in 9%11; our finding that these DRMs are significantly more common in subtype-C (18%) than in other non-B subtypes is novel (a previous study of this mutation in several large databases showed a difference in frequency between subtype-C and subtype-B, but not between C and non-B subtypes).23 The H221Y mutation (conferring rilpivirine resistance7 and with Y181C possibly etravirine resistance24) has been noted in patients with non-B subtypes failing first-line nevirapine or efavirenz-based regimens in a predominantly subtype-G and CRF02 Nigerian cohort and subtype-C South African cohort.25,26 However, our study is the first to report differences between non-B subtypes for this mutation (more common in subtype-C). To our knowledge, the differences we found in the other NNRTI DRMs, K101E (rilpivirine and etravirine resistance), Y181C (resistance to all NNRTIs), V108I (efavirenz and nevirapine resistance), and P225H (efavirenz resistance) have not been reported previously.
Arising from these DRM differences, viral subtype significantly affected the probability of resistance to drugs that could be used in second- or third-line regimens under the public health approach. The much higher zidovudine resistance in subtype-C may be of practical importance, given that zidovudine is currently recommended for use in second-line therapy after failure of a tenofovir-based regimen. However, participants were mainly on zidovudine/stavudine first line, and the impact of subtype on zidovudine susceptibility may differ for failure on tenofovir-based first-line regimens (although we adjusted for this). Although we found no significant difference in the proportion with tenofovir resistance between subtypes, only a minority of our participants were taking tenofovir at the time of failure. An analysis of patients failing a tenofovir-based first-line regimen in Western European cohorts showed a higher rate of tenofovir resistance in subtype-C compared with non-C subtypes,27 although subtype does not appear to affect long-term outcomes on tenofovir-based regimens.28 The higher rates of etravirine and rilpivirine resistance seen in subtype-C might also be an important consideration in region-specific program policy (although overall rates are high regardless of subtype, see below). We confirmed the effects of specific first-line NRTIs on resistance to second-line NRTI drugs, which form the basis for the recommendations in the WHO algorithm. We also confirmed the independent association between first-line nevirapine use and etravirine and rilpivirine resistance.29–31 Increasing use of efavirenz over nevirapine in first-line therapy may preserve second-generation NNRTIs for potential third-line regimens, but the high rates of resistance seen even in participants failing on efavirenz (40% etravirine, 51% rilpivirine) suggests that these are unlikely to remain sufficiently active to be used in standardized regimens in the public health approach. Furthermore, rates of etravirine and rilpivirine resistance have been shown to increase when tested with deep sequencing.26 Higher VL and lower CD4 counts independently predicted resistance to all potential second-line NRTI drugs, as previously observed.12 The marginal associations between VL and etravirine and rilpivirine resistance support this arising quickly after virological failure.
Study strengths are the large sample size, the setting within representative sub-Saharan African programs using the public health approach, and well-defined first-line failure. Limitations are lack of data on CD4/VL monitoring during first-line ART and the few participants using the currently recommended tenofovir-based regimen for first line. There is potential for residual confounding by setting, since viral subtypes are strongly clustered with countries, and countries used different drug regimens (subtype-C virus predominated in Malawi and Zimbabwe, with more stavudine and less tenofovir or zidovudine use at failure; NNRTI use broadly similar). Also, the duration of virological failure on first-line ART is unknown, may be only imperfectly adjusted for by CD4 and VL at failure, and first-line monitoring approaches did differ between countries. Nevertheless, our large sample size allowed us to adjust for different regimens and patient characteristics at failure and identify some strong independent associations between subtype and specific DRMs. The levels of statistical significance seen likely indicate a true effect rather than an artifact of the multiple comparisons performed. Although we did not adjust for these multiple comparisons, this does not affect the magnitude of observed intersubtype differences but rather requires caution in the interpretation of borderline differences.
Our study provides important information on the prevalence of DRMs in patients failing NNRTI-based first-line ART in these settings, allowing a more robust comparison between viral subtypes than hitherto possible. We found substantial differences between subtypes, with particular disadvantages for subtype-C, but these are likely to have limited impact on the selection of standardized second-line regimens for use in the public health approach.
Sequences have been deposited with GenBank accession numbers KY061369–KY062155.
ACKNOWLEDGMENTS
The authors thank all the participants and staff from all the centers participating in the EARNEST trial. Members of the EARNEST Trial Team: Participating Sites: Uganda; JCRC Kampala (African trial co-ordinating centre; 231): E Agweng, P Awio, G Bakeinyaga, C Isabirye, U Kabuga, S Kasuswa, M Katuramu, C Kityo, F Kiweewa, H Kyomugisha, E Lutalo, P Mugyenyi, D Mulima, H Musana, G Musitwa, V Musiime, M Ndigendawan, H Namata, J Nkalubo, P Ocitti Labejja, P Okello, P Olal, G Pimundu, P Segonga, F Ssali, Z Tamale, D Tumukunde, W Namala, R Byaruhanga, J Kayiwa, J Tukamushaba, S Abunyang, D Eram, O Denis, R Lwalanda, L Mugarura, J Namusanje, I Nankya, E Ndashimye, E Nabulime, D Mulima, and O Senfuma; IDI, Kampala (216): G Bihabwa, E Buluma, P Easterbrook, A Elbireer, A Kambugu, D Kamya, M Katwere, R Kiggundu, C Komujuni, E Laker, E Lubwama, I Mambule, J Matovu, A Nakajubi, J Nakku, R Nalumenya, L Namuyimbwa, F Semitala, B Wandera, and J Wanyama; JCRC, Mbarara (97): H Mugerwa, A Lugemwa, E Ninsiima, T Ssenkindu, S Mwebe, L Atwine, H William, C Katemba, S Abunyang, M Acaku, P Ssebutinde, H Kitizo, J Kukundakwe, M Naluguza, K Ssegawa, Namayanja, F Nsibuka, P Tuhirirwe, and M Fortunate; JCRC Fort Portal (66): J Acen, J Achidri, A Amone, M. Chamai, J Ditai, M Kemigisa, M Kiconco, C Matama, D Mbanza, F Nambaziira, M Owor Odoi, A Rweyora, and G Tumwebaze; San Raphael of St Francis Hospital, Nsambya (48): H Kalanzi, J Katabaazi, A Kiyingi, M Mbidde, M Mugenyi, R Mwebaze, P Okong, and I Senoga; JCRC Mbale (47): M Abwola, D Baliruno, J Bwomezi, A Kasede, M Mudoola, R Namisi, F Ssennono, and S Tuhirwe; JCRC Gulu (43): G Abongomera, G Amone, J Abach, I Aciro, B Arach, P Kidega, J Omongin, E Ocung, W Odong, and A Philliam; JCRC Kabale (33): H Alima, B Ahimbisibwe, E Atuhaire, F Atukunda, G Bekusike, A Bulegyeya, D Kahatano, S Kamukama, J Kyoshabire, A Nassali, A Mbonye, T M Naturinda, Ndukukire, A Nshabohurira, H Ntawiha, A Rogers, and M Tibyasa; JCRC Kakira (31): S Kiirya, D Atwongyeire, A Nankya, C Draleku, D Nakiboneka, D Odoch, L Lakidi, R Ruganda, R Abiriga, M Mulindwa, F Balmoi, S Kafuma, and E Moriku. Zimbabwe; University of Zimbabwe Clinical Research Centre, Harare (265): J Hakim, A Reid, E Chidziva, G Musoro, C Warambwa, G Tinago, S Mutsai, M Phiri, S Mudzingwa, T Bafana, V Masore, C Moyo, R Nhema, and S Chitongo. Malawi; Department of Medicine, University of Malawi College of Medicine and the Malawi–Liverpool–Wellcome Trust Clinical Research Programme, University of Malawi College of Medicine (92): Robert Heyderman, Lucky Kabanga, Symon Kaunda, Aubrey Kudzala, Linly Lifa, Jane Mallewa, Mike Moore, Chrissie Mtali, George Musowa, Grace Mwimaniwa, Rosemary Sikwese, Joep van Oosterhout, and Milton Ziwoya; Mzuzu Central Hospital, Mzuzu (19): H Chimbaka, B Chitete, S Kamanga, T Kayinga E Makwakwa, R Mbiya, M Mlenga, T Mphande, C Mtika, G Mushani, O Ndhlovu, M Ngonga, I Nkhana, and R Nyirenda. Kenya; Moi Teaching and Referral Hospital (52): P Cheruiyot, C Kwobah, W Lokitala Ekiru, M Mokaya, A Mudogo, A Nzioka, A Siika, M Tanui, S Wachira, and K Wools-Kaloustian. Zambia; University Teaching Hospital (37): P Alipalli, E Chikatula, J Kipaila, I Kunda, S Lakhi, J Malama, W Mufwambi, L Mulenga, P Mwaba, E Mwamba, A Mweemba, and M Namfukwe. The Aids Support Organisation (TASO), Uganda: E Kerukadho, B Ngwatu, and J Birungi. MRC Clinical Trials Unit: N Paton, J Boles, A Burke, L Castle, S Ghuman, L Kendall, A Hoppe, S Tebbs, M Thomason, J Thompson, S Walker, J Whittle, H Wilkes, and N Young. Monitors: C Kapuya, F Kyomuhendo, D Kyakundi, N Mkandawire, S Mulambo, and S Senyonjo. Clinical Expert Review Committee: B Angus, A Arenas-Pinto, A Palfreeman, F Post, and D Ishola. European Collaborators: J Arribas (Hospital La Paz, Madrid, Spain), R Colebunders (Institute of Tropical Medicine, Antwerp, Belgium), M Floridia (ISS, Italy), M Giuliano (ISS, Italy), P Mallon (University College Dublin, Ireland), P Walsh (University College Dublin, Ireland), M De Rosa (CINECA, Italy), and E Rinaldi (CINECA, Italy). Trial Steering Committee: I Weller (Chair), C Gilks, J Hakim, A Kangewende, S Lakhi, E Luyirika, F Miiro, P Mwamba, P Mugyenyi, S Ojoo, N Paton, S Phiri, J van Oosterhout, A Siika, S Walker, and A Wapakabulo. Data Monitoring Committee: T Peto (Chair), N French, and J Matenga. Pharmaceutical companies: G Cloherty, J van Wyk, M Norton, S Lehrman, P Lamba, K Malik, J Rooney, W Snowden, and J Villacian.
Footnotes
The EARNEST trial was funded by the European and Developing Countries Clinical Trials Partnership (EDCTP, Grant Code: IP.2007.33011.003) with contributions from the Medical Research Council, United Kingdom; Institito de Salud Carlos III, Spain (Grant A107/90015); Irish Aid, Ireland; Swedish International Development Cooperation Agency (SIDA), Sweden; Instituto Superiore di Sanita (ISS), Italy; World Health Organisation; and Merck, USA. Substantive in-kind contributions were made by the Medical Research Council Clinical Trials Unit, United Kingdom; CINECA, Bologna, Italy; Janssen Diagnostics, Beerse, Belgium; GSK/ViiV Healthcare Ltd., United Kingdom; and Abbott Laboratories, USA. Trial medication was donated by AbbVie, Merck, Pfizer, GSK, and Gilead. The Malawi–Liverpool–Wellcome Trust Clinical Research Programme, University of Malawi College of Medicine, receives core funding from the Wellcome Trust, United Kingdom.
C.K., J.T., I.N., A.H., E.N., C.W., I.M., J.J.v.O., K.W.-K., P.J.E., P.M., A.S.W. and N.I.P. report grants from EDCTP, non-financial support from AbbVie, Janssen laboratories, Abbott Laboratories and Gilead, grants and non-financial support from GSK/ViiV and Merck, grants from WHO, non-financial support from Gilead during the conduct of the study. A.S.W. reports money paid to institution from Janssen and Gilead Sciences outside the submitted work. N.I.P. reports personal fees from AbbVie, Janssen and Roche outside the submitted work. S.B. reports no conflicts.
The authors A.S.W. and N.P. contributed equally to this article.
Members of the EARNEST Trial Team are listed in the Acknowledgments.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.Bhargava M, Cajas JM, Wainberg MA, et al. Do HIV-1 non-B subtypes differentially impact resistance mutations and clinical disease progression in treated populations? Evidence from a systematic review. J Int AIDS Soc. 2014;17:18944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilks CF, Crowley S, Ekpini R, et al. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006;368:505–510. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organisation. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. Geneva, Switzerland: World Health Organisation; 2016. [PubMed] [Google Scholar]
- 4.Paton NI, Kityo C, Hoppe A, et al. Assessment of second-line antiretroviral regimens for HIV therapy in Africa. N Engl J Med. 2014;371:234–247. [DOI] [PubMed] [Google Scholar]
- 5.Kyeyune F, Nankya I, Metha S, et al. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. AIDS. 2013;27:1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods CK, Brumme CJ, Liu TF, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wensing AM, Calvez V, Gunthard HF, et al. 2014 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 8.Stanford HIV Drug Resistance Database. 2014. Available at: http://hivdb.stanford.edu/. Accessed November 27, 2016. [Google Scholar]
- 9.de Oliveira T, Deforche K, Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. [DOI] [PubMed] [Google Scholar]
- 10.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23:1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etiebet MA, Shepherd J, Nowak RG, et al. Tenofovir-based regimens associated with less drug resistance in HIV-1-infected Nigerians failing first-line antiretroviral therapy. AIDS. 2013;27:553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallis CL, Aga E, Ribaudo H, et al. Drug susceptibility and resistance mutations after first-line failure in resource limited settings. Clin Infect Dis. 2014;59:706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigaloff KC, Ramatsebe T, Viana R, et al. Accumulation of HIV drug resistance mutations in patients failing first-line antiretroviral treatment in South Africa. AIDS Res Hum Retroviruses. 2012;28:171–175. [DOI] [PubMed] [Google Scholar]
- 14.Cozzi-Lepri A, Paredes R, Phillips AN, et al. The rate of accumulation of nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance in patients kept on a virologically failing regimen containing an NNRTI. HIV Med. 2012;13:62–72. [DOI] [PubMed] [Google Scholar]
- 15.Cozzi-Lepri A, Phillips AN, Martinez-Picado J, et al. Rate of accumulation of thymidine analogue mutations in patients continuing to receive virologically failing regimens containing zidovudine or stavudine: implications for antiretroviral therapy programs in resource-limited settings. J Infect Dis. 2009;200:687–697. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann CJ, Charalambous S, Sim J, et al. Viremia, resuppression, and time to resistance in human immunodeficiency virus (HIV) subtype C during first-line antiretroviral therapy in South Africa. Clin Infect Dis. 2009;49:1928–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamers RL, Sigaloff KC, Wensing AM, et al. Patterns of HIV-1 drug resistance after first-line antiretroviral therapy (ART) failure in 6 sub-Saharan African countries: implications for second-line ART strategies. Clin Infect Dis. 2012;54:1660–1669. [DOI] [PubMed] [Google Scholar]
- 18.Manasa J, Lessells RJ, Skingsley A, et al. High-levels of acquired drug resistance in adult patients failing first-line antiretroviral therapy in a rural HIV treatment programme in KwaZulu-Natal, South Africa. PLoS One. 2013;8:e72152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang MW, Rhee SY, Bertagnolio S, et al. Nucleoside reverse transcriptase inhibitor resistance mutations associated with first-line stavudine-containing antiretroviral therapy: programmatic implications for countries phasing out stavudine. J Infect Dis. 2013;207(suppl 2):S70–S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaplin B, Eisen G, Idoko J, et al. Impact of HIV type 1 subtype on drug resistance mutations in Nigerian patients failing first-line therapy. AIDS Res Hum Retroviruses. 2011;27:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orrell C, Walensky RP, Losina E, et al. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther. 2009;14:523–531. [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford KW, Njeru D, Maswai J, et al. Occurrence of etravirine/rilpivirine-specific resistance mutations selected by efavirenz and nevirapine in Kenyan patients with non-B HIV-1 subtypes failing antiretroviral therapy. AIDS. 2014;28:442–445. [DOI] [PubMed] [Google Scholar]
- 23.Sluis-Cremer N, Jordan MR, Huber K, et al. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antivir Res. 2014;107:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiga AI, Descamps D, Morand-Joubert L, et al. Resistance-associated mutations to etravirine (TMC-125) in antiretroviral-naive patients infected with non-B HIV-1 subtypes. Antimicrob Agents Chemother. 2010;54:728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taiwo B, Chaplin B, Penugonda S, et al. Suboptimal etravirine activity is common during failure of nevirapine-based combination antiretroviral therapy in a cohort infected with non-B subtype HIV-1. Curr HIV Res. 2010;8:194–198. [DOI] [PubMed] [Google Scholar]
- 26.Casadella M, Noguera-Julian M, Sunpath H, et al. Treatment options after virological failure of first-line tenofovir-based regimens in South Africa: an analysis by deep sequencing. AIDS. 2016;30:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TenoRes Study Group. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis. 2016;16:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White E, Smit E, Churchill D, et al. No evidence that HIV-1 subtype C infection compromises the efficacy of tenofovir-containing regimens: cohort study in the United Kingdom. J Infect Dis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anta L, Llibre JM, Poveda E, et al. Rilpivirine resistance mutations in HIV patients failing non-nucleoside reverse transcriptase inhibitor-based therapies. AIDS. 2013;27:81–85. [DOI] [PubMed] [Google Scholar]
- 30.van Zyl GU, van der Merwe L, Claassen M, et al. Antiretroviral resistance patterns and factors associated with resistance in adult patients failing NNRTI-based regimens in the Western Cape, South Africa. J Med Virol. 2011;83:1764–1769. [DOI] [PubMed] [Google Scholar]
- 31.Kiertiburanakul S, Wiboonchutikul S, Sukasem C, et al. Using of nevirapine is associated with intermediate and reduced response to etravirine among HIV-infected patients who experienced virologic failure in a resource-limited setting. J Clin Virol. 2010;47:330–334. [DOI] [PubMed] [Google Scholar]