Abstract
The original motivation of the article is to give a systematic investigation on the protocol of combining computer simulation and accurate synthesis of serial peptide protected gold clusters for potent tumor targeting therapy. Glutathione peroxidase-1 (GPx-1) is a crucial antioxidant selenoenzyme that regulates cellular redox level, thus becomes a potential target in cancer treatment. We firstly utilize molecular dynamic (MD) simulation to rationally design and screen serial peptide-Au cluster compounds with special peptide sequences and precise gold atoms, which can recognize and bind specific domain of GPx-1 with high affinity. The theoretical simulations were further verified by the following peptide-Au clusters synthesis and GPx-1 activity suppression studies in buffer and cells, respectively. Further cytological experiments corroborated that peptide-Au clusters are promising nanoparticles inducing tumor cells apoptosis by suppressing GPx-1 activity and increasing higher cellular reactive oxygen species level to initiate tumor cell apoptosis through intrinsic mitochondrial pathway.
Introduction
After two decades of studies, nanoparticles are still be described by shape, size, composition and surface coating, etc. The versatile physicochemical and biological properties of nanoparticles are dependent on these basic parameters, therefore tailoring the parameters of nanoparticles would provide potential advanced materials in catalysis, energy and biomedicine field1–3. To date, however, nanoparticles could not be described as chemical molecules with precise molecular formula, uniform compositions and exact properties. Nowadays, it is widely accepted that precise synthesis of nanoparticles with well-defined molecular structure and consistent properties is the priority to ensure their intrinsic properties and relevant applications4–6. Although great success has been achieved in past two decades, most of synthetic strategies of nanoparticles refer to “random” and “try-and-error” processes. Meanwhile, nanoparticles are normally approximately characterized by aforementioned parameters and further applied in catalysis, energy, and biomedicine, leading to the versatile rather than uniform results in literatures7, 8. For instance in nanomedicine field, unlike natural macromolecules or organic molecules, nanoparticles are not capable of precisely targeting to the therapeutic aim in cells9. Because it is difficult to synthesize and describe these nanoparticles with precise molecular formula and consistent properties, thus these nanoparticles could not strictly recognize and bind the active site of the biomolecules following ‘lock and key’ molecular interaction mode10, 11. To explore nanoparticles as efficient inhibitors of therapeutic target, it is highly urgent to rationally design and elaborately synthesize nanoparticles with strict molecular structure, uniform chemical composition and consistent properties.
Metal clusters are molecular species composed of several to a few hundreds of metal atoms, which are intermediate states of matter between isolated molecules and large nanoparticles12, 13. Peptide-protected metal clusters possess a great deal of exceptional advantages such as accurate chemical formula, nanometer dimension, high photostability, good biocompatibility and scalable production14, 15. Nevertheless, it is a great challenge to achieve peptides with rational composition, sequence and length for precise synthesis of metal clusters with desirable structure, size, charge and targeting ability. Molecular dynamics (MD) simulations have been performed to study the precise conformations of molecules and interaction mechanism between specific molecules16, 17. It could be expected that the combination of computer simulation and accurate preparation would be a highly efficient approach to fabricate peptide-conjugated metal clusters for potent targeted therapy.
Glutathione peroxidase-1 (GPx-1), a crucial antioxidant selenoenzyme in mammalian cells, is selected as a model drug target for tumor treatment18. Suppression of intracellular GPx-1 activity induces the accumulation of hydrogen peroxide (reactive oxygen species, ROS), which would render tumor cells susceptible to ROS-induced apoptosis19, 20. Recent reports have witnessed gold-contained compounds such as auranofin and gold thiomalate can inhibit GPx-1 activity through interacting with selenocystein (Sec) by forming a stable gold-Sec complex to block the active site of the protein21, 22. In light of the well defined structure of mammalian GPx-1 consisting of four identical subunits23 and Sec is surrounded by some positively charged amino acid residues. With the aid of MD simulations, we attempt to design and synthesize a class of Au clusters with well-defined molecular structure consisting of exact number of peptides and Au atoms. It is anticipated that the peptides would recognize and bind the domain around the Sec site of GPx-1, thus Au cores would be more prone to interact with active Sec to suppress the enzyme activity with high efficiency.
Herein, we firstly designed a series of negative charged peptide-Au clusters to recognize and bind the domain around the Sec of GPx-1 by MD approach. After MD optimizations, these peptide-Au clusters can be well scored via comparing their recognition and bind affinity to GPx-1 throughout salt bridges, hydrogen bonds and hydrophobic interactions between the peptides and GPx-1. Next, these peptide-cluster compounds were chemically synthesized and their suppression activities were verified by studying GPx-1 activity in buffer solution and tumor cells, respectively. At last, the optimized peptide-Au clusters inducing lung tumor cells apoptosis via GPx-1 triggered intrinsic mitochondrial pathway was disclosed in vitro.
Results
Design of peptide-Au clusters
In the molecular structure design of peptide-Au clusters, we mainly considered the peptide sequences and the definite Au cluster size by checking the surficial electrostatic potential distribution and the concave structure around the Sec of GPx-1, respectively (Figure S1). For the peptide sequence design, we tried to choose highly negatively charged peptide with E (Glu) and D (Asp) to push Au cluster to approach the Sec of GPx-1 by strong electrostatic interaction24, this because the positive electrostatic potential mainly distributed around the active sites. We also introduced V (Val) and P (Pho) hydrophobic residues around C-terminal to bind with the uncharged surficial residues distributed about 15.0 Å away from the Sec of GPx-1. Meanwhile, CCY (Cys-Cys-Tyr) was added in peptide N-terminal to mineralize Au(III) and anchored Au cluster in situ 25. The P and G (Gly) were introduced into sequence to enhance the peptide flexibility to benefit peptide-Au cluster well matching and complex with GPx-1. Considering all these factors, we proposed three sequences including P1 = CCYGGPEEEEEVG (−5e), P2 = CCYGGPDDDEDVG (−5e) and P3 = CCYGGPEEVEEVG (−4e). In these candidates, the electrostatic and hydrophobic interaction are the main screen mechanisms, i.e. P1 and P2 are designed with different charges from P3, and the hydrophobicity of P1 and P2 are regulated by the different hydrophobic side chain lengths of E and D. For the Au cluster size selection, we built a serial of peptide protected Au clusters as Au10Peptide5, Au25Peptide9 and Au40Peptide12 with different sizes according to the typical thiolate coated Au clusters26–28. After measuring the dynamic diameters of Au clusters, we found the average diameters of Au clusters are 13.7 Å, 18.5 Å and 23.0 Å for Au10Peptide5, Au25Peptide9 and Au40Peptide12, respectively (see Figure S2). Thus, we figure out Au25Peptide9 as the molecular composition for the designed peptide protected Au cluster with diameter of 18.5 Å, which could match and dock the 20.7 Å concave width of Se active site well. Because the Au/S ratio is 25:18 in the coated Au25 cluster29, there should be nine peptides with 18 thiols coating one Au25 cluster in the above peptide-Au cluster design. The molecular structures can be built into Au25P19, Au25P29 and Au25P39.
Molecular dynamic simulation of molecular interactions
Subsequently, to screen the most effective peptide-Au cluster structrure, we investigated the dynamic behaviors of the three peptide-Au cluster candidates during their molecular interactions with the active site of GPx-1. Utilizing the molecular docking method, we investigated the possible interaction sites of Au25P19, Au25P29 and Au25P39 in their representative configurations (Figure S3) and found that they all can approach to the Sec active site of GPx-1. We took the Sec active site in subunit A as the targeting site and set the same initial center of mass (COM) distance as 20.0 Å between Se atom of Sec45 (center of active site) and Au25 core in peptide-Au cluster candidates, respectively. The subunits in upper left, lower left, upper right and lower right of GPx-1 were denoted as A, B, C and D. Then, we performed MD simulations of the interaction dynamic procedures between peptide coated Au25 candidates and the active site to screen their targeting abilities. After the above screening procedures, we filtered Au25P19 as the valid targeted compound with a stable binding configuration to block the Sec active site of GPx-1 (Fig. 1a). The targeted binding abilities of candidates can be clearly compared by their COM distance to Se atom in Sec active site center during the interaction dynamics (Fig. 1b). The candidates start from the same location, but evolve into different ways during the simulations. In detail, Au25P19 (black) quickly approaches to the center of active site and stably binds to the active site with 14.5 Å COM distance. Au25P29 (red) fluctuates around the starting location in the first 10 ns, then moves near to the active site at about 16.5 Å COM distance. As for Au25P39 (blue), it diffuses away from the active site region during the first 50 ns, and locates on the protein surface around 22.0 Å COM distance to the active site. Therefore, it is clear to figure out the best targeted agent is Au25P19. To better understand the effective targeting ability of Au25P19, we took a close look at its salt bridge interaction mode in the stable binding configuration at the 126th ns (Fig. 1c). There are 10 salt bridges formed between four coating peptides (denoted as 1 to 4) of Au25P19 and GPx-1 surface, i.e. Glu408-Arg173, Glu406-Arg173 between peptide 1 and GPx-1 subunit A; Glu410-Arg18, Glu408-Lys84, C-terminal Gly412-Lys84, C-terminal Gly412-Lys110 between peptide 2 and GPx-1 subunit B; Glu406-Lys144 between peptide 3 and Gpx-1 subunit A; Glu406-Lys117 between peptide 3 and GPx-1 subunit B; Glu407-Arg178 between peptide 4 and GPx-1 subunit A; Glu410-Arg10 between peptide 4 and GPx-1 subunit C. In Fig. 1d, the average numbers of salt bridges and hydrogen bonds are 8 and 13 between Au25P19 and GPx-1 surface during the stable binding (50–130 ns), respectively. The correlation contact atoms averagely reach 153. As a result, the designed Au25P19 can stably bind to the active site of GPx-1 due to the sufficient and stable salt bridge and hydrogen bond interactions. The detail binding simulations of Au25P29 and Au25P39 candidates were analyzed in Figure S4. As the screening mechanism proposed above, we can filter Au25P19 and Au25P29 with approaching to charged active site by electrostatic interaction and further figure out Au25P19 with longer hydrophobic side chain of E interact with uncharged residues distributed around the Sec active site (see Figure S5).
Figure 1.
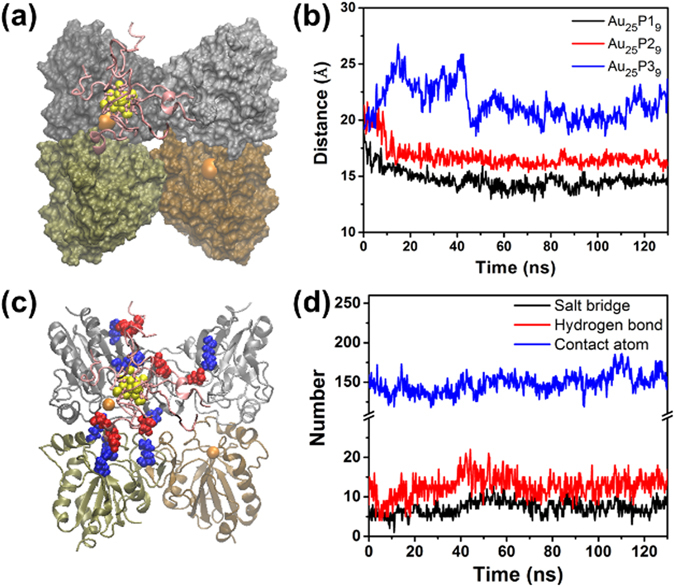
Comparison of GPx-1 binding abilities of Au25P19, Au25P29 and Au25P39 candidates. (a) The stable configuration of designed Au25P19 bound to the active site in subunit A of GPx-1 (at the 126th ns in MD simulation). (b) Distance between the Se atom of Sec45 (active site in subunit A) and the Au25 core COM of Au25P19 (black), Au25P29 (red) and Au25P39 (blue) during their binding dynamics to GPx-1. (c) Salt bridge distribution between designed Au25P19 and GPx-1 surface in the stable configuration (at the 126th ns in MD simulation). Positive and negative charged residues are in blue and red, respectively. (d) The number of salt bridges (black), hydrogen bonds (red) and contact atoms (blue) between designed Au25P19 and GPx-1 surface.
Synthesis and characterization of peptide-Au clusters
To verify the results of molecular simulations, these peptide-Au clusters were precisely synthesized by chemical method. The optical properties of these peptide-Au clusters are similar to previously reported Au25 clusters25, 30–34. As shown in Figure S7, peptide-Au clusters depict obvious peptide absorption peaks at around 275 nm and are with a gradient absorption from 350 to 600 nm ascribed to Au cluster. All the fluorescence spectra of peptide-Au clusters present a maximum excitation peak at 550 nm and a maximum emission peak at 650 nm, whose red emission can be used to track their location in tumor cells in an expedient manner (Fig. 2a, Figures S6 and S8). HRTEM images show that they are well dispersed with diameter of ~1.15 nm (Fig. 2b and Figure S9), whose size are close to Fermi wavelength thus cause these unique optical properties35. To determine the accurate molecular formula of these peptide-Au clusters, matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) was further employed. In Fig. 2c and Figure S10, the mass spectra of these peptide-Au clusters are composed of a series of peaks (black arrows) between 5.0–12.5 k m/z and the spacing between adjacent peaks is equal to one peptide with two thiol groups missing, this is attributed to the C–S bond can be easily broken during the desorption/ionization laser processing36. Since the strongest peak of these peptide-Au clusters locate at about 5501 m/z and the m/z of the main peaks match the formula Au25S18-2mPm (where P is peptide, m = 0–4), the molecular formula of synthesized peptide-Au clusters can be assigned to Au25P9. The identification of molecular formula indicates that these peptide-Au clusters we designed and optimized by MD simulations have been successfully synthesized. In the following quantitative analysis, the concentration of these peptide-Au clusters can be measured by inductively coupled plasma mass spectrometry (ICP-MS) (see calibration curve of Au standards in Figure S11).
Figure 2.
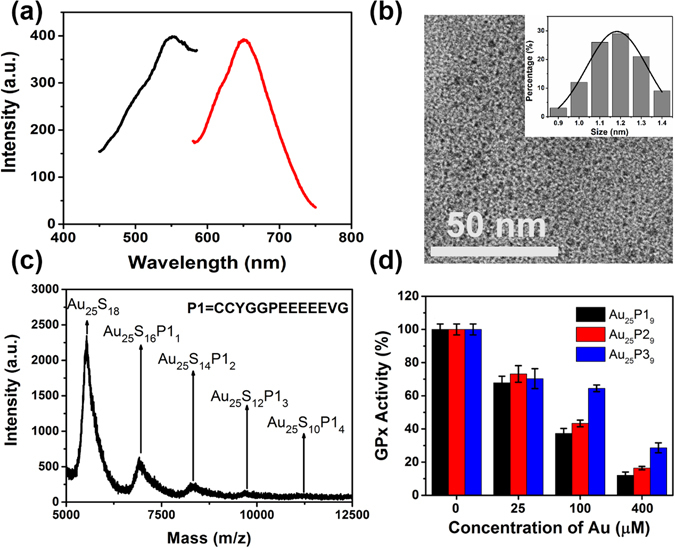
(a) Fluorescence excitation (black line) and emission (red line) spectra of Au25P19. (b) HRTEM images of Au25P19. Inset shows diameter distribution from statistic results of 100 particles. (c) MALDI-TOF MS spectra of Au25P19. (d) GPx-1 activity suppressed by peptide-Au clusters in a dose dependent manner in buffer.
GPx-1 enzyme activity suppression by peptide-Au clusters in buffer
The peptide-Au clusters suppress GPx-1 enzyme activity were studied in buffer solution. The experimental results reveal that pure GPx-1 activity is suppressed by peptide-Au clusters in dose dependent manner (Fig. 2d and Figure S12). At an Au dose of 400 μM, GPx-1 activity is 12%, 16% and 29% when treated by Au25P19, Au25P29 and Au25P39, respectively. The experiments match the MD simulations exactly where the Au25P19 exhibits the best suppression capacity for GPx-1.
GPx-1 enzyme activity suppression and ROS generation in cells
As the peptide-Au clusters could well suppressing the GPx-1 activity in buffer solution, we further check if the peptide-Au clusters could be taken into human non-small cell lung carcinoma cells (A549 cells), suppress the GPx-1 and further up-regulate the ROS level in cell. As shown in Fig. 3a,b and Figure S13, cellular uptake of Au25P19 follow a dosage dependent manner and their red emission can be observed in the cytoplasm of live A549 cells by confocal laser scanning microscopy (CLSM). These results imply that Au25P19 located in cell may interact with GPx-1 therein. To verify this assumption, we firstly detected the GPx-1 expression level in A549 cells after treated with Au25P19 for 48 h. The result of western blotting analysis show that GPx-1 level remains stable when cells treated with different doses of Au25P19 (Figure S17). Meanwhile, the cellular GPx-1 activities of these samples were measured. The cellular GPx-1 activity significantly decreases as Au25P19 dose increase in cell cytosol (Fig. 3c). In particular, cellular GPx-1 activity decreases to 27% after cells are treated with 800 μM of Au. For the control experiments, Au25P19 was added into the cell lysate containing GPx-1. As shown in Figure S14, the GPx-1 activity is also suppressed via an Au dose dependent manner.
Figure 3.
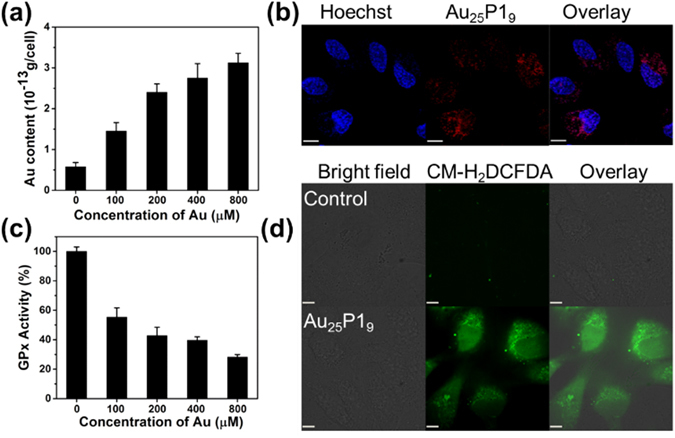
(a) Quantification of Au25P19 uptake by A549 cells (b) CLSM images of A549 cells treated with 800 µM of Au25P19 for 48 h and the cell nuclei were stained with Hoechst 33342. (c) GPx-1 activity suppressed by Au25P19 in a dose dependent manner in A549 cells. (d) CLSM images of cellular ROS level of A549 cells treated with Au25P19 for 48 h, Scale bar is ~11 µm.
Considering the suppression of cytoplasm GPx-1 activity could enhance the accumulation of ROS, we investigated intracellular ROS level in Au25P19 treated A549 cells by CLSM and flow cytometry. As shown in CLSM images (Fig. 3d), compared to control, Au25P19 treated cells stained by CM-H2DCFDA exhibit obvious green fluorescence, a sign of increased intracellular ROS. Quantitatively, when cells were treated by Au25P19 with Au dose of 800 μM, the intracellular ROS level increases to ~210% by flow cytometry analysis (Figure S15). Such higher ROS level is expected to trigger cell apoptosis.
Cell apoptosis
Generally, the significant increase of intracellular ROS level could further induce cell apoptosis via intrinsic mitochondrial pathway37, 38. In mitochondrial pathway, ROS would decline mitochondrial membrane potential and initiate a cascade process that series of biomolecules will be released from mitochondria into the cytosol to activate downstream biomolecules including caspase-3, followed by the activated caspase-3 cleaving poly(ADP-ribose) polymerase (PARP) to induce cell apoptosis39. To prove this, the change of mitochondrial membrane potential in Au25P19 treated A549 cells was firstly analyzed by live cell fluorescent dye 5, 5′, 6′, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1). As shown in Fig. 4a, a red to green emission shift can be observed in most cells after treated with Au25P19, indicating mitochondria depolarization. For further studying, A549 cells were treated with Au25P19 for 48 h at different doses, respectively. Then the cells were lysed and caspase-3 and PARP were extracted for immunoblotting studies. The results in Fig. 4b disclose that Au25P19 lead to an Au dose-dependent increase of cleaved caspases-3 and cleaved PARP levels in A549 cells. In addition, the activation of caspase-3 was also monitored and verified by the strong Nucview488 emission in cell imaging by CLSM (Figure S16). Above mentioned results indicate cell apoptosis via mitochondrial pathway happens when tumor cells are treated with Au25P19. Further, we investigated the cell apoptosis ratio by Annexin V-FITC/PI dual staining assay. The data show that the cell apoptosis continuously increases from 5.8% to 15.2% when the Au25P19 dose increases from 100 μM to 800 μM (Fig. 4c and Figure S18). In addition, Au25P19, rather than free peptides, can suppress cell viability which further supports peptide-Au clusters induced cell apoptosis (Figure S19a and S19b). We also considered the cell toxicity differences of Au25P19 in other two tumor cell types as a control. Hela and HCT-8 cell lines were treated with Au25P19 under the same condition as that of A549 cells line. The change of the cell viability of two cell lines was almost negligible (Figure S19c). This result indicates that the gold clusters were more effective to induce A549 cell apoptosis, which may result from the different susceptibility to oxidative stress40, 41. As hypothesis, Au25P19 induce A549 cells apoptosis via intrinsic mitochondrial pathway, because Au25P19 suppress GPx-1 activity and induce ROS level significant enhancement in cytosol.
Figure 4.
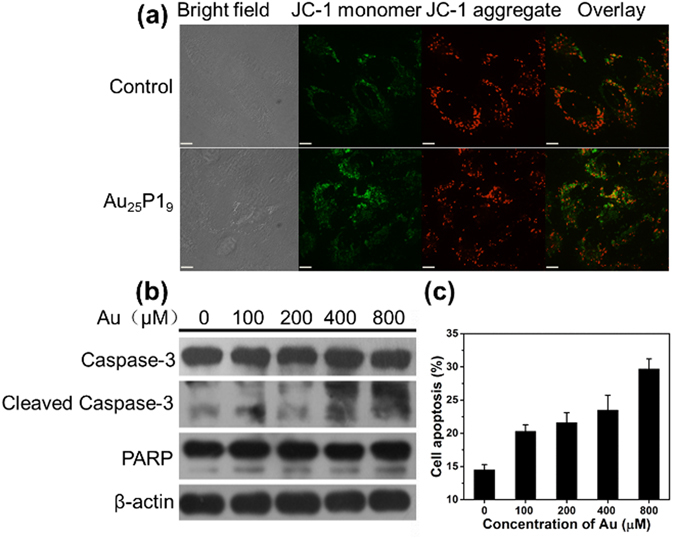
(a) CLSM images of mitochondrial membrane potential (JC-1 staining) in A549 cells pre-treated with Au25P19 at an Au dose of 800 μM for 48 h, Scale bar is ~11 μm. (b) The express level of caspase-3, PARP and β-actin extracted from A549 cells pre-treated with serial doses of Au25P19 for 48 h. (c) A549 cells apoptosis induced by Au25P19 in a dose-dependent manner.
Discussion
Computer-Aided Drug Design is a greatly important method to develop candidate drug for potent targeting therapy. In the molecular structure design of peptide-Au cluster, we mainly considered the peptide sequences and definite Au cluster size by checking the surficial electrostatic potential distribution and the concave structure around the Sec of GPx-1. Through MD simulations of molecular interactions between three peptide-Au cluster candidates and the active site of GPx-1, we filtered Au25P19 as the valid targeted compound with stable binding to the active site of GPx-1, which could be mainly attributed to their efficient electrostatic and hydrophobic interaction. In the following experimental studies, we successfully synthesized peptide-Au clusters and found Au25P19 was the most effective candidate for suppression of GPx-1 activity in buffer and cells.
In summary, under the guidance of MD stimulations, we rationally design and facilely synthesize peptide-Au clusters with strict molecular structure and consistent properties as potential anticancer drugs targeting GPx-1. As optimization by MD simulations and following experimental studies, the peptide shell in the peptide-Au clusters can recognize and bind GPx-1 with high affinity, thus the Au core with matched size could be more prone to interact with active Sec to efficiently suppress its activity. As a result, peptide-Au clusters as conceived and prepared can mediate tumor cells apoptosis via ROS induced mitochondrial pathway. The strategy paves a new route to develop other anticancer drugs targeting specific proteins following “lock and key” molecular interaction mode.
Methods
Materials
The peptides (N2H-CCYGGPEEEEEVG-COOH (named P1), NH2-CCYGGPDDDEDVG-COOH (named P2) and NH2-CCYGGPEEVEEVG-COOH (named P3)) were chemically synthesized by a solid phase method (China Peptides Co. Ltd, Purity: 98%). Hydrogen tetrachloroaurate (III) (HAuCl4·4H2O) was purchased from Sinopharm Chemical Reagent Co. Ltd. Sodium hydroxide (NaOH), nitric acid (HNO3), hydrochloric acid (HCl) and hydrogen peroxide (H2O2) were purchased from Beijing Chemical Reagent Co., China. Human non-small cell lung carcinoma cells (A549 cells), Henrietta Lacks strain of cancer cells (Hela cells) and human colon adenocarcinoma cells (HCT-8 cells) were from Cancer Institute and Hospital, Chinese Academy of Medical Sciences. Fetal bovine serum (FBS) and trypsin-EDTA were obtained from Gibco, USA. Penicillin-streptomycin solution, cell culture medium DMEM and RPMI 1640 were purchased from Hyclone. Hoechst 33342, LysoTracker®Green DND-26, and CM-H2DCFDA were from Molecular Probes, USA. NucView™ 488 Caspase-3 Assay Kit was bought from Biotium, USA. 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1), cell lysis buffer for Western and IP, phenylmethanesulfonyl fluoride (PMSF) and BCA Protein Assay Kit were obtained from Beyotime Institute of Biotechnology, China. Protease inhibitor cocktail tablets were from Roche Diagnostics, GmbH, Mannheim, Germany. Cell counting kit-8 (CCK-8) reagents and Annexin V-FITC/Propidium (PI) Apoptosis Detection Kit were purchased from Dojindo Laboratories, Japan. GPx from bovine erythrocytes from rat liver was provided by Sigma Aldrich, USA. GPx Activity Colorimetric Assay Kit was provided by Biovision, USA. Anti-GPx-1 antibody was offered by Abcam, UK. PARP antibody, caspase-3 antibody and β-actin antibody were offered by Cell Signaling Technology, USA. Amicon ultra centrifugal filter devices (Merck Millipore, MWCO: 10 kDa) were used for purification steps. Ultrapure Millipore (Mini-Q) water (18.2 MΩ) was used throughout the experiments. All glassware was thoroughly washed with aqua regia (conc. HNO3: conc. HCl, volume ratio = 1:3), rinsed with ultrapure water and ethanol, and then dried in an oven prior to use. All other materials were commercially available and used as received unless otherwise mentioned.
Molecular simulation methods
(1) Molecular dynamics (MD) simulation. The structure of GPx-1 was obtained from the Protein Data Bank (PDB ID: 1GP1)42. The relaxation system for GPx-1 tetramer contained ~120 000 atoms in 105 Å × 105 Å × 112 Å periodic simulation box. Each of the peptide-Au cluster candidates was respectively relaxed in the dimension as 81 Å × 81 Å × 90 Å containing ~60 000 atoms. We studied the binding dynamics of Au25P19, Au25P29 and Au25P39 candidates to GPx-1 in 121 Å × 121 Å × 130 Å dimension containing ~180 000 atoms, respectively. The TIP3P water was used in all systems at a neutral physiological ionic concentration as 0.15 M. After energy minimization, the binding interactions between Au25P19, Au25P29, Au25P39 candidates and GPx-1 were simulated with the 130 ns production run for each binding system. These MD calculations were performed in NPT ensemble at 1 bar and 310 K. The pressure and temperature were maintained by the Parrinello−Rahman barostat and the velocity rescaling thermostat. The AMBER99SB force field in CHARMM format was employed in this study. The parameter of Se was referenced from the previous study43. The particle-mesh Ewald (PME) method was used to calculate long-range electrostatic interactions. A typical 12 Å cutoff distance was applied to calculate van der Waals and real-space Coulomb interactions. A time step of 2 fs was utilized. All the simulations were carried out using NAMD 2.844. (2) Molecular docking simulation. We used ZDOCK package45 to study the possible interaction sites of Au25P19, Au25P29 and Au25P39 in their representative configurations (Figure S3) binding to GPx-1 surface. All the peptide-Au cluster candidates can approach to the Sec active site of GPx-1. The ZDOCK sever is at the website as http://zdock.umassmed.edu.
Synthesis of peptide-Au clusters
Peptide-Au clusters were synthesized by a previous method with a slight modification25. The peptide (2.5 mg) was dissolved in NaOH solution (16.61 mM, 1.505 mL). Subsequently, an aqueous solution of HAuCl4 (25 mM, 75 μL) was slowly added under vigorous stirring and then NaOH (0.5 M, 125 μL) was introduced for another 5 min. Finally, the mixture was stored in dark for 24 h to produce the peptide-Au clusters products. The as-synthesized products were purified through ultrafiltration tube (Millipore, MWCO: 10 KDa) to remove free ions and peptide. The purified products were stored in dark at 4 °C for further experiments.
Characterization of peptide-Au clusters
UV-Vis absorption spectra of peptide-Au clusters were recorded by a spectrophotometer (Shimadzu UV-1800, Japan). Fluorescence spectra of peptide-Au clusters were obtained by a fluorescence spectrophotometer (Shimadzu RF-5301, Japan). The size distribution of synthesized peptide-Au clusters was characterized by HRTEM. Samples were prepared by casting and evaporation a droplet of water solution on a 300-mesh holey carbon-coated copper grid (Electron Microscopy Sciences, Washington, USA). Then the high resolution images were acquired using a HRTEM (TECNAI F20, USA) with an operating voltage of 200 kV. The molecular formula of peptide-Au clusters was analyzed by MOLDI-TOF MS (UltrafleXtreme, Germany) on an ABI MALDI-TOF system in positive ion linear mode using α-cyano-4-hydroxycinnamic acid (CHCA) as the matrix.
Concentration of peptide-Au clusters analyzed by ICP-MS
Au concentration was measured by ICP-MS (Thermo Elemental X7, USA). The prepared and purified 50 µL peptide-Au clusters solutions was predigested by HNO3 and H2O2 at a volume ratio of 3:1 for 12 h and further digested by aqua regia at 160 °C. When evaporated to 0.5 mL, the sample was diluted to 5000-fold by 1% HCl and 2% HNO3. Calibration plots for standard aqueous Au were obtained by injecting a series of standard aqueous Au solutions (0.5, 1, 5, 10, 50 μg/L containing 1% HCl and 2% HNO3) into the ICP-MS system. A 20 ng/mL bismuth standard solution containing 1% HCl and 2% HNO3 was injected as an internal standard. The completely digested and diluted sample was injected to measure the Au content. The experiment was carried out in triplicate. The Au concentration could be accurately quantified by calibration curve.
Activity assay of GPx-1 in buffer solution
Pure GPx (lyophilized powder, ≥300 units/mg protein) were diluted by GPx sample buffer and incubated with peptide-Au clusters (Au25P19, Au25P29 and Au25P39) at a series of Au dose (0, 25, 100, 400 μM) for 10 min, respectively. Then the GPx-1 activity was assayed spectrophotometrically by GPx Activity Colorimetric Assay Kit following the manufacturer’s instructions. The GPx activity of untreated samples was set as 100%, and that of peptide-Au clusters treated samples were expressed as a relative percentage of untreated ones.
Au25P19 uptake determined by ICP-MS
The cells were cultured in medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in a culture flask (25 cm2), reseeded every 2–4 days to maintain subconfluency and kept in humidified atmosphere of 95% air and 5% CO2 at 37 °C. After cells reaching 80% confluence during the exponential growth in culture flask, the cells were harvested and subcultured on 6-well plate (at the density of 1 × 105 cells/well) for 24 h. Then the medium was discarded and fresh culture medium with Au25P19 at different doses was added to make cells exposed to final Au doses at 0, 100, 200, 400 and 800 μM, respectively. After another 48 h incubation, the cells were treated with trypsin and collected, and then washed thrice with PBS. 5 × 105 cells of each sample counted by flow cytometry were predigested by HNO3 and H2O2 at a volume ratio of 3:1 for 12 h and further digested by aqua regia at 160 °C, and then diluted to 10 mL aqueous solution with 1% HCl and 2% HNO3. Finally, the amount of Au25P19 taken into cells was measured by ICP-MS with Au calibration curve.
Cellular location of Au25P19 observed by CLSM
A549 cells were seeded on glass-bottom dishes (35 mm, MatTek Corporation) for confocal laser scanning microscopy (CLSM) observation. After 24 h, Au25P19 was added to culture medium and the final Au dose was 800 µM. The A549 cells were next exposed to Au25P19 for 48 h. Then the cells were washed three times with PBS and incubated with 10 µg/mL Hoechst 33342 at 37 °C under darkness for 10 min. Before observation, the culture medium was discarded and the cells were washed three times with PBS, supplemented with fresh culture medium. Finally, in A549 cells, the cell nucleus and Au25P19 were imaged under Nikon Ti-e microscope with excitation at wavelengths of 405 nm and 560 nm, respectively. To identify the accurate location of Au25P19 in cytoplasm of A549 cells, lysotracker green, a lysosome probe, was added in the medium after cells were incubated with Au25P19 for 48 h. The cells were then observed by CLSM with excitation at wavelengths of 488 nm and 560 nm, respectively.
Activity assay of GPx-1 in A549 cells and cells lysate
A549 cells were prior seeded on 6-well plate (at the density of 1 × 105 cells/well) for 24 h. Then the medium was discarded and fresh culture medium with Au25P19 at different doses was added to make cells exposed to final Au doses at 0, 100, 200, 400 and 800 μM, respectively. After another 48 h incubation, the cells were washed thrice with PBS and lysed by cell lysis buffer containing 1 mM PMSF for 15 min on a shaker at 4 °C. The lysed cells were collected and centrifuged for 20 min at 12,000 g to get total protein of cells. The protein dose of each sample was quantified by BCA Protein Assay Kit and the GPx-1 activity of peptide-Au clusters treated A549 cells was measured by GPx Activity Colorimetric Assay Kit, respectively. For the control experiments, a series of Au doses of Au25P19 at 0, 5, 25, 100, 400 μM were incubated with the GPx-1 containing cell lysate of A549 cells for 10 min and the GPx-1 activity of theses samples was also measured. The GPx-1 activity of untreated samples was set as 100%, and that of peptide-Au clusters treated samples were expressed as a relative percentage of untreated ones.
Intracellular ROS level analyzed by CLSM and flow cytometry
For CLSM observation, cells were prior cultured on glass-bottom dishes. Au25P19 was added to culture medium and the final Au dose was 800 µM. After 48 h. the cells were washed three times with PBS and incubated with 5 µM CM-H2DCFDA, at 37 °C under darkness for 30 min. Before CLSM observation, the culture medium was discarded and the cells were washed three times with PBS, supplemented with fresh culture medium. Finally, ROS in cells were highlighted by green fluorescence emission of CM-H2DCFDA, imaged under Nikon Ti-e microscope with excitation at wavelengths of 488 nm. For flow cytometry analysis, cells were prior seeded on 6-well plate (at the density of 1 × 105 cells/well) for 24 h. Then the medium was discarded and fresh culture medium containing Au25P19 with Au dose at 800 μM was added. For another 48 h incubation, the cells were washed thrice with PBS, and incubated with 5 µM CM-H2DCFDA at 37 °C under darkness for 30 min. After washing with PBS, the cells were collected and suspended in ice cold PBS. Fluorescence intensity of 20,000 events was recorded using an Accuri C6 flow cytometer and the level of intracellular ROS was analyzed by Cflow software.
Mitochondrial membrane potential (ΔΨm) assay
Disruption of ΔΨm is an important step in the induction of cellular apoptosis. JC-1, a sensitive mitochondrial dye, was used to evaluate the ΔΨm. JC-1 can aggregates in the mitochondrial matrix to yield red fluorescence (λem = 590 nm) at high mitochondrial membrane potential, while JC-1 monomer has green fluorescence at low mitochondrial membrane potential (λem = 527 nm). Therefore, using JC-1 easily signals the loss of ΔΨm, an event of early cell apoptosis. After treated with 800 µM of Au25P19 for 48 h on glass-bottom dish, cells were stained with JC-1 at 37 °C for 20 min under darkness. Then, cells were washed twice with cold buffer solution and JC-1 fluorescence inside cells was immediately observed by CLSM. JC-1 monomers and JC-1 aggregates were observed with excitation at wavelengths of 488 nm.
Caspase-3 activity assay
A549 cells were seeded on glass-bottom dishes for 24 h, and the Au25P19 was added to culture medium and final Au dose was 800 µM. A549 cells were next exposed to Au25P19 for 48 h. Then the cells were washed three times with PBS and incubated with 5 µM Nucview488 caspase-3 substrate (which can be cleaved by caspase-3 and release the DNA dye to stain DNA with bright green fluorescence during apoptosis) at room temperature for 30 min. Finally, cells were washed with PBS and observed by CLSM with excitation at wavelengths of 488 nm.
Expression of GPx-1, caspase-3 and PARP analyzed by western blotting
The expression of GPx-1, caspase-3 and PARP from Au25P19 treated A549 cells were analyzed by western blotting. A549 cells were prior seeded on 6-well plate (at the density of 1 × 105 cells/well) for 24 h. Then the medium was discarded and fresh culture medium with Au25P19 at different doses was added to make cells exposed to final Au doses at 0, 100, 200, 400 and 800 μM, respectively. After another 48 h incubation, the cells were harvested and lysed by cell lysis buffer for Western and IP (containing 20 mM Tris-HCl, pH7.5, 150 mM NaCl, 1% Triton X-100, sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4 and leupeptin), protease inhibitor cocktail tablet and 1 mM PMSF for 15 min on a shaker at 4 °C. The lysed cells were collected and centrifuged for 15 min at 12,000 g to get total protein of cells. The protein dose of each sample was prior quantified, and then loading buffer was added. After boiling for 5 min, the protein samples with equal amount were separated on 12% SDS polyacrylamide gel electrophoresis and then transferred onto the polyvinylidene difluoride (PVDF) membrane. Membrane was blocked by 5% nonfat dried milk prepared in tris(hydroxymethyl)amin omethane-NaCl-Tween 20 (TBST) at room temperature for 1 h, and then incubated with primary antibodies specific to β-actin, Caspase-3, PARP and GPx-1 at 4 °C overnight, respectively. After that, horseradish peroxidase-conjugated secondary antibody was added at room temperature for 1 h, and then the protein bands from antibody-antigen reactions were visualized using Amersham ECL TM Prime Western Blotting Detection Reagent (GE healthcare, UK).
Apoptosis evaluation
Apoptosis detection with Annexin V-FITC/PI fluorescence dual staining was carried out by flow cytometry. Briefly, about 2 mL of 1 × 105 cells/mL of A549 cells were seeded in 6-well plate for 24 h before treatment with Au25P19 at different Au doses. The cells were treated with Au25P19 (Au dose at 0, 100, 200, 400 and 800 μM) for 48 h, respectively. Then culture medium and cells in each well were collected and centrifuged at 1200 rpm for 3 min. The supernatant was discarded and the cells were washed twice with PBS. 195 μL Annexin V-FITC binding buffer was added to re-suspend the cells at a dose of ~5 × 105 cells/mL in 1.5 mL Eppendorf tube. 5 μL Annexin V-FITC and 10 μL PI stains were added into each cell suspension, respectively. The tubes were incubated at room temperature under darkness for exactly 15 min. The fluorescence of the cells was determined immediately with flow cytometer. Cells at the early stage of apoptotic process were stained with the Annexin V-FITC alone. Necrotic cells were stained by both the Annexin V-FITC and PI. Live cells showed no staining by either Annexin V-FITC or PI. So, the percentage of apoptotic and necrosis cells can be analyzed by Cflow software.
Cell viability assay
Briefly, cells were prior cultured on 96-well plates (at the density of 5 × 103 cells/well) for 24 h to allow cells attachment. The cells were treated with different doses of these free peptides (0, 1, 4, 16, 64, 256, 1024 µM) and peptide-Au clusters (0, 50, 100, 200, 400, 800 µM in Au dose) for 48 h, respectively. The cells were washed three times with PBS and then incubated with fresh growth medium containing 10% (v/v) CCK-8 reagent for 1 h at 37 °C for cell viability assay. The absorbance was measured at 450 nm by using a microplate reader (SpectraMAX M2, Sunnyvale, California). Cell viability of untreated cells was set as 100%, and that of treated cells were expressed as a relative percentage of untreated cells. All data were shown as mean percentages ± standard deviation from three independent experiments.
Electronic supplementary material
Acknowledgements
We acknowledge financial support by National Key Basic Research Program of China (2013CB932703 and 2013CB933704) and National Natural Science Foundation of China (31500815, 21425522, 21390414, 11404333 and 31571026). We also acknowledge the Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (the second phase).
Author Contributions
L. Zhao, P. Zhang, Y. Zhao, and X. Gao help to finish the theoretical simulations research. M. Liu, L. Gao, J. He, Q. Yuan, and X. Gao help to finish the experimental studies. What is more, the data processing and manuscript writing are finished by M. Liu, L. Zhao, L. Gao and X. Gao. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00278-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang Gao, Email: gaoliang@ihep.ac.cn.
Lina Zhao, Email: linazhao@ihep.ac.cn.
Xueyun Gao, Email: gaoxy@ihep.ac.cn.
References
- 1.Xie X, Li Y, Liu Z, Haruta M, Shen W. Low-temperature oxidation of CO catalysed by Co3O4 nanorods. Nature. 2009;458:746–9. doi: 10.1038/nature07877. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Shen W. Morphology-dependent nanocatalysts: rod-shaped oxides. Chem. Soc. Rev. 2014;43:1543–74. doi: 10.1039/C3CS60296F. [DOI] [PubMed] [Google Scholar]
- 3.Parak WJ. Controlled interaction of nanoparticles with cells. Science. 2016;351:814–5. doi: 10.1126/science.aaf0751. [DOI] [PubMed] [Google Scholar]
- 4.Xia Y, Xiong Y, Lim B, Skrabalak SE. Shape-controlled synthesis of metal nanocrystals: simple chemistry meets complex physics? Angew. Chem., Int. Ed. 2009;48:60–103. doi: 10.1002/anie.200802248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling D, Lee N, Hyeon T. Chemical synthesis and assembly of uniformly sized iron oxide nanoparticles for medical applications. Acc. Chem. Res. 2015;48:1276–85. doi: 10.1021/acs.accounts.5b00038. [DOI] [PubMed] [Google Scholar]
- 6.Smith AM, Nie S. Next-generation quantum dots. Nat. Biotechnol. 2009;27:732–3. doi: 10.1038/nbt0809-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami T, et al. Photodynamic and photothermal effects of semiconducting and metallic-enriched single-walled carbon nanotubes. J. Am. Chem. Soc. 2012;134:17862–65. doi: 10.1021/ja3079972. [DOI] [PubMed] [Google Scholar]
- 8.He X, et al. Quantifying the biodistribution of nanoparticles. Nat. Nanotechnol. 2011;6:755. doi: 10.1038/nnano.2011.219. [DOI] [PubMed] [Google Scholar]
- 9.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug. Discov. 2010;9:615–27. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 10.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–7. doi: 10.1016/S1748-0132(08)70014-8. [DOI] [Google Scholar]
- 11.Mahmoudi M, et al. Protein-nanoparticle interactions: opportunities and challenges. Chem. Rev. 2011;111:5610–37. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 12.Wilcoxon JP, Abrams BL. Synthesis, structure and properties of metal nanoclusters. Chem. Soc. Rev. 2006;35:1162–94. doi: 10.1039/b517312b. [DOI] [PubMed] [Google Scholar]
- 13.Qian h, Zhu M, Wu Z, Jin R. Quantum sized gold nanoclusters with atomic precision. Acc. Chem. Res. 2012;45:1470–79. doi: 10.1021/ar200331z. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Q, et al. Peptide protected gold clusters: chemical synthesis and biomedical applications. Nanoscale. 2016;8:12095–104. doi: 10.1039/C6NR02750D. [DOI] [PubMed] [Google Scholar]
- 15.Goswami N, Zheng K, Xie J. Bio-NCs-the marriage of ultrasmall metal nanoclusters with biomolecules. Nanoscale. 2014;6:13328–47. doi: 10.1039/C4NR04561K. [DOI] [PubMed] [Google Scholar]
- 16.De La Rica R, et al. A designer peptide as a template for growing Au nanoclusters. Chem. Commun. 2014;50:10648–50. doi: 10.1039/C4CC03240C. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, et al. Computational design of peptide-Au cluster probe for sensitive detection of alpha(IIb)beta3 integrin. Nanoscale. 2016;8:4203–8. doi: 10.1039/C5NR09175F. [DOI] [PubMed] [Google Scholar]
- 18.Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox. Signal. 2011;15:1957–97. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dokic I, Hartmann C, Herold-Mende C, Régnier-Vigouroux A. Glutathione peroxidase 1 activity dictates the sensitivity of glioblastoma cells to oxidative stress. Glia. 2012;60:1785–1800. doi: 10.1002/glia.22397. [DOI] [PubMed] [Google Scholar]
- 20.Gouaze V, et al. Glutathione peroxidase-1 protects from CD95-induced apoptosis. J. Biol. Chem. 2002;277:42867–74. doi: 10.1074/jbc.M203067200. [DOI] [PubMed] [Google Scholar]
- 21.Roberts JR, Frank Shaw C. Inhibition of Erythrocyte Selenium-Glutathione Peroxidase by Auranofin Analogues and Metabolites. Biochem. Pharmacol. 1998;55:1291–99. doi: 10.1016/S0006-2952(97)00634-5. [DOI] [PubMed] [Google Scholar]
- 22.Bhabak KP, Mugesh G. A synthetic model for the inhibition of glutathione peroxidase by antiarthritic gold compounds. Inorg. Chem. 2009;48:2449–55. doi: 10.1021/ic8019183. [DOI] [PubMed] [Google Scholar]
- 23.Günzler WA, et al. The amino-acid sequence of bovine glutathione peroxidase. Physiol. Chem. 1984;365:195–212. doi: 10.1515/bchm2.1984.365.1.195. [DOI] [PubMed] [Google Scholar]
- 24.An D, et al. A peptide-coated gold nanocluster exhibits unique behavior in protein activity inhibition. J. Am. Chem. Soc. 2015;137:8412–8. doi: 10.1021/jacs.5b00888. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Bifunctional peptides that precisely biomineralize Au clusters and specifically stain cell nuclei. Chem. Commun. 2012;48:871–3. doi: 10.1039/C1CC15926G. [DOI] [PubMed] [Google Scholar]
- 26.Shao N, Pei Y, Gao Y, Zeng X. Onset of double helical structure in small-sized homoleptic gold thiolate clusters. J. Phys. Chem. A. 2009;113:629–32. doi: 10.1021/jp810447k. [DOI] [PubMed] [Google Scholar]
- 27.Zhu M, Aikens CM, Hollander FJ, Schatz GC, Jin R. Correlating the crystal structure of a thiol-protected Au25 cluster and optical properties. J. Am. Chem. Soc. 2008;130:5883–5. doi: 10.1021/ja801173r. [DOI] [PubMed] [Google Scholar]
- 28.Malola S, et al. Au40(SR)24 cluster as a chiral dimer of 8-electron superatoms: structure and optical properties. J. Am. Chem. Soc. 2012;134:19560–3. doi: 10.1021/ja309619n. [DOI] [PubMed] [Google Scholar]
- 29.Zhu M, Aikens CM, Hollander FJ, Schatz GC, Jin R. Correlating the crystal structure of a thiol-protected Au25. J. Am. Chem. Soc. 2008;130:5883. doi: 10.1021/ja801173r. [DOI] [PubMed] [Google Scholar]
- 30.Liu R, et al. The Au clusters induce tumor cell apoptosis via specifically targeting thioredoxin reductase 1 (TrxR1) and suppressing its activity. Chem. Commun. 2014;50:10687–90. doi: 10.1039/C4CC03320E. [DOI] [PubMed] [Google Scholar]
- 31.Yuan X, et al. Balancing the rate of cluster growth and etching for gram-scale synthesis of thiolate-protected Au(25) nanoclusters with atomic precision. Angew. Chem., Int. Ed. 2014;53:4623–7. doi: 10.1002/anie.201311177. [DOI] [PubMed] [Google Scholar]
- 32.Zhu M, Lanni E, Garg N, Bier ME, Jin R. Kinetically controlled, high-yield synthesis of Au-25 clusters. J. Am. Chem. Soc. 2008;130:1138–9. doi: 10.1021/ja0782448. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Gayathri C, Gil RR, Jin R. Probing the Structure and Charge State of Glutathione-Capped Au-25(SG)(18) Clusters by NMR and Mass Spectrometry. J. Am. Chem. Soc. 2009;131:6535–42. doi: 10.1021/ja900386s. [DOI] [PubMed] [Google Scholar]
- 34.Xie J, Zheng Y, Ying JY. Protein directed synthesis of highly fluorescent gold nanoclusters. J. Am. Chem. Soc. 2009;131:888–9. doi: 10.1021/ja806804u. [DOI] [PubMed] [Google Scholar]
- 35.Zheng J, Nicovich PR, Dickson RM. Highly fluorescent noble-metal quantum dots. Annu. Rev. Phys. Chem. 2007;58:409–31. doi: 10.1146/annurev.physchem.58.032806.104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tracy JB, et al. Electrospray ionization mass spectrometry of uniform and mixed monolayer nanoparticles: Au25[S(CH2)2Ph]18 and Au25[S(CH2)2Ph]18-x(SR)x. J. Am. Chem. Soc. 2007;129:16209–15. doi: 10.1021/ja076621a. [DOI] [PubMed] [Google Scholar]
- 37.Yu H, et al. Chamaejasmine induces apoptosis in human lung adenocarcinoma A549 cells through a Ros-mediated mitochondrial pathway. Molecules. 2011;16:8165–80. doi: 10.3390/molecules16108165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W, et al. Hispolon induces apoptosis in human gastric cancer cells through a ROS-mediated mitochondrial pathway. Free. Radic. Biol. Med. 2008;45:60–72. doi: 10.1016/j.freeradbiomed.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer. Biol. Ther. 2014;4:147–171. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 40.Kang YH, et al. Role of reactive oxygen species in the induction of apoptosis by alpha-tocopheryl succinate. Int. J. Cancer. 2004;112:385–92. doi: 10.1002/ijc.20424. [DOI] [PubMed] [Google Scholar]
- 41.Raj L, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–4. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Epp, O., Ladenstein, R. & Wendel, A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2‐nm resolution. Eur. J. Biochem. 133 (1983). [DOI] [PubMed]
- 43.Zhao l, et al. Molecular modeling and in vitro activity of an HIV-1-encoded glutathione peroxidase. P. Natl. Acad. Sci. USA. 2000;97:6356–61. doi: 10.1073/pnas.97.12.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierce BG, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–3. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


