Abstract
Cutaneous squamous cell carcinoma (cSCC) is the second most common cancer among Caucasians in the United States, with rising incidence over the past decade. Treatment for non-melanoma skin cancer, including cSCC, in the United States was estimated to cost $4.8 billion in 2014. Thus, an understanding of cSCC pathogenesis could have important public health implications. Immune function impacts cSCC risk, given that cSCC incidence rates are substantially higher in patients with compromised immune systems. We report a systematic review of published associations between cSCC risk and the human leukocyte antigen (HLA) system. This review includes studies that analyze germline class I and class II HLA allelic variation as well as HLA cell-surface protein expression levels associated with cSCC risk. We propose biological mechanisms for these HLA-cSCC associations based on known mechanisms of HLA involvement in other diseases. The review suggests that immunity regulates the development of cSCC and that HLA-cSCC associations differ between immunocompetent and immunosuppressed patients. This difference may reflect the presence of viral co-factors that affect tumorigenesis in immunosuppressed patients. Finally, we highlight limitations in the literature on HLA-cSCC associations, and suggest directions for future research aimed at understanding, preventing and treating cSCC.
Keywords: HLA, immunosuppression, cutaneous SCC, HPV
1. INTRODUCTION
cSCC is the second most common cancer in the United States [1], particularly affecting Caucasians. cSCCs present as an uncontrolled growth of abnormal keratinocytes, mostly arising on sun-exposed anatomic sites. If left untreated, cSCCs can penetrate underlying tissues and metastasize. cSCC is a major public health concern due to its high incidence and associated medical costs [2]. Risk factors for cSCC can be classified as genetic (family history, pigmentation) and environmental (ultraviolet radiation (UVR), human papillomavirus (HPV) infection, immunosuppression, cigarette smoking) [3].
The immune system impacts cSCC susceptibility and pathogenesis, as evidenced by the substantially higher incidence of cSCC in immunocompromised patients (e.g. solid organ transplant recipients who undergo iatrogenic long-term immunosuppressive therapy and patients infected with human immunodeficiency virus (HIV)) [3,4]. Furthermore, susceptibility to the effects of UVR is known to be genetically determined [3–5]. Variations in immunological makeup of human hosts may influence their ability to recruit immune responses needed to prevent cSCC development [6].
The human leukocyte antigen (HLA) system comprises genes that encode the major histocompatibility complex (MHC) proteins. Variation in the expression pattern of these proteins, which are involved in the presentation of tumor antigens to T lymphocytes, has been implicated in multiple cancers by influencing host defenses against tumorigenesis [6]. Class I HLA genes (A, B, and C) encode proteins expressed on the surface of all nucleated cells, which present intracellular peptides to CD8+ T lymphocytes. Class II HLA genes (DR, DQ, DP, DM, DOA, and DOB) encode proteins expressed only on the surface of antigen-presenting cells, which serve as important restriction elements for the induction and proliferation of CD4+ T lymphocytes.
This paper provides a systematic review of the associations between the HLA system and cSCC risk. It suggests mechanistic explanations for these associations based on known mechanisms of HLA involvement in other diseases, as well as further directions for research. Since the literature on HLA and cSCC risk is restricted to class I and class II HLA genes, we focus on these two classes. Both classes contain highly polymorphic genes, which greatly increases the number of possible interactions with antigenic peptides such as tumor antigens. This allows for effective immune surveillance, as tumor antigens presented by class I and class II HLA proteins activate CD8+ and CD4+ T lymphocytes for antitumor immunity. However, cancer cells are able to evade this normal surveillance, leading to unrestricted growth.
We review studies that address associations between cSCC risk and three features of the HLA system: (1) germline HLA allele polymorphisms in immunocompetent and immunosuppressed patients; (2) HLA mismatching and homozygosity in organ transplant patients undergoing immunosuppression; and (3) cell-surface expression levels of HLA proteins in cSCC tumor lesions versus in healthy skin of both immunocompetent and immunosuppressed cSCC patients. Evaluating such associations provides an understanding of the immunogenetic risk factors and immune mechanisms involved in cSCC pathogenesis, which could illuminate novel approaches to the prevention and treatment of these cutaneous neoplasms [2,3].
2. MATERIAL AND METHODS
2.1 Selection Criteria
Study selection criteria are illustrated in Figure 1. We included observational association studies between the HLA system and cSCC risk in immunocompetent and immunosuppressed patients. Inclusion criteria were: (1) publications between January 1, 1980 and June 30, 2016; (2) study population of Caucasian patients only (3) histologic confirmation of at least one cSCC. In addition, studies of immunosuppressed patients were limited to those analyzing cSCCs that developed after immune suppression, to isolate the effects of immunosuppression on HLA-cSCC associations.
Fig 1.
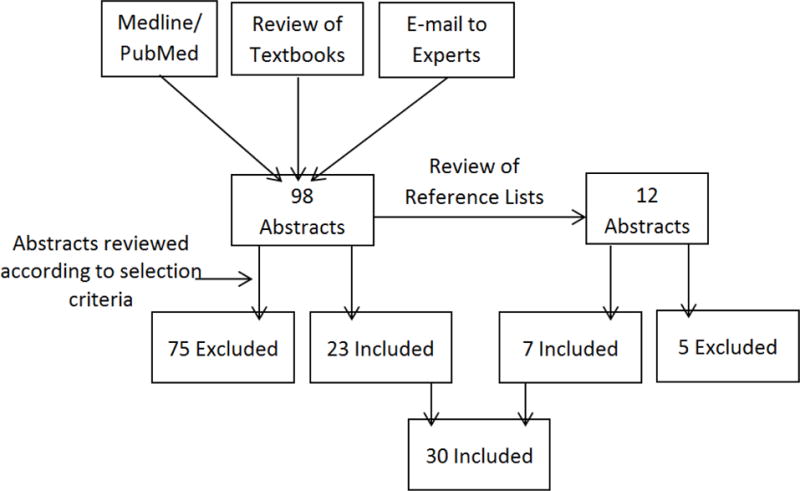
Diagram of Study Selection
2.2 Data Sources
Studies that met the selection criteria were identified using the following four methods:
(1) Computer Search
Medline/PubMed were queried for articles in English using one or more of the following Medical Subject Headings (MeSH) terms: “carcinoma, squamous cell,” “Bowen’s disease,” “skin neoplasms,” “major histocompatibility complex,” “histocompatibility antigens,” “HLA antigens,” “genome-wide association study,” and “cutaneous.”
(2) Review of Specialized Textbooks
Dermatology textbooks [7–9] were identified using the Elsevier Dermatology Books database and were selected for further review if they contained information on cSCC [7–9].
(3) Contact with Experts
Experts were selected by querying the NIH RePORTER database for investigators funded to study cSCC or HLA, and were contacted for knowledge of unpublished or ongoing studies.
(4) Review of Reference Lists
The reference lists of all articles identified were reviewed, and full texts of potentially eligible studies were examined.
3. RESULTS
In the following section, we summarize published HLA-cSCC associations in immunocompetent and immunosuppressed individuals and then propose putative biological mechanisms for the observed data. HLA-cSCC association studies identified through systematic review were organized into three categories: (1) studies associating cSCC risk with particular germline HLA allelic polymorphisms by comparing allelic frequencies between cSCC patients and healthy controls, (2) studies linking cSCC risk with HLA mismatching and homozygosity in immunosuppressed patients, and (3) studies analyzing cell-surface HLA protein expression levels in cSCC tumors versus normal skin from cSCC patients. In the studies reviewed, an alpha value of 0.05 was used as the threshold for significance.
3.1 Summary of Observational Data
3.1.1 cSCC Risk is Associated with Particular Germline HLA Allelic Variants
HLA alleles differ by at least one single nucleotide polymorphism [10]. The majority of the peptide sequence variation encoded by different HLA alleles occurs within domains involved in antigen binding and T-cell receptor interaction [11]. Most studies looking at germline HLA allelic variants between cSCC cases and healthy controls only specified allele groups and not individual alleles, given that serological typing was used. These association studies typed for HLA-A, B, C, DR, and/or DQ allele groups using blood samples from cSCC patients and healthy controls in either immunosuppressed or immunocompetent Caucasian populations. HLA allele groups tested for association with cSCC risk are reported in Tables 1–2. These tables show that multiple HLA genes and allele groups have been associated with cSCC risk. These associations vary by the immunosuppression status of the study population, as summarized in Sections 3.1.1a and 3.1.1.b.
Table 1.
HLA-cSCC Associations in Immunocompetent Patients
| HLA ALLELE | STUDY RESULTS
|
||||
|---|---|---|---|---|---|
| Population (Latitude) | No. of Cases | No. of Controls | Association (p-value) | Reference | |
| Class I HLA-cSCC Associations | |||||
|
| |||||
| B*58 | Italy (41.9°N) | 13 | 220 | negative (0.024) | [74] |
| Southern Australia (30.0°S) | 34 | 201 | null | [75] | |
|
| |||||
| Bw4a | United States (43.2°N) | 238 | 398 | negative (<0.001) | [76] |
|
| |||||
| Class II HLA-cSCC Associations | |||||
|
| |||||
| DRB1*01 | Italy (41.9°N) | 13 | 220 | positive (0.005) | [74] |
| United Kingdom (55.4°N) | 12 | 108 | nullb (0.09) | [6] | |
| Tropical Island, Saba (17°N) | 14 | 91 | null | [77] | |
|
| |||||
| DRB1*07 | Southern Australia (30.0°S) | 34 | 201 | positive (<0.01) | [75] |
|
| |||||
| DRB1*15 | United Kingdom (55.4°N) | 12 | 108 | nullc | [6] |
|
| |||||
| DRB1*16 | United Kingdom (55.4°N) | 12 | 108 | nullc | [6] |
|
| |||||
| DQB1 | United States (38.8°N) | 6579 | 280558 | positive (<10−5) | [12] |
|
| |||||
| DQA1 | United States (38.8°N) | 7701 | 60166 | positive (<5*10−8) | [13] |
Note: p-values for associations that were significant or borderline-significant at the α=0.05 level are reported
Bw4 is a serological epitope defined by amino acid residues at positions 77–83. All HLA-B alleles containing the Bw4 epitope are included here (including B*58).
Positive association with borderline significance at the α=0.05 level
Nonsignificant trend in favor of a negative association (α=0.05)
Table 2.
HLA-cSCC Associations in Immunosuppressed Patients
| HLA ALLELE | STUDY RESULTS
|
||||
|---|---|---|---|---|---|
| Population (Latitude) | No. of Cases | No. of Controls | Association (p-value) | Reference | |
| Class I HLA-cSCC Associations | |||||
|
| |||||
| A*03 | Netherlands (52.1°N) | 49 | 631 | nulla (0.10) | [20,78] |
| United Kingdom (55.4°N) | 27 | 220 | positiveb (<0.05) | [6] | |
| Spain (43°N) | 9 | 91 | positive (0.02) | [23] | |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
|
| |||||
| A*11 | Netherlands (52.1°N) | 49 | 631 | negative (0.01) | [20,78] |
| United Kingdom (55.4°N) | 27 | 220 | negative (0.01) | [14] | |
| United Kingdom (55.4°N) | 37 | 292 | negative (0.01) | [79] | |
| Australia (38.0°S) | 30 | 201 | null | [80] | |
| United Kingdom (55.4°N) | 27 | 220 | null | [6] | |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
| Spain (43°N) | 9 | 91 | null | [23] | |
| Australia (27.5°S) | 219 | 827 | positive (0.009) | [19] | |
|
| |||||
| B*27 | Netherlands (52.1°N) | 49 | 631 | positive (0.001) | [20,78] |
| Australia (38°S) | 30 | 201 | positive (<0.005) | [80] | |
| United Kingdom (55.4°N) | 27 | 220 | null | [6] | |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
| Spain (43°N) | 9 | 91 | null | [23] | |
| Australia (27.5°S) | 219 | 827 | null | [19] | |
|
| |||||
| Class II HLA-cSCC Associations | |||||
|
| |||||
| DRB1*01 | United Kingdom (55.4°N) | 37 | 292 | null | [79] |
| Australia (38.0°S) | 30 | 201 | null | [80] | |
| United Kingdom (55.4°N) | 27 | 220 | null | [6] | |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
|
| |||||
| DRB1*07 | Netherlands (52.1°N) | 55 | 505 | nullc (0.35) | [20] |
| Australia (38.0°S) | 30 | 201 | positive (<0.025) | [80] | |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
| Australia (27.5°S) | 219 | 827 | nullc (0.15) | [19] | |
|
| |||||
| DQA1*01 | United Kingdom (55.4°N) | 27 | 220 | positive (0.001) | [6] |
| Canada (56.1°N) | 16 | 444 | null | [16] | |
| United States (38.8°N) | 7701 | 60166 | positive (<5*10−8) | [13] | |
Note: p-values for associations that were significant or borderline-significant at the α=0.05 level are reported
Positive association with borderline significance at the α=0.05 level
Compared to immunocompetent controls
Nonsignificant trend in favor of a positive association (α=0.05)
3.1.1.a Summary of Associations between Germline HLA Variants and cSCC Risk in Immunocompetent Patients
Table 1 summarizes class I and class II HLA genes and allele groups associated with cSCC risk in immunocompetent patients. Table 1 suggests that the association between cSCC and HLA-DRB1*01 is not statistically significant in populations closer to the equator. This trend may reflect interaction with UV exposure: in populations near the equator, subjects’ cumulative UV exposures may overpower associations between cSCC and HLA-DRB1*01. Two GWAS are also included in Table 1. Chahal et al. [12] identified a locus with high imputation quality, 6p21.32 (rs28993540) that was positively associated with cSCC risk. This locus lies 35 kb upstream of HLA-DQB1, a gene also associated with squamous cell cervical cancer risk [12]. Asgari et al. [13] found that HLA locus 6p21 reached genome-wide significance and was strongly associated with cSCC risk. This locus is associated with expression levels of HLA-DQA1, HLA-DQB1, and HLA-DRB5.
3.1.1.b Summary of Associations between Germline HLA Variants and cSCC Risk in Immunosuppressed Patients
Table 2 reports class I and class II HLA allele groups associated with cSCC in immunosuppressed patients. Table 2 includes HLA locus 6p21 identified by Asgari et al. [13], as it was strongly associated with cSCC risk and the risk ratio per allele was not seen to vary by immunosuppression status. Table 2 also summarizes a reanalysis by Glover et al. in 1993 [6] of the data reported in their 1991 study [14], but with some controls reclassified as cSCC cases upon more detailed screening. They no longer observed the previously-reported negative association between HLA-A*11 and cSCC. Nevertheless, this data may indicate some protective capacity for HLA-A*11, as patients with HLA-A*11 may have fewer or less aggressive tumors [6]. Additionally, Table 2 shows that multiple studies found no association between HLA-DRB1*01 and cSCC in immunosuppressed patients, in contrast to the positive association reported in immunocompetent patients (Table 1). Factors such as immunosuppression and HPV infection may have masked the effects of particular HLA allele groups in immunosuppressed patients, which could explain the lack of association with HLA-DRB1*01 in this population, in contrast to the immunocompetent population [15]. HPV infection in immunosuppressed patients is further discussed in Section 3.2.1.b.
Studies of solid organ transplant patients in Nova Scotia [16], Sweden [17], and Norway [18] were unable to corroborate the associations in Table 2, though some of these studies may have been underpowered to detect an association [16]. Bouwes Bavinck et al. [19] studied renal transplant patients in Queensland, Australia, and also failed to support previously-reported HLA-cSCC associations. In fact, they reported a reversal of association between HLA-A*11 and cSCC, in contrast to multiple previous studies in the Netherlands and the United Kingdom. Possible explanations for these contrasting findings are discussed in Section 3.2.1.b.
3.1.2 HLA Mismatching/Homozygosity may be Associated with cSCC Risk in Immunosuppressed Patients
Bouwes Bavinck et al. [20] reported a significant association between cSCC and the extent of HLA-B mismatching between Dutch renal transplant patients and their renal grafts (p=0.025), as well as a positive association between HLA-DR homozygosity and cSCC risk that reached borderline significance (p=0.06). The association between HLA-B mismatching and cSCC was not thought to be confounded by cumulative doses of immunosuppressive drugs. This is because neither the extent of HLA-B mismatching nor the occurrence of cSCC was associated with the cumulative dose of immunosuppressive drug administered. Additionally, the HLA-B mismatching and cSCC association was not thought to be confounded by lifetime sun exposure. The presence of one mismatched HLA-B antigen was also associated with increased cSCC risk (p < 0.001) in a study of Swedish transplant patients [21]. In contrast, Roeger et al. [22] found no association between HLA-B mismatching and cSCC in transplant patients. Similarly, a follow-up study by Bouwes Bavinck et al. [19] in Australia reported no association between HLA-DR homozygosity or HLA-B mismatching and cSCC risk. A study of heart transplant patients in Spain [23] also reported no association between HLA-DR homozygosity or HLA-B mismatching and cSCC risk, but this study may have been underpowered to detect an association with only 9 cSCC patients. Biological mechanisms for these associations between cSCC and HLA mismatching/homozygosity are discussed in Section 3.2.2.
3.1.3 Abnormal Cell Surface Expression of HLA Protein is seen in cSCC Tumors
Aberrant expression of both class I and class II HLA proteins on the surface of cSCC cancer cells is reported in immunocompetent and immunosuppressed patients [24–28].
3.1.3.a cSCC Tumors have Heterogeneous Cell Surface Expression of Class I HLA
Complete loss of class I HLA protein expression has been observed in malignant skin cancers (melanoma, basal cell carcinoma (BCC)) and breast cancer [29]. However, unlike these cancers, cSCCs often partially express class I HLA proteins, creating a heterogeneous cell surface expression pattern [24–28]. Tumors from immunocompetent cSCC patients have higher expression of class I HLA proteins and β2-microglobulin (a component of HLA class I proteins) than tumors from patients with BCC or melanoma, though expression of class I HLA proteins was not observed consistently on all cSCC tumors or even on all cells within a given cSCC tumor [24]. The differences in class I HLA protein expression among cSCC patients were not associated with histopathological type or degree of tumor infiltration or differentiation, which are clinical pathological parameters related to cSCC prognosis [24,30]. This finding contrasts with what is seen in head and neck SCC (HNSCC) and suggests that cell surface expression of class I HLA proteins is not important for cSCC prognosis, but this lack of association may reflect the limited sample size of the study [24,30]. In terms of specific class I HLA protein expression, a study from France reported that HLA-G cell-surface expression was positively associated with cSCC in immunosuppressed patients (p < 0.02), and that HLA-G expression was largely restricted to cSCC tumors as compared to nonmalignant cutaneous lesions in the transplant recipients [31]. Potential biological mechanisms for the association between class I HLA protein expression levels and cSCC are discussed in Section 3.2.3.a.
3.1.3.b Class II HLA Protein is Overexpressed on the Surface of cSCC Tumors
Most solid tumors acquire aberrant cell surface expression of HLA class II proteins, allowing cancer cells that express class II HLA proteins to act as antigen presenting cells [32]. Multiple studies have shown that cSCC tumors also demonstrate high cell surface expression of class II HLA proteins [24–28]. Specifically, cSCC tumors most often express HLA-DR, though HLA-DP and DQ expression have also been reported [24,26]. Peripheral areas of cSCC tumors near inflammatory infiltrates show highest expression of class II HLA proteins [24–28]. Class II HLA protein expression is correlated with histopathological type (p < 0.05) and cellular differentiation (p < 0.01) in cSCC patients, with high levels of class II HLA expression seen on more undifferentiated tumors [24].
3.2 Proposed Mechanisms Suggested by Observational Data
3.2.1 cSCC Risk is Associated with Particular Germline HLA Allelic Variants
3.2.1.a Structural Differences in HLA Alleles May Underlie Associations between Germline HLA Variants and cSCC Risk
We propose that the associations in Tables 1–2 could be due to structural differences in HLA alleles that alter their ability to present cSCC tumor antigens to T lymphocytes. UVR, the main causal factor for cSCC, is known to induce specific UVR fingerprint mutations in particular genes, creating specific tumor antigens [33]. Thus, certain HLA alleles, as a result of the unique structure of their binding cleft, could have a higher binding affinity for these tumor-specific antigens and can better present them to T cells.
UVR-induced fingerprint mutations are found in more than 50% of cSCC cases and produce cSCC-specific tumor antigens [33]. Genes known to be mutated in cSCC include TP53, PTCH, and NOTCH [34]. Antigenic peptides resulting from UVR-mediated damage to such genes are presented by HLA molecules, and cells presenting these tumor antigens are recognized by the T cells and targeted for destruction [35]. Thus, immune response against UVR-damaged cells mediated by the detection of these neoantigens can prevent tumorigenesis.
HLA alleles typically differ at their antigen binding cleft and may have different binding affinities for tumor antigens and T cell receptors. For example, the discriminating site of HLA-A*03 as compared to HLA-A*11 is at codon 9 in the floor of the antigen-binding groove, where HLA-A*03 has a phenylalanine residue while HLA-A*11 has a tyrosine- two amino acids that differ only by a hydroxy group. HLA-A*03 is positively associated with cSCC in immunosuppressed patients from the Netherlands, while HLA-A*11 is negatively associated [36]. HLA alleles that are positively associated with cSCC could be those which are not as efficient in presenting cSCC tumor antigens (e.g. mutant p53). In this way, these tumor cells could escape immune surveillance. On the other hand, HLA alleles negatively associated with cSCC could be those which prevent cSCC tumorigenesis by effectively presenting cSCC tumor antigens to T lymphocytes.
3.2.1.b β-HPV Infection May Influence Associations between Germline HLA Variants and cSCC Risk in Immunosuppressed cSCC Patients
Immunosuppressed patients have increased susceptibility to HPV-induced cutaneous warts and abnormally high incidence of cSCC, which are seen to co-localize on sun-exposed sites [6]. Cutaneous HPV is classified into alpha, beta, and gamma types. β-HPV is thought to be a cofactor alongside UVR in cSCC pathogenesis in immunosuppressed patients- many studies have detected DNA from multiple β-HPV types in cSCC lesions, with β-HPV species 2 identified in particular as a high-risk subtype [37,38]. β-papillomaviruses are thought to have an early role in cSCC tumorigenesis, through a “hit and run” mechanism or by possibly altering cell cycle progression, DNA repair, and immune surveillance, thus facilitating clonal expansion of keratinocytes with UVR-induced DNA damage [38–40]. Furthermore, different β-papillomavirus types are known to exert different carcinogenic effects, so infection by multiple β-HPV types could compound cSCC risk [40]. Certain α-HPV types have also been implicated in cSCC [41]. For instance, HPV77, an α-papillomavirus thus far only detected in cutaneous lesions of immunosuppressed patients, contains a p53-DNA binding site. Once activated by UVR, p53 is thought to stimulate HPV77 promoter activity, leading to the production of E6 and E7 proteins that deregulate p53 and Rb tumor suppressor pathways [42]. In all of these cases, HPV in conjunction with UVR may promote cSCC pathogenesis. However, the exact role of HPV in cutaneous oncogenesis remains to be elucidated because HPV DNA has been found in normal skin samples from cSCC patients, though differential detection of β-HPV in tumors suggests that certain high-risk HPV types may be involved in cSCC pathogenesis [38,42]. HLA allele groups positively associated with cSCC in immunosuppressed patients may encode HLA protein with less efficiency in presenting tumor or HPV antigens, and conversely, HLA allele groups negatively associated with cSCC may encode HLA protein with greater efficiency in presenting tumor or HPV antigens. A good example is HLA-DRB1*07, an allele group thought to be associated with impaired presentation of the L1 antigen of HPV8 to CD4+ T lymphocytes, leading to an ineffective Th2-mediated humoral immune response [36,43]. HLA-DRB1*07 has a significant positive association with or a trend favoring a positive association with cSCC in three out of four case-control studies in Table 2.
The inconsistency seen by Bouwes Bavinck et al. [19] may result from differing environmental factors affecting renal transplant patients in northern Europe versus Australia, because genetic background, as characterized by the frequency of HLA antigens, did not differ greatly between the two populations [19]. In the Dutch population, DNA from HPV types known to be associated with cSCC (HPV-15, -20, and -38) was found in more than 80% of cSCC tumors [19,44], so HPV infection may have been a cofactor in cSCC pathogenesis. The Australian population, on the other hand, had a great excess of UV exposure as compared to the Dutch patients, and thus neoantigens produced by UVR-induced DNA mutations may have played a much larger role in the etiology of cSCC in these patients. Such underlying differences in tumor antigens may have altered cSCC and HLA-A*11 associations in each population.
3.2.2 HLA Mismatching/Homozygosity May Impair Immune Recognition of cSCC Tumor Antigens
The association between HLA-B mismatching and increased cSCC risk may be a result of tolerance to tumor antigens induced in patients with poorly-matched grafts. Patients may develop tolerance to alloantigens of the transplanted kidney, leading to cross-reactive tolerance to cSCC tumor antigens [20]. Such a mechanism is supported by the literature- transplantation is known to promote acquired tolerance of foreign antigens, and the immune system has been shown to develop tolerance to tumor antigens in particular [45,46]. To understand the contradictory results found by Roeger et al. [22], the length of time post-transplant could be considered. Bouwes Bavinck et al. [20] looked at a long-surviving transplant patient group (transplanted between 1967 and 1981), in whom stable tolerance to the mismatched graft could have been induced, while Roeger et al. [22] studied patients with a shorter time since transplant (transplanted between 1981 and 1989). The patients in the Bouwes Bavinck et al. [20] study may have had more time for cross-reactive tolerance to cSCC tumor antigens to develop, making them more susceptible to cSCC than patients in the Roeger et al. study [22,36].
HLA homozygosity is a known risk factor for various cancers [47]. The association between HLA-DR homozygosity and cSCC may be explained by the reduced diversity of HLA-DR proteins. This may result in fewer chances for HLA proteins to interact with antigenic peptides, diminishing recognition of tumor antigens [47].
The lack of association between HLA-DR homozygosity or HLA-B mismatching and cSCC risk seen in the follow up study by Bowes Bavinck et al. [19] could be attributed to the study location. The follow up study was done in Australia, and the patients surveyed had excessive UVR exposure as compared to the first study population from the Netherlands. This could have masked the effects of HLA mismatching or homozygosity on cSCC risk [19].
3.2.3 Abnormal Cell Surface Expression of HLA Protein is seen in cSCC Tumors
3.2.3.a Heterogeneous Cell Surface Expression of Class I HLA Protein in cSCC Tumors Could Facilitate Immune Evasion
Downregulation of class I HLA protein expression prevents presentation of tumor antigens to CD8+ T lymphocytes, thus inhibiting T lymphocyte-mediated destruction of cSCC cells [48]. However, such downregulation also puts cSCC cells at increased risk of lysis by natural killer (NK) cells, which have receptors that negatively regulate NK cell activity upon interaction with particular class I HLA proteins. We propose that the heterogeneous class I HLA protein expression profile presents a mechanistic advantage for cSCC cancer cells, especially if it is a result of selective downregulation, as is seen in HNSCCs [49]. Selective downregulation could mean that expression of class I HLA proteins which efficiently present tumor antigens to CD8+ T lymphocytes is downregulated while expression of class I HLA proteins that inhibit NK activity is retained, given that NK cells are known to have preferential interactions with specific HLA subclasses [50]. Multiple studies suggest a role for immunoselection in carcinogenesis via HLA class I protein expression abnormalities [50,51]. Selective downregulation would allow cSCC cells to escape recognition by CD8+ T cells while also avoiding NK-mediated lysis. Additionally, the overall reduction in class I HLA proteins on the cell surface may significantly improve the ability of cSCC cells to avoid immune destruction [51].
The heterogeneous expression of class I HLA proteins in cSCC may also explain why immunosuppression increases cSCC risk 65-fold, but BCC risk only 10-fold [52]. Immunosurveillance may have greater control over cSCC pathogenesis due to the partial expression of class I HLA proteins in cSCC, as compared to BCC, where class I HLA proteins are often completely absent [53]. Therefore, a loss of immunosurveillance due to immunosuppression would impact cSCC pathogenesis more than that of BCC, and cSCC cells may proliferate much more rapidly than BCC cells given a diminished adaptive immune response.
In immunosuppressed patients, we propose that the aberrant expression of HLA-G protein on the surface of cSCC cancer cells allowed for evasion of immune surveillance. HLA-G expression has been detected in various cancers (melanoma, breast, colon, lung, and renal), and melanoma cell lines expressing HLA-G isoforms have shown inhibited cytotoxic responses from NK and T-cells [54]. The immunomodulatory effects of HLA-G under normal physiological conditions are also well documented: HLA-G, expressed in embryonic tissues, adult immune privileged organs, and in hematopoietic cells, is known to provide inhibitory signals to NK and T cells [50,55]. It is thus reasonable to postulate that HLA-G expression on cSCC tumors could allow cSCC cells to negatively regulate NK and T lymphocyte-mediated destruction. Furthermore, HLA-G positive antigen presenting cells have been shown to inhibit CD4+ T cells and induce the differentiation of these cells into regulatory T cells [55]. As previously mentioned, cSCC cells can aberrantly express class II HLA proteins and serve as antigen presenting cells, and thus HLA-G expression on such cSCC cells could promote tolerance to cSCC tumor antigens.
3.2.3.b Overexpression of Class II HLA Protein in cSCC Tumors May Inhibit Immune Response to Tumor Antigens
We propose that aberrant expression of class II HLA proteins on cSCC cancer cells enables tumor escape from host defense mechanisms, as has been shown in other cancers such as HNSCC and acute myeloid leukemia [56]. Class II HLA proteins are needed for the activation of CD4+ T lymphocytes, which directly mediate cytotoxicity against tumor cells. cSCC cells expressing class II HLA proteins act as antigen presenting cells and present tumor antigens. However, these cells lack the B7 family co-stimulatory molecules required to completely activate CD4+ T cells. Binding of CD4+ T lymphocytes to class II HLA proteins on cancer cells in the absence of co-stimulatory molecules can result in an immune-suppressed or anergic CD4+ T lymphocyte, thus promoting tolerance to tumor antigens [57]. HLA class II proteins expressed by cancer cells can further inhibit the immune response through their interaction with regulatory T cells, which modulate the immune system. Regulatory T cells do not require costimulation for functional activation. Thus, after interacting with class II HLA on cSCC cells presenting tumor antigens, regulatory T cells act as powerful nonspecific suppressors of immune response against the cancer by preventing expansion of CD8+ and CD4+ T lymphocytes as well as NK cells [58].
4. LIMITATIONS & FUTURE WORK
Summarizing the literature on HLA-cSCC associations highlights important limitations. The inter-study inconsistency seen in Tables 1–2 could reflect a lack of allelic specificity; most of these studies only report HLA serotypes since they were carried out before more sophisticated methods of HLA typing were developed. Within a HLA serotype or allele group, individual HLA alleles can differ greatly in terms of structure. Thus, in a given HLA serotype, particular alleles could predispose an individual to cSCC while other alleles could protect against cSCC, leading to the conflicting results [59]. Additionally, differences in HLA-cSCC associations among these studies could reflect differences in patient population beyond what was previously discussed (i.e. geography), methodology (i.e. composition of the control group), immunosuppression protocols (azathioprine has been reported to increase cSCC risk), or other unidentified factors [60,61]. Here we outline future work to address some of these limitations.
4.1 More Precise Typing of HLA Alleles Associated with cSCC
As previously discussed, most of the HLA-cSCC association studies use serological typing, which cannot completely distinguish between different HLA alleles. High-resolution and high-throughput HLA DNA typing would allow researchers to identify the exact HLA allele associated with cSCC. Such precision is needed given that different alleles even within the same allele group can differ in efficacy of cSCC tumor antigen presentation [11,62].
4.2 Functional Analyses of HLA-cSCC Associations
Most HLA association studies focus on HLA proteins as single units. Marthandan et al. [63] describe a novel genetic association analysis approach, where HLA genes and proteins are subdivided into smaller, biologically-relevant sequence features. Sequence features are amino acid sequences categorized by structural (such as α-helices and β-sheets) and functional information (such as peptide antigen binding sites). The sequence feature variant analysis method would elucidate the biological nature of associations between cSCC risk and germline HLA allelic variants, by allowing researchers to better pinpoint molecular determinants of risk underlying these associations. Such an analysis would focus on molecular variants between HLA alleles that are likely of biological importance, unlike traditional analyses that focus on individual polymorphic amino acids without considering their structural and functional context. Additionally, sequence feature variant analysis can improve statistical power of small data sets. HLA-cSCC association studies may produce nonsignificant results if the HLA allele in question is rare. Rare alleles could be appropriately grouped with alleles that present similar antigens and interact with T-cell receptors in a similar manner, increasing the statistical power for this particular sequence feature variant type. The sequence feature variant type method has been validated in association studies between particular HLA alleles and autoimmune diseases [63,64]. Understanding the nature of HLA amino acid residues involved in cSCC risk can improve understanding and prediction of cSCC tumor antigens. This would help to further understand cSCC pathogenesis and could help direct cSCC immunotherapy.
4.3 Further Evaluation of Tumor Antigens Involved in cSCC Pathogenesis
cSCCs are characterized by cellular heterogeneity and are some of the most highly mutated human cancers [34]. Further investigation of tumor antigens involved in cSCC pathogenesis is necessary, especially given that there is currently no targeted treatment option for cSCC widely in use [65]. Some cSCC tumor antigens in immunocompetent patients have been identified, such as mutated TP53, HRAS, CDKN2A, PTCH, KNSTRN, CARD11, NOTCH1, and NOTCH2 gene products, overexpressed HER-2, MUC-1, EGFR, Fyn, and Ep-CAM gene products, as well as cancer-testis antigens [34,35,53,65–68]. Additional tumor antigens in cSCC driver genes have been described among immunosuppressed patients, including FAT1 [69,70]. Given that immunosuppressed patients face increased risk of cSCC and are more prone to aggressive, metastatic disease, cSCC tumor antigens in this particular patient group should also be further investigated [71]. Studies have identified peptide epitopes from various diseases associated with HLA, which has informed analysis of the specific HLA residues involved in antigen binding and presentation [72]. Similar analysis of cSCC tumor antigens would be very informative regarding HLA-cSCC associations, especially because more than 90% of cSCC patients samples at least partially express HLA class I [73]. Thus, an important part of cSCC pathogenesis may involve presentation of tumor antigens to T-cells, warranting further identification of the antigens themselves.
5. DISCUSSION
Having reviewed the literature for HLA-cSCC associations and discussed HLA-cSCC associations in immunocompetent versus immunosuppressed populations, we now highlight implications of the reported associations for cSCC pathogenesis. The reported heterogeneous expression of class I HLA proteins on the surface of cSCC cells reinforces the idea that immunogenetic regulation is involved in cSCC pathogenesis. Unlike BCC and melanoma, cSCC tumors often retain partial expression of class I HLA proteins, suggesting that cell surface expression of these proteins may play an important biological role. cSCC tumors also display aberrant surface expression of class II HLA proteins, which may serve as a mechanism for immune escape. Particular HLA genetic variants are associated with cSCC in immunocompetent and immunosuppressed patients, with more evidence for class I HLA-cSCC associations in immunosuppressed patients than in immunocompetent patients. Class I HLA could play a more important role in cSCC in immunosuppressed patients because HPV may be a co-factor in tumorigenesis- class I HLA proteins present intracellular peptide antigens, including viral proteins degraded into peptides.
In summary, we report a systematic review of associations between HLA and cSCC risk, both in terms of how germline HLA allelic variants modulate cSCC risk and how cell-surface HLA protein expression levels as well as HLA matching during transplantation are associated with cSCC risk. The observed data are used to propose putative mechanisms for these HLA-cSCC associations. This review contributes to the growing understanding of cSCC pathogenesis from an immunological standpoint, which can aid in effective prevention and treatment of cSCC. This review also highlights future work that could further illuminate the immunogenetic etiology of cSCC.
Acknowledgments
This work was supported by the National Cancer Institute [grant R01CA166672].
Abbreviations
- HLA
human leukocyte antigen
- cSCC
cutaneous squamous cell carcinoma
- HPV
human papillomavirus
- UVR
ultraviolet radiation
- MHC
major histocompatibility complex
- GWAS
genome wide association study
- BCC
basal cell carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jennings L, Schmults CD. Management of high-risk cutaneous squamous cell carcinoma. J Clin Aestheti Dermatol. 2010;3:39–48. [PMC free article] [PubMed] [Google Scholar]
- 2.Dlugosz A, Merlino G, Yuspa SH. Progress in cutaneous cancer research. J Invest Dermatol Symposium Proceedings. 2002;7:17–26. doi: 10.1046/j.1523-1747.2002.19631.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindström LS, Yip B, Lichtenstein P, Pawitan Y, Czene K. Etiology of familial aggregation in melanoma and squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers & Prev. 2007;16:1639–1643. doi: 10.1158/1055-9965.EPI-07-0047. [DOI] [PubMed] [Google Scholar]
- 4.Silverberg MJ, Leyden W, Warton EM, Quesenberry CP, Engels EA, Asgari MM. HIV infection status, immunodeficiency, and the incidence of non-melanoma skin cancer. J Natl Cancer Inst. 2013;105:350–360. doi: 10.1093/jnci/djs529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshikawa T, Rae V, Bruins-Slott W. Susceptibility to effects of UVB radiation on induction of contact hypersensitivity as a risk factor for skin cancer in humans. J Invest Dermatol. 1990;95:530–536. doi: 10.1111/1523-1747.ep12504877. [DOI] [PubMed] [Google Scholar]
- 6.Glover MT, Brown J, Navarrete C, Kwan JTC, Bodmer J, Bodmer W, Kennedy LJ, Leigh IM. HLA antigen frequencies in renal transplant recipients and immunocompetent patients with non-melanoma skin cancer. Eur J Cancer. 1993;29:520–524. doi: 10.1016/s0959-8049(05)80143-1. [DOI] [PubMed] [Google Scholar]
- 7.Burns T, Breathnach S. Rook’s Textbook of Dermatology. Vol. 4. Blackwell Scientific Publications; London: 1992. [Google Scholar]
- 8.Rigel DS, Robinson J, Ross M, Friedman R, Cockerell C, Lim H, Stockfleth E. Cancer of the Skin. second. Saunders; 2011. [Google Scholar]
- 9.Habif TP. Clinical Dermatology: A Color Guide to Diagnosis and Therapy. sixth. Saunders; Philadelphia: 2015. [Google Scholar]
- 10.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:423–431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffbrand AV, Higgs DR, Keeling DM, Mehta AB. Postgraduate Haematology. John Wiley & Sons; oboken, NJ: 2015. [Google Scholar]
- 12.Chahal HS, Lin Y, Ransohoff KJ, Hinds DA, Wu W, Dai H, Quereshi AA, Li W, Kraft P, Tang JY, Han J, Sarin K. Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nat Commun. 2016;7 doi: 10.1038/ncomms12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asgari MM, Wang W, Ioannidis NM, Itnyre J, Hoffmann T, Jorgenson E, Whittemore AS. Identification of susceptibility loci for cutaneous squamous cell carcinoma. J Invest Dermatol. 2016;136:930–937. doi: 10.1016/j.jid.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glover MT, Kennedy LJ, Bodmer JG, et al. HLA associations in renal transplant recipients and in non-immunosuppressed patients with epidermal tumours. Br J Dermatol. 1991;125:34–35. [Google Scholar]
- 15.Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- 16.Dyall-Smith D, Ross JB. Cutaneous malignancies in renal transplant recipients from Nova Scotia, Canada. Australasian J Dermatol. 2007;36:79–82. doi: 10.1111/j.1440-0960.1995.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 17.Ingvar A, Ekstrom Smedby K, Lindelof B, Fernberg P, Bellocco R, Tufveson G, Hoglund P, Adami J. No association between infections, HLA type and other transplant-related factors and risk of cutaneous squamous cell carcinoma in solid organ transplant recipients. Acta Derm Venereol. 2012;92:609–614. doi: 10.2340/00015555-1271. [DOI] [PubMed] [Google Scholar]
- 18.Jensen P, Hansen S, Moller B, Leivestad T, Pfeffer P, Geiran O, Fauchald P, Simonsen S. Skin cancer in kidney and heart transplant recipients and different long-term immunosuppressive therapy regimens. J Am Acad Dermatol. 1999;40:177–186. doi: 10.1016/s0190-9622(99)70185-4. [DOI] [PubMed] [Google Scholar]
- 19.Bavinck JNB, Claas FH, Hardie DR, Green A, Vermeer BJ, Hardie IR. Relation between HLA antigens and skin cancer in renal transplant recipients in Queensland, Australia. J Invest Dermatol. 1997;108:708–711. doi: 10.1111/1523-1747.ep12292086. [DOI] [PubMed] [Google Scholar]
- 20.Bavinck JNB, Vermeer BJ, van der Woude FJ, Vandenbroucke JP, Schreuder GMT, Thorogood J, Claas FH. Relation between skin cancer and HLA antigens in renal-transplant recipients. N Engl J Med. 1991;325:843–848. doi: 10.1056/NEJM199109193251203. [DOI] [PubMed] [Google Scholar]
- 21.Ingvar A, Smedby KE, Lindelof B, Fernberg P, Bellocco R, Tufveson G, Hoglund P, Adami J. Immunosuppressive treatment after solid organ transplant and risk of post-transplant cutaneous squamous cell carcinoma. Nephrol Dial Transplant. 2010;25:2764–2771. doi: 10.1093/ndt/gfp425. [DOI] [PubMed] [Google Scholar]
- 22.Roeger LS, Sheil AGR, Disney APS, Mathew TH, Amiss N. Risk factors associated with the development of squamous cell carcinomas in immunosuppressed renal transplant recipients. Clin Transplantation. 1992;6:202–211. [Google Scholar]
- 23.Espana A, Redondo P, Fernandez AL, Zabala M, Herreros J, Llorens R, Quintanilla E. Skin cancer in heart transplant recipients. J Am Acad Dermatol. 1995;32:458–465. doi: 10.1016/0190-9622(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Plata D, Mozos E, Carrasco L, Solana R. HLA molecule expression in cutaneous SCC: an immunopathological study and clinical-immunohistopathological correlations. Histol Histopathol. 1993;8:219–226. [PubMed] [Google Scholar]
- 25.Hua L, Kacen CN, Carpenter RJ, Goltz RW. HLA and β2-microglobulin expression in basal and squamous cell carcinomas of the skin, Int. J Dermatol. 1985;24:660–663. doi: 10.1111/j.1365-4362.1985.tb05719.x. [DOI] [PubMed] [Google Scholar]
- 26.Markey AC, Churchill LJ, MacDonald DM. Altered expression of major histocompatibility complex (MHC) antigens by epidermal tumors. J Cutan Pathol. 1990;17:65–71. doi: 10.1111/j.1600-0560.1990.tb00058.x. [DOI] [PubMed] [Google Scholar]
- 27.Mauduit G, Turbitt M, Mackie RONA. Dissociation of HLA heavy chain and light chain (β2 microglobulin) expression on the cell surface of cutaneous malignancies. Br J Dermatol. 1983;109:377–381. doi: 10.1111/j.1365-2133.1983.tb04611.x. [DOI] [PubMed] [Google Scholar]
- 28.Natali PG, Viora M, Nicotra MR, Giacomini P, Bigotti A, Ferrone S. Antigenic heterogeneity of skin tumours of non-melanocytic origin: analysis with monoclonal antibodies to tumor-associated antigens. J Natl Cancer Inst. 1983;71:439–447. [PubMed] [Google Scholar]
- 29.Bubeník J. Tumour MHC class I downregulation and immunotherapy. Oncol Reports. 2003;10:2005–2008. [PubMed] [Google Scholar]
- 30.Grandis JR, Falkner DM, Melhem MF, Gooding WE, Drenning SD, Morel PA. Human leukocyte antigen class I allelic and haplotype loss in squamous cell carcinoma of the head and neck: clinical and immunogenetic consequences. Clin Cancer Research. 2000;6:2794–2802. [PubMed] [Google Scholar]
- 31.Aractingi S, Kanitakis J, Euvrard S, Le Danff C, Carosella ED. Selective expression of HLA-G in malignant and premalignant skin specimens in kidney transplant recipients. Int J Cancer. 2003;106:232–235. doi: 10.1002/ijc.11217. [DOI] [PubMed] [Google Scholar]
- 32.Donia M, Andersen R, Kjeldsen JW, Fagone P, Munir S, Nicoletti F, Svane IM. Aberrant expression of MHC class II in melanoma attracts inflammatory tumor-specific CD4+ T-Cells, which dampen CD8+ T-cell antitumor reactivity. Cancer Res. 2015;75:3747–3759. doi: 10.1158/0008-5472.CAN-14-2956. [DOI] [PubMed] [Google Scholar]
- 33.Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin, Photochem. Photobiol. 2008;84:539–549. doi: 10.1111/j.1751-1097.2007.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dotto GP, Rustgi AK. Squamous cell cancers: A unified perspective on biology and genetics. Cancer Cell Review. 2016;29:622–637. doi: 10.1016/j.ccell.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratushny V, Gober MD, Hick R, Ridky RW, Seykora JT. From keratinocyte to cancer: The pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122:464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bavinck JNB, Claas FH. The role of HLA molecules in the development of skin cancer. Hum Immunol. 1994;41:173–179. doi: 10.1016/0198-8859(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 37.Harwood CA, Surentheran T, McGregor JM, Spink PJ, Leigh IM, Breuer J, Proby CM. Human papillomavirus infection and non-melanoma skin cancer in immunosuppressed and immunocompetent individuals. J Med Virol. 2000;61:289–297. doi: 10.1002/1096-9071(200007)61:3<289::aid-jmv2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 38.Asgari MM, Kiviat NB, Critchlow CW, Stern JE, Argenyi ZB, Raugi GJ, Berg D, Odland PB, Hawes SE, de Villiers E. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol. 2008;128:1409–1417. doi: 10.1038/sj.jid.5701227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viarisio D, Decker KM, Aengeneyndt B, Flechtenmacher C, Gissmann L, Tommasino M. Human papillomavirus type 38 E6 and E7 act as tumour promoters during chemically induced skin carcinogenesis. J Gen Virol. 2013;94:749–752. doi: 10.1099/vir.0.048991-0. [DOI] [PubMed] [Google Scholar]
- 40.Bouwes Bavinck JN, Plasmeijer EI, Feltkamp MCW. β-papillomavirus infection and skin cancer. J Invest Dermatol. 2008;128:1355–1358. doi: 10.1038/jid.2008.123. [DOI] [PubMed] [Google Scholar]
- 41.Aldabagh B, Angeles JGC, Cardones AR, Arron ST. Cutaneous squamous cell carcinoma and human papillomavirus: Is there an association? Dermatol Surg. 2014;39:1–23. doi: 10.1111/j.1524-4725.2012.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Aldabagh B, Yu J, Arron ST. Role of human papillomavirus in cutaneous squamous cell carcinoma: a meta-analysis. J Am Acad Dermatol. 2014;70:621–629. doi: 10.1016/j.jaad.2014.01.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bavinck JN, Gissmann L, Claas FH, Van der Woude FJ, Persijn GG, Ter Schegget J, Vermeer BJ, Jochmus I, Muller M, Steger G, et al. Relation between skin cancer, humoral reponses to human papillomaviruses, and HLA class II molecules in renal transplant recipients. J Immunol. 1993;151:1579–1586. [PubMed] [Google Scholar]
- 44.Bavinck JB, Boer A, Vermeer BJ, Hartevelt MM, Woulde FVD, Claas FHJ, Vandenbroucke JP. Sunlight, keratotic skin lesions and skin cancer in renal transplant recipients. Br J Dermatol. 1993;129:242–249. doi: 10.1111/j.1365-2133.1993.tb11841.x. [DOI] [PubMed] [Google Scholar]
- 45.Turka LA, Lechler RI. Towards the identification of biomarkers of transplantation tolerance. Nat Rev Immunol. 2009;9:521–526. doi: 10.1038/nri2568. [DOI] [PubMed] [Google Scholar]
- 46.Ugel S, Peranzoni E, Desantis G, Chioda M, Walter S, Weinschenk T, Ochando JC, Cabrelle A, Mandruzzato S, Bronte V. Immune tolerance to tumor antigens occurs in a specialized environment of the spleen. Cell Reports. 2012;2:628–639. doi: 10.1016/j.celrep.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Dausset J, Colombani J, Hors J. Major histocompatibility complex and cancer, with special reference to human familiar tumors (Hodgkin’s disease and other malignancies) Cancer Surv. 1982;1:119–147. [Google Scholar]
- 48.Urosevic M, Dummer R. Immunotherapy for nonmelanoma skin cancer. Cancer. 2002;94:477–485. doi: 10.1002/cncr.10178. [DOI] [PubMed] [Google Scholar]
- 49.Chang CC, Campoli M, Ferrone S. Classical and nonclassical HLA class I antigen and NK cell–activating ligand changes in malignant cells: current challenges and future directions. Adv Cancer Res. 2005;93:189–234. doi: 10.1016/S0065-230X(05)93006-6. [DOI] [PubMed] [Google Scholar]
- 50.Rees RC, Mian S. Selective MHC expression in tumours modulates adaptive and innate antitumour responses. Cancer Immunol Immunother. 1999;48:374–381. doi: 10.1007/s002620050589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang CC, Campoli M, Ferrone S. HLA class I defects in malignant lesions: what have we learned? Keio J Med. 2003;52:220–229. doi: 10.2302/kjm.52.220. [DOI] [PubMed] [Google Scholar]
- 52.Pfister H. Human papillomavirus and skin cancer. J Natl Cancer Inst Monogr. 2003;31:52–56. doi: 10.1093/oxfordjournals.jncimonographs.a003483. [DOI] [PubMed] [Google Scholar]
- 53.Walter A, Barysch MJ, Behnke S, Dziunycz P, Schmid B, Riter E, Gnjatic S, Kristiansen G, Moch H, Knuth A, Dummer R, van den Broek M. Cancer-Testis antigens and immunosurveillance in human cutaneous squamous cell and basal cell carcinomas. Clin Cancer Res. 2010;16:3652–3570. doi: 10.1158/1078-0432.CCR-09-3136. [DOI] [PubMed] [Google Scholar]
- 54.Riteau B, Menier C, Khalil-Daher I, Martinozzi S, Pla M, Dausset J, Carosella ED, Rouas-Freiss N. HLA-G1 co-expression boosts the HLA class 1-mediated NK lysis inhibition. Int Immunol. 2001;13:193–201. doi: 10.1093/intimm/13.2.193. [DOI] [PubMed] [Google Scholar]
- 55.Amiot L. Biology of HLA-G in cancer: A candidate molecule for therapeutic intervention? Cell Mol Life Sci. 2011;68:417–431. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thibodeau J, Bourgeois-Daigneault MC, Lapointe R. Targeting the MHC class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology. 2012;1:908–916. doi: 10.4161/onci.21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knutson KL, Disis ML. Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother. 2005;54:721–728. doi: 10.1007/s00262-004-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campoli M, Ferrone S. Tumor escape mechanisms: potential role of soluble HLA antigens and NK cells activating ligands. Tissue Antigens. 2008;72:321–334. doi: 10.1111/j.1399-0039.2008.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karp DR, Long EO. Identification of HLA-DR1 beta chain residues critical for binding staphylococcal enterotoxins A and E. J Exp Med. 1992;175:415–424. doi: 10.1084/jem.175.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bock A, Bliss RL, Matas A, Little JA. Human leukocyte antigen type as a risk factor for nonmelanomatous skin cancer in patients after renal transplant. Transplantation. 2004;78:775–558. doi: 10.1097/01.tp.0000131666.17216.11. [DOI] [PubMed] [Google Scholar]
- 61.Sanders ML, Karnes JH, Denny JC, Roden DM, Ikizler TA, Birdwell KA. Clinical and genetic factors associated with cutaneous squamous cell carcinoma in kidney and heart transplant recipients. Transplant Direct. 2015;1 doi: 10.1097/TXD.0000000000000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herman A, Croteau G, Sekaly RP, Kappler J, Marrack P. HLA-DR alleles differ in their ability to present staphylococcal enterotoxins to T cells. J Exp Med. 1990;172:709–717. doi: 10.1084/jem.172.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marthandan N, Karp DR, Arnett F, Waller MJ, Guidry P, Feolo M, Marsh SGE, Scheuermann RH. HLA genetic association analysis using the sequence feature variant type approach. J Immunol. 2009;182:136. [Google Scholar]
- 64.Thomson G, Marthandan N, Hollenbach JA, Mack SJ, Erlich HA, Single RM, Waller MJ, Marsh SG, Guidry PA, Karp DR, Scheuermann RH, Thompson SD, Glass DN, Helmberg W. Sequence feature variant type (SFVT) analysis of the HLA genetic association in juvenile idiopathic arthritis. Pac Symp Biocomput. 2010;10:359–370. doi: 10.1142/9789814295291_0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Rohil RN, Tarasen AJ, Carlson JA, Wang K, Johnson A, Yelensky R, Lipson D, Elvin JA, Vergilio J, Ali SM, Suh J, Miller VA, Stephens PJ, Ganesan P, Janku F, Karp DD, Subbiah V, Mihm MC, Ross JS. Evaluation of 122 advanced-stage cutaneous squamous cell carcinomas by comprehensive genomic profiling opens the door for new routes to targeted therapies. Cancer. 2016;122:249–257. doi: 10.1002/cncr.29738. [DOI] [PubMed] [Google Scholar]
- 66.Urosevic M, Dummer R. Immunotherapy for Nonmelanoma skin cancer. Cancer. 2002;94:477–485. doi: 10.1002/cncr.10178. [DOI] [PubMed] [Google Scholar]
- 67.Lee CS, Bhaduri A, Mah A, Johnson WL, Ungewickell A, Aros CJ, Nguyen CB, Rios EJ, Siphrashvili Z, Straight A, Kim J, Aasi SZ, Khavari PA. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nature Genetics. 2014;46:1060–1062. doi: 10.1038/ng.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watt SA, Purdie KJ, den Breems NY, Dimon M, Arron ST, McHugh AT, Xue DJ, Dayal JHS, Proby CM, Harwood CA, Leigh IM, South AP. Novel CARD11 mutations in human cutaneous squamous cell carcinoma lead to aberrant NF-kB regulation. Am J Pathol. 2015;185:2354–2363. doi: 10.1016/j.ajpath.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perrett CM, Harwood CA, McGregor JM, Warwick J, Cerio R, Karran P. Expression of DNA mismatch repair proteins and MSH2 polymorphisms in nonmelanoma skin cancers of organ transplant recipients. Br J Dermatol. 2010;162:732–742. doi: 10.1111/j.1365-2133.2009.09550.x. [DOI] [PubMed] [Google Scholar]
- 70.South AP, Purdie KJ, Watt SA, Haldenby S, den Breems NY, Dimon M, Arron ST, Kluk MJ, Aster JC, McHugh A, Xue DJ, Dayal JHS, Robinson KS, Hasan Riszvi SM, Proby CM, Harwood CA, Leigh IM. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. 2014;134:2630–2638. doi: 10.1038/jid.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert SR, Harwood CA, Purdie KJ, Gulati A, Matin RN, Romanowska M, Cerio R, Kelsell DP, Leigh IM, Proby CM. Metastatic cutaneous squamous cell carcinoma shows frequent deletion in the protein tyrosine phosphatase receptor type D gene. Int J Cancer. 2012;131:E216–E226. doi: 10.1002/ijc.27333. [DOI] [PubMed] [Google Scholar]
- 72.Karp DR, Marthandan N, Marsh SG, Ahn C, Arnett FC, DeLuca DS, Guidry PA, et al. Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis. Hum Mol Genet. 2010;19:707–719. doi: 10.1093/hmg/ddp521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urosevic M, Dummer R. Immunotherapy for nonmelanoma skin cancer. Cancer. 2002;94:477–485. doi: 10.1002/cncr.10178. [DOI] [PubMed] [Google Scholar]
- 74.Cerimele D, Contu L, Carcassi C, Costa G, La Nasa G, Sanna E, Campus GV. HLA and multiple skin carcinomas. Dermatologica. 1988;176:176–181. doi: 10.1159/000248700. [DOI] [PubMed] [Google Scholar]
- 75.Czarnecki DB, Lewis A, Nicholson I, Tait B. Multiple nonmelanoma skin cancer associated with HLA DR7 in southern Australia. Cancer. 1991;68:439–440. doi: 10.1002/1097-0142(19910715)68:2<439::aid-cncr2820680238>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 76.Vineretsky KA, Karagas MR, Christensen BC, Kuriger-Laber JK, Perry AE, Storm CA, Nelson HH. Skin cancer risk is modified by KIR/HLA interactions that influence the activation of natural killer immune cells. Cancer Res. 2016;76:370–376. doi: 10.1158/0008-5472.CAN-15-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bavinck JNB, Bastiaens MT, Marugg ME, Beckers RC, Westendorp RG, Vermeer BJ, Claas FH. Further evidence for an association of HLA-DR7 with basal cell carcinoma on the tropical island of Saba. Arch Dermatol. 2000;136:1019–1022. doi: 10.1001/archderm.136.8.1019. [DOI] [PubMed] [Google Scholar]
- 78.Bavinck JNB, Kootte AM, Van Der Woude FJ, Vandenbroucke JP, Vermeer BJ, Claas FH. On a possible protective effect of HLA-A11 against skin cancer and keratotic skin lesions in renal transplant recipients. J Invest Dermatol. 1991;97:269–272. doi: 10.1111/1523-1747.ep12480376. [DOI] [PubMed] [Google Scholar]
- 79.McGregor JM, Reddi G, MacDonald D, Vaughan RW, Welsh KI. HLA-A11 in renal allograft recipients with skin cancer. J Invest Dermatol. 1992;98:261–262. doi: 10.1111/1523-1747.ep12556128. [DOI] [PubMed] [Google Scholar]
- 80.Czarnecki D, Lewis A, Nicholson I, Tait B, Nash C. HLA-DR1 is not a sign of poor prognosis for the development of multiple basal cell carcinomas. J Am Acad Dermatol. 1992;26:717–719. doi: 10.1016/0190-9622(92)70099-2. [DOI] [PubMed] [Google Scholar]


