Abstract
A combination of biological and chemical methods was applied in the present study to evaluate the removal of arsenic (As) from contaminated soil. The treatment involved As-oxidizing microbes aimed of transforming the more toxic As (III) to less toxic As (V) in the soil. FeCl3 was added at three different concentrations (1, 2, and 3%) to stabilize the As (V). Leaching of the treated soil was investigated by making a soil column and passing tap water through it to determine solubility. Experimental results indicated that the bacterial activity had a pronounced positive effect on the transformation of As, and decreased the soluble exchangeable fraction from 50 to 0.7 mg/kg as compared to control and from 50 to 44 mg/kg after 7 days of treatment. FeCl3 also played an indispensable role in the adsorption/stabilization of As in the soil; 1 and 2% FeCl3 strongly influenced the adsorption of As (V). The soil leachate contained negligible amount of As and trace metals, which indicates that combining an efficient microbe with a chemical treatment is very effective route for the removal and stabilization of As from contaminated soil in the environment.
Keywords: Soil remediation, Pseudomonas sp. XS4, Oxidizing microbes, FeCl3 treatment, Soil leachate, Contaminated sites, Environments
Introduction
Rapid industrialization and extraction of natural resources has resulted in large-scale environmental contamination. A large amount of the toxic waste generated is dispersed in thousands of contaminated sites spread across the world exposing people to these toxic contaminants. The effect of environmental contamination on human health is increasing rapidly and there is evidence that mixtures of pollutants are contributing to global epidemics and other degenerative diseases and cancer. Arsenic (As) is one of the most toxic elements present in water and soil. Over the recent decades, a significant amount of arsenic has been used in agriculture. Industrial products such as pesticides, fertilizers, and wood preservatives are being released, as well as waste products of mining and burning of coal. The most abundant arsenic species are inorganic As (III) and As (V). As (III) is approximately 100 times more toxic than As (V) (Cervantes et al. 1994). Toxicity of As (III) is due to its binding to the sulfhydryl groups in proteins (Gebel 2002) whereas As (V) is a toxic analog of inorganic phosphate and interferes in phosphorylating metabolism (Cervantes et al. 1994).
Several chemical and electrochemical treatment technologies exist for the remediation of metal contaminated soils like subsurface barriers, immobilization, solidification/stabilization, soil replacement extraction, etc., but these technologies affect the texture of the soil, but they do not necessarily reduce the risk. They are also very expensive. Therefore, there is renewed interest in using bioremediation to treat and remove complex metals from soil and waste streams. Bioremediation processes entail natural attenuation, bio-augmentation, and bio-transformation (Dua et al. 2002). Microorganisms play an important role in the environmental fate of As with a multiplicity of mechanisms affecting transformation between soluble and insoluble forms. Though As-oxidizing bacteria have been reported, there is a need to select efficient microbes with strong As (III) oxidizing capability and improved rate of activity to improve bioremediation of As (III) contaminated sites (Karn and Pan 2016a). Shah and Jha (2013) found Alishewanella sp. to be an efficient As (III) oxidizer which tolerated up to 15 mM of As (III) possibly due to the presence of membrane-bound arsenite oxidase. Davolos and Pietrangeli (2011) isolated members of the phylum Proteobacteria, Firmicutes, and Bacteroidetes that were capable of oxidizing As (III) due to an As (III)-resistant efflux membrane protein. Dey et al. (2016) identified Bacillus sp. and Aneurinibacillus aneurinilyticus capable of removing 51.45 and 51.99% of As (III), respectively, in the culture media. Karn and Pan (2016b) reported Pseudomonas sp. which exhibited high resistance to As (III) and transforms up to 6400 mg/l in culture medium.
The present challenge is to develop innovative and cost-effective solutions to decontaminate polluted sites, to make them safe for human habitation and consumption, and to protect the ecosystem. For this, it is essential first to transform more toxic As (III) to less toxic As (V) by an efficient microbe and further stabilize/fix it. In the present investigation, both biological and chemical treatments were used to remove the As from the soil by isolating and using an efficient indigenous bacterial strain with strong As-oxidizing activity for the transformation of As (III) to As (V) followed by reacting the As (V) with FeCl3 preparing a column with the reacted soil and leaching the As (V) from the soil using tap water.
Materials and methods
Soil sample collection and characterization
A soil sample was collected from a gold mine tailing area, Karamy (45°36′N 84°53′E), Xinjiang, China. The sample was characterized for pH, salinity, cation exchange capacity (CEC), available forms of nutrients, content of mineral N, organic carbon (OC), and total organic carbon (TOC), sulphate and chloride, by standard protocols. Analyses of the Na, K, Mg, Ca, Cd, Cu, Pb, and Hg in the soil sample were determined according to APHA (1999). Metals from the digest sample were analyzed using an inductively coupled plasma mass spectroscopy (ICP-MS) (ELANDRC II; Perkin Elmer). Hydride forming metal was analyzed using a generator atomic flame spectroscopy (AFS).
Isolation and enrichment of microorganisms
Isolation and enrichment of cultivable bacteria for As resistance were done by the standard dilution-plating method. The collected sample had an As concentration of about 200 mg/kg. Five grams of soil sample was added to sucrose low phosphate medium (SLP) and was incubated on a shaker for 5 days at 30 °C. The SLP had the following composition sucrose (1%), (NH4)2SO4 (0.1%), K2HPO4 (0.5%), MgSO4 (0.05%), NaCl (0.01%), and yeast extract (0.05%), and the medium pH was adjusted to 7.0 with HCl and NaOH. To solidify the medium, 1.5% agar was added. The enrichments were subcultured twice into SLP medium containing 200 mg/l As (III). After the second subculture, enrichment was serially diluted. A series of dilutions (10−3, 10−4 and 10−5) was prepared using sterile water. Aliquots of 0.1 ml from the different dilutions were spread on the sucrose low phosphate medium without and with sodium arsenite As (III). For each dilution, three replicate plates were prepared. Morphologically different colonies were isolated and purified by repeated sub culturing on SLP agar medium with 200 mg/l As (III). As (III)-resistant strain obtained, were further screened for their As (III) oxidizing ability using the silver nitrate (AgNO3) screening method described by (Simeonova et al. 2004). Furthermore, As (III) oxidizing bacteria were screened and one efficient As (III) oxidizing strain was selected for further study and designated as strain XS4. This isolate showed a minimum inhibitory concentration (MIC) of about 800 mg/l (10 mM).
As transformation in soil by strain XS4
As transformation in soil was observed by inoculating (about 106 cells/ml) (freshly grown in Luria broth (LB) medium containing 100 mg/l of As (III), culture was grown overnight on a shaker at 30 °C in 100 grams sterilized soil in a beaker supplemented with 50 mg/kg of NaAsO2 further beaker was incubated at 30 °C. As transformation ability in soil was observed by collecting soil from inoculated and uninoculated soil sample with strain XS4 every 24 h and analyzing for a soluble exchangeable fraction of As by the Tessier et al. (1979) extraction method for 7 days. Simultaneously, the growth of strain XS4 was also monitored in soil by checking CFU (colony forming unit) in soil samples. As analysis was carried out using hydride generation atomic fluorescence spectroscopy (AFS) (Jitian AFS-820, Beijing, China) as described by Glaubig and Goldberg (1988) and Voth-Beach and Shrader (1985). The AFS supplied with a continuous flow hydride generation system for the detection of arsenic. The Instrumental parameters were as follows: lamp current (60 mA), negative high voltage photomultiplier (270 V), atomizer height (8 mm), and carrier argon flow (300 ml/min). A known concentration of As standard was analyzed before and a calibration curve was determined during the analysis.
Immobilization and leaching of arsenic
After As transformation, further adsorption or immobilization of As was carried out by supplementing FeCl3 at concentrations of 1, 2, and 3% into the soil sample treated with strain XS4 mixed thoroughly and incubated for 1 week. Analysis of As immobilization in the soil was determined by five-stage sequential extractions: soluble/exchangeable, bound carbonate, bound Fe–Mn, bound organic, and the residual fraction as described by (Tessier et al. 1979).
Next, the leaching experiment was carried out to treat the soil sample in a glass column. A standard glass column was taken and filled with the treated soil, and normal tap water was poured into the column to check the solubility of As at different time intervals from the soil. A water sample was collected from the column at 1, 2, 4, 8, 12, and 24 h. Soluble As was determined from the collected water using AFS analysis.
Arsenic-resistance gene (aioA gene) and identification by 16Sr RNA gene sequencing
Genomic DNA was isolated from the bacterial isolate using a simple lysis protocol (Sambrook et al. 1989). PCR reaction contained 10 ng of genomic DNA, 1X PCR buffer; 200 µM of dNTPs; 1.5 mM MgCl2, 0.1 µM of each primer, and 2.5 Units of Taq DNA polymerase (Invitrogen, USA) in a final volume of 100 µl sterile MQ waters. The primers were (Forward primer 5′GTSGGBTGYGGMTAYCABGYCTA3′; Reverse primer 5′TTGTASGCBGGNCGRTTRTGRAT′) as described by Inskeep et al. (2007). Following 35 PCR cycles each of 94 °C for 1 min, 49 °C for 1 min, and 72 °C for 2 min, the reaction product was analyzed using agarose gel electrophoresis. The PCR product was purified and cloned into a pGEM-T easy vector and sequenced (Sambrook et al. 1989). The sequences were compared against the available DNA sequences in GenBank using BLASTN (to compare the gene sequence) and BLASTX to deduce the amino-acid sequence of NCBI database. The deduced amino acid sequences were aligned using the MAFFT (http://mafft.cbrc.jp/alignment/server/), program and the alignments were manually corrected, and a phylogenetic tree was constructed using the MEGA5 (Tamura et al. 2007). To identify bacteria isolates, we used 16S rRNA gene amplification. Genomic DNA extraction and sequence analysis were performed as described in Karn et al. (2010). The 16S rRNA gene sequence and aioA gene determined in this study have been deposited in GenBank under the accession numbers JX569766, JX569770, respectively.
Statistical analysis
All experiments were performed in three replicates and statistical analysis was performed using GraphPad Prism (version 4.03) software (GraphPad, CA, USA).
Results and discussion
Soil characteristics
The physicochemical characteristics of the soil samples were analyzed and it was found to have very stable chemical composition. The pH of the soil was 7.5 ± 0.2. The organic matter was 16.5 g/kg, and total nitrogen was about 0.799 mg/kg. Predominantly, potassium, sodium, and magnesium were major constituents of the soil sample. Anion analysis of the soil sample revealed the presence of different anions such as chloride at 0.393 mg/kg and sulphate at 0.157 mg/kg. Furthermore, other heavy metals including copper, cadmium, chromium, lead, mercury, and arsenic were also analyzed and the result is shown in Table 1.
Table 1.
Physiochemical characteristics of soil sample
| Serial no. | Parameter | Concentration |
|---|---|---|
| 1 | pH | 7.6 |
| 2 | Organic matter | 16.58 g/kg |
| 3 | Total carbon | 1.75% |
| 4 | Nitrogen | 0.799 g/kg |
| 5 | Chloride | 0.393 mg/kg |
| 6 | Sulphate | 0.157 mg/kg |
| 7 | Cation exchange capacity (CEC) | 0.2789 coml./kg |
| 8 | Na | 49 mg/kg |
| 9 | K | 643 mg/kg |
| 10 | Mg | 306 mg/kg |
| 11 | Ca | 20 mg/kg |
| 12 | Cu | 1.3 mg/kg |
| 13 | Cr | 0.5 mg/kg |
| 14 | Cd | 0.11 mg/kg |
| 15 | Pb | 0.83 mg/kg |
| 16 | As | 0.205 mg/kg |
Arsenic transformation
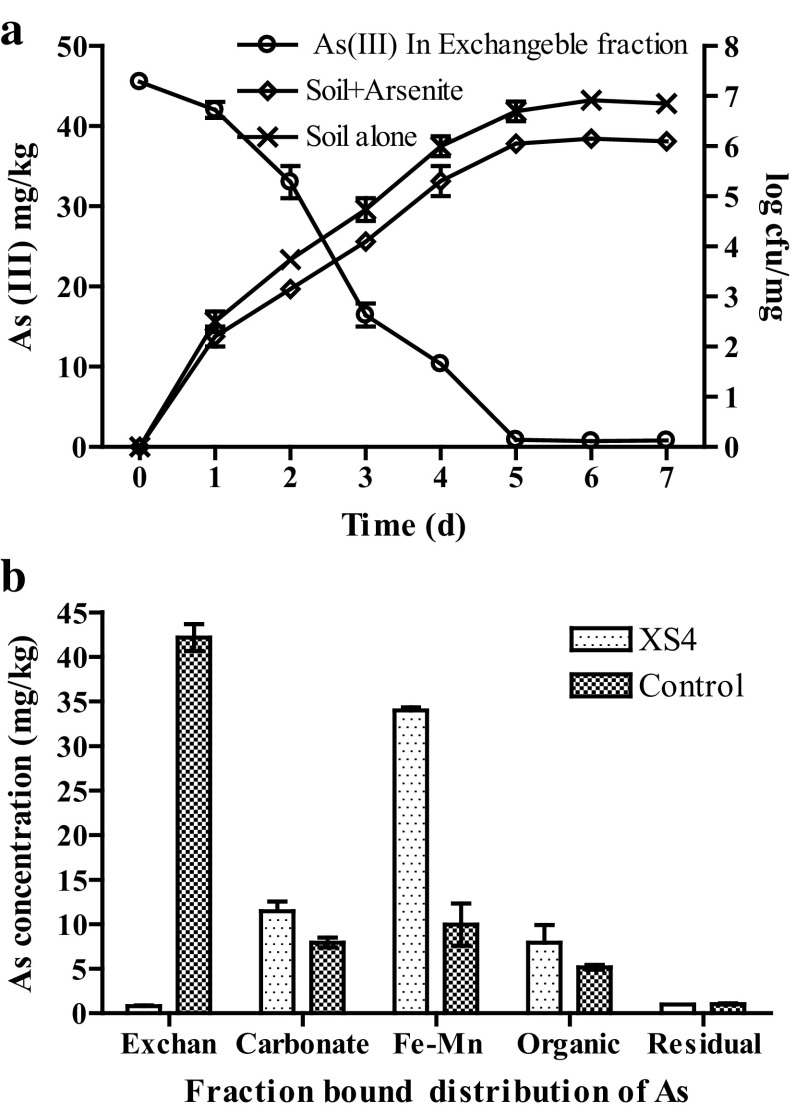
A good bioremediation approach involves strategic use of native microbes to achieve the best possible detoxification level. An advantage of inoculation of indigenous bacteria in a contaminated site is that it may not show a deterrent effect on the indigenous microbial community. Commonly, As (III) in the soil is present as free metal ions, soluble metal complexes, exchangeable metal ions which are easily available to the biological system, and the toxicity of metals in the soil depends on their bio-availability. Therefore, it is of great environmental concern to cause a biological threat and a need to develop an appropriate and easy technique which can reduce the soluble exchangeable fraction part of the metals from the soil. Strain XS4 has been shown potentially efficient to transform As (III) in the soil without any extra manipulation or additional nutrients. After addition of strain XS4, results indicated that a significant amount of the soluble exchangeable fraction of As was decreased in the soil. From the 1st day of inoculations, the exchangeable fraction started to decrease significantly and left 0.07 mg/kg in the soil on the 5th day of incubation, as shown in Fig. 1a, indicating that strain XS4 can be prospective bacteria for arsenite transformation, whereas control or non-remediated soil sample had a high concentration of available arsenite (43.34 mg/kg). Previously, As (III) resistant bacteria were found by Anderson and Cook (2004) who isolated 17 arsenic-resistant bacteria that belong to genera Acinetobacter, Aeromonas, Bacillus, Exiguobacterium, Escherichia, and Pseudomonas, and all the bacteria showed tolerance up to 0-20 mM As (III). Shivaji et al. (2005) obtained an arsenic-resistant bacterium Bacillus arsenicus from arsenic sediments of West Bengal, India that could grow in the presence of 0.5 mM As (III). Jackson et al. (2005) isolated bacteria that belonged to the phyla Proteobacteria, Bacteroidetes, and the Firmicutes, and some of these bacteria were capable of growing at high levels of arsenate up to 275 mM, although As (III) tolerance of bacteria was much lower, about 10 mM. Rajdeep et al. (2009) isolated Planococcus from an arsenic-infested bore-well of West Bengal, India and the bacterium was resistant to As (III) at 20 mM. Therefore, from the previous results, we found that some bacteria are able to grow at high concentration and some are able to tolerate at high concentration, but the transformation ability of these microbes is very limited consequently for the remediation purpose. Finding a bacteria having high transforming ability in the presence of arsenic is important and strain XS4 showed this successfully.
Fig. 1.
a Arsenite transformation ability of strain XS4 in the soil and growth, observed by forming colony forming unit (CFU) in soil supplemented with arsenite and soil alone. b Distribution pattern of total arsenic in five different fractions of control and bioremediated sample by strain XS4
Furthermore, the growth of strain XS4 increased significantly in the soil. From the 1st day of inoculation, CFU was about 2.4 × 102 and gradually increased to 5 × 106 on the 7th day of incubation, which shows that strain XS4 has not any constrain to grow well in the soil. Complete observation of the day-to-day decrease of the soluble exchangeable fraction part of As (III) and increasing of CFU into the soil is shown in Fig. 1a. Microbial growth and activity in soil is often very limited due to the availability of complex mixtures of pollutants, which include both organic contaminants and heavy metals (Kovalick 1992). However, if the metals in soils are toxic to the microbes, removal of pollutants may be slowed or prevented. Many reports have shown that the short-term response to toxic metals is a large reduction in microbial activity (Kuperman and Carreiro 1997; Shivaji et al. 2005). However, from the present study and analysis, we establish that strain XS4 grew well in the soil and increased in CFU day after day, which indicates the high potential survival of this organism in soil. Strain XS4 has strong transforming ability for As (III) in the soil, which has strong potential for bioremediation of As-contaminated soil.
Figure 1b shows the distribution of As after treatment with strain XS4, we can see that in bioremediated soil, the highest fraction of As was in Fe–Mn bound, and then carbonate bound and organic bound which stabilize the metals to become not easily bio-available, whereas in non-remediated or control samples having the highest concentration of exchangeable fraction, which was 43.34 mg/kg, and least in the residual and the organic fraction. In the present report, the soil also has been characterized for various parameters described in result, and observed a significant amount of Fe, Al, and Mn, which determine the soil’s ability to retain and immobilize heavy metals.
After transformation of As (III) to As (V), we used FeCl3 as an adsorbing or immobilizing agent for stabilizing As in the soil. We found that 1 and 2% of FeCl3 significantly increased the adsorbing capacity of As, as compared to control (without FeCl3) of bacteria treated soil. However, 3% of FeCl3 had no positive effect on the adsorption of As Table 2. The results show that up to 2% FeCl3 had a better effect on adsorption and can be used as an adsorption agent from the soil. Earlier researchers have also used Fe compound to adsorb or immobilize the various metals from the soil (Zhang et al. 2013).
Table 2.
Adsorption capacity of FeCl3 in the bacteria treated soil observed by the addition of 1, 2, and 3%. All the concentrations are represented in terms of mg/kg
| Exchangeable | Carbonate | Fe–Mn | Organic | Residual |
|---|---|---|---|---|
| Adsorption by FeCl3 (1%) | ||||
| 0.42 ± 0.4 | 1.1 ± 0.1 | 45.81 ± 2 | 1.42 ± 0.5 | 2.25 ± 0.2 |
| Adsorption by FeCl3 (2%) | ||||
| 0.47 ± 0.2 | 1.91 ± 0.3 | 44.58 ± 3 | 3.26 ± 0.4 | 1.67 ± 0.2 |
| Adsorption by FeCl3 (3%) | ||||
| 0.51 ± 0.3 | 2.27 ± 0.3 | 39.38 ± 2 | 2.73 ± 0.3 | 4.74 ± 0.5 |
| No adsorption by FeCl3 | ||||
| 0.44 ± 0.2 | 6.58 ± 0.3 | 35.89 ± 2 | 3.38 ± 0.6 | 4.8 ± 0.7 |
| Soil without bacteria and FeCl3 | ||||
| 39.22 ± 3 | 1.92 ± 2 | 5.47 ± 4 | 1.01 ± 2 | 2.27 ± 2 |
In addition, we carried out a leaching experiment in a glass column of this treated soil to measure the solubility of the As from the soil, because in nature after treatment, soil kept in the natural condition may be in contact with water including rain water. Sometime researchers observed that leaching of heavy metals including As, Fe, Pb, and Cd from the contaminated soil, which can be harmful in even small amounts to the organisms and can alter the biodiversity (Zhang et al. 2013). Therefore, a standard glass column was taken and filled with the treated soil and normal tap water was passed in the column and water was collected at different time intervals 1, 2, 4, 8, 12, and 24 h and further collected water analyzed for As. We found that the As is stably bound to the soil and there is no available As into the collected water. Therefore, from the present investigation, we found the strain XS4 successfully transformed the As in the soil and FeCl3 also played an important role to immobilize the metal in the soil which prevents the metal to become bio-available.
aioA gene and 16s rRNA gene amplification
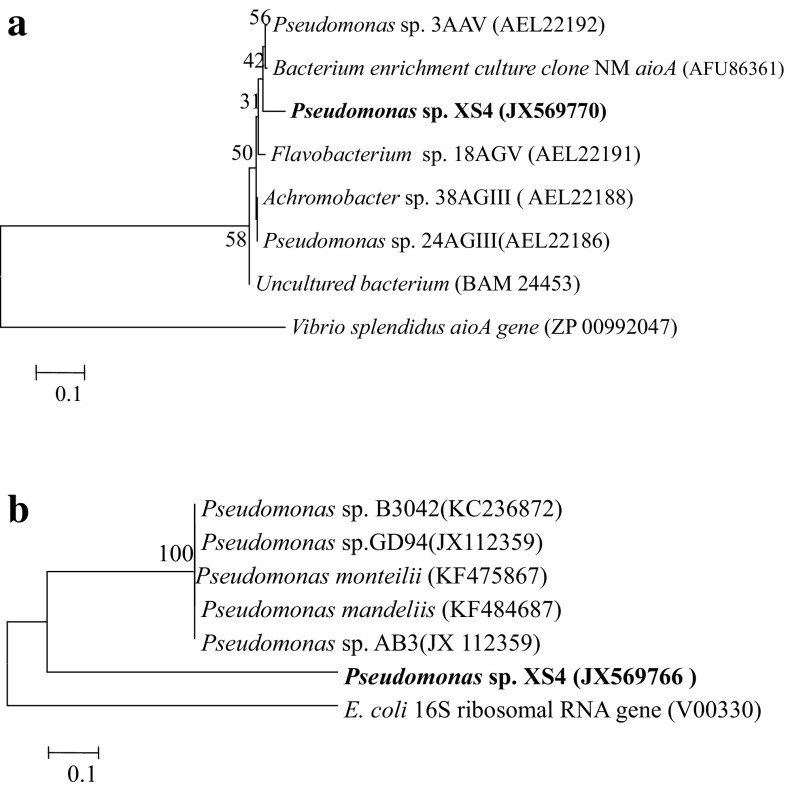
The result showed that strain XS4 has a genetic basis of its As resistance, because we amplified the 500 bp long aioA gene fragment from genomic DNA. Furthermore, this fragment was cloned, sequenced and compared with the aioA gene to the NCBI database, which showed 99% similarity with previously reported aioA gene of various bacterial isolates. The putative amino-acid sequences of aioA gene were compared with the closely related aioA genes from other bacterial species from the GenBank using BlastX program (Altschul et al. 1997). The sequences were aligned using Multalin program. The evolutionary distance was calculated by Kimura 2 parameters and the phylogenetic tree was constructed by a neighbor-joining method and Vibrio splendidus aio gene (ZP00992047) used as the out-group. Bootstrap analysis was based on 1500 resamplings using the MEGA 4.0 package (Tamura et al. 2007). Phylogenetic analysis of aioA gene with related bacterial species showed that maximum 58 and 56% similarity with two previously reported Pseudomonas sp. 24 AGIII (Accession No. AEL22186) and Pseudomonas sp. 3AAV (Accession no. AEL22192) as shown in (Fig. 2a). Furthermore, the aioA gene showed similarity with Flavobacterium sp., Acromobacter sp., and Uncultured clones. The alignment of deduced amino-acid sequences of aioA gene with other known bacterial aioA showed that bacterial aioA genes bear the Molybdopterin-Binding (MopB) domain or Mo-pterin large fragment protein signature.
Fig. 2.
a Dendrogram showing the percentage similarity among the bacterial isolates. The tree was constructed on the basis of aioA gene translated amino-acid sequences. b Phylogeny of strain XS4 generated from the neighbor-joining method of 16s rRNA gene sequences rooted with Escherichia coli
Microorganisms have developed various strategies to counteract As toxicity in the environment (Tsai et al. 2009). Briefly, these include; first, As (III) extrusion from cells (Oremland and Stolz 2000); second, intracellular chelation with As (III) by various metal binding peptides, such as GSH, phytochelatins (PC) and metallothioneins (MT) (Rosen 2002); third, transformation which is less toxic (Qin et al. 2006). Bacteria have also developed the molecular machinery to overcome the toxicity of As which is also considered the genetic basis of tolerance. The aioA gene may also produce some peptide which helps the bacteria to cope with toxicity and help in the transformation of Arsenite. Arsenite-oxidizing bacteria perform arsenite oxidation by arsenite oxidase, an enzyme belongs to dimethylsulfoxide reductase family of the molybdopterin containing protein and a Fe-Rieske protein (Ellis et al. 2001). Genes encoding these protein subunits have been successfully amplified in the As (III)-oxidizing bacteria Herminiimonas arsenicoxydans, Agrobacterium tumefaciens 5A (Muller et al. 2003; Kashyap et al. 2006). Zhang et al. (2015) observed microbe-mediated As bio-transformation in paddy soils and revealed an abundance of (aioA) arsenite oxidase genes, and another gene involved in the bio-transformation which determines the availability and the fate of As in soils. In the current study, we also found the aioA gene in the bacterial isolate which is consistent with the bacteria having a genetic basis to its As resistance system. Furthermore, the translated sequence of about 200 amino acids showed a clear identity with the Mo-pterin-binding protein in the previously reported bacteria from the database. This Mo-pterin protein has played a major role in the resistance of As-oxidizing bacteria. Therefore, future studies are needed to find the complete genetic machinery regulation and understand their role in the As transformation to acquire more information and practical use in bioremediation of As-contaminated soils.
For identification, 16s rRNA gene was amplified and cloned into E. coli DH5α cells, and further selected clones were sequenced. The sequence was compared to the similarity in the GeneBank using BlastN. Pairwise alignment revealed that 16s rRNA gene of the bacterial isolates had 99% similarity with Pseudomonas sp. 16s rRNA sequence of bacterial isolates and similar sequences were aligned using the Clustal W and dendrogram which were generated by the neighbor-joining distance methods using MEGA 4.0 software (Fig. 2b) (Tamura et al. 2007). Numbers are percent gap value after 1000 bootstrap relationship. Phylogenetic analysis revealed that strain XS4 grouped with Pseudomonas sp., and hence, it designated as a Pseudomonas sp.
Conclusion
A combined approach was applied to remove As from contaminated soil using As (III) oxidizing strain XS4 to transform As (III) to As (V) followed by immobilization of As (V) by FeCl3. Strain XS4 potentially grew well in soil and oxidized 95.4% of As (III) in the soil. Furthermore, 1 and 2% FeCl3 enhanced the adsorption capacity by about 20% and stabilized the As as compared to control. In treated soil leachate, also no mobile As was observed. Presently, studies on using strain XS4 in actual contaminated sites are being undertaken.
Acknowledgements
This work was supported by the Program of Chinese Academy of Sciences Fellowships for Young International Scientists (Grant No. 2011Y2ZB06).
Compliance with ethical standards
Conflict of interest
The authors have declared no conflict of interest.
Contributor Information
Santosh Kumar Karn, Email: santoshkarn@gmail.com.
Xiangliang Pan, Phone: +86-991-7823156, Email: xlpan@ms.xjb.ac.cn.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Cook GM. Isolation and characterization of arsenate-reducing bacteria from arsenic contaminated sites in New Zealand. Curr Microbiol. 2004;48:341–347. doi: 10.1007/s00284-003-4205-3. [DOI] [PubMed] [Google Scholar]
- APHA . Standard methods for the examination of water and wastewater. Washington, DC: American Public Health Association of American Water Association; 1999. [Google Scholar]
- Cervantes C, Ji G, Ramirez JL, Silver S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol Rev. 1994;15:355–367. doi: 10.1111/j.1574-6976.1994.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Davolos D, Pietrangeli B. Phylogenetic analysis on the arsenic-resistant bacteria isolated from three different freshwater environments. Chem Ecol. 2011;27:79–87. doi: 10.1080/02757540.2010.536157. [DOI] [Google Scholar]
- Dey U, Chatterjee S, Mondal NK. Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol Rep. 2016 doi: 10.1016/j.btre.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua M, Singh A, Sethunathan N, Johri AK. Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol. 2002;59:143–152. doi: 10.1007/s00253-002-1024-6. [DOI] [PubMed] [Google Scholar]
- Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 angstrom and 2.03 angstrom. Structure. 2001;9:125–132. doi: 10.1016/S0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Gebel TW. Arsenic methylation is a process of detoxification through accelerated excretion. Int J Hyg Environ Health. 2002;205:05–508. doi: 10.1078/1438-4639-00177. [DOI] [PubMed] [Google Scholar]
- Glaubig RA, Goldberg S. Determination of inorganic arsenic (III) and arsenic (III plus V) using automated hydride-generation atomic absorption spectrometry. Soil Sci Soc Am J. 1988;52:536–537. doi: 10.2136/sssaj1988.03615995005200020044x. [DOI] [Google Scholar]
- Inskeep WP, Macur RE, Hamamura N, Warelow TP, Ward SA, Santini JM. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ Microbiol. 2007;9:934–943. doi: 10.1111/j.1462-2920.2006.01215.x. [DOI] [PubMed] [Google Scholar]
- Jackson CR, Dugas SL, Harrison KG. Enumeration and characterization of arsenate-resistant bacteria in arsenic free soils. Soil Biol Biochem. 2005;37:2319–2322. doi: 10.1016/j.soilbio.2005.04.010. [DOI] [Google Scholar]
- Karn SK, Pan X. Role of Acinetobacter sp. in arsenite As(III) oxidation and reducing its mobility in soil. Chem Ecol. 2016;32:460–471. doi: 10.1080/02757540.2016.1157174. [DOI] [Google Scholar]
- Karn SK, Pan X. Bacterial oxidation and stabilization of arsenite As(III) in soil. Environ Eng Sci. 2016;34(3):158–164. doi: 10.1089/ees.2015.0390. [DOI] [Google Scholar]
- Karn SK, Chakrabarty SK, Reddy MS. Characterization of pentachlorophenol degrading Bacillus strains from secondary pulp and paper industry sludge. Int Biodeter Biodegrad. 2010;64:609–613. doi: 10.1016/j.ibiod.2010.05.017. [DOI] [Google Scholar]
- Kashyap DR, Botero LM, Franck WL, Hassett DJ, McDermott TR. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J Bacteriol. 2006;188:1081–1088. doi: 10.1128/JB.188.3.1081-1088.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalick W (1992) Perspectives on risks of soil pollution and experience with innovative remediation technologies, In: Strategies 2000. Proceedings 4th World Congress Chemical Engineering. Dechema, Frankfurt, Germany, pp 281–295
- Kuperman RG, Carreiro MM. Soil heavy metal concentrations, microbial biomass and enzyme activities in a contaminated grassland ecosystem. Soil Biol Biochem. 1997;29:179–190. doi: 10.1016/S0038-0717(96)00297-0. [DOI] [Google Scholar]
- Muller D, Lievremont D, Simeonova DD, Hubert JC, Lett MC. Arsenite oxidase aox genes from a metal-resistant b-Proteobacterium. J Bacteriol. 2003;185:135–141. doi: 10.1128/JB.185.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R, Stolz J. Dissimilatory reduction of selenate and arsenate in nature. In: Lovley DR, editor. Environmental microbe-metal interactions. Washington, DC: American Society Microbiology; 2000. pp. 199–224. [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang GJ, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajdeep C, Sen AK, Karak P, Chatterjee R, Giri AK, Chaudhuri K. Isolation and characterization of an arsenic-resistant bacterium from a bore-well in West Bengal, India. Ann Microbiol. 2009;59:253–258. doi: 10.1007/BF03178325. [DOI] [Google Scholar]
- Rosen BP. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp Biochem Physiol. 2002;133:689–693. doi: 10.1016/S1095-6433(02)00201-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd ed. N.Y., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, p 1659. ISBN 0-87969-309-6
- Shaha R, Jha S. Alishewanella sp. strain GIDC-5, Arsenite hyper-tolerant bacteria isolated from industrial effluent of South Gujarat, India. Chem Ecol. 2013;29:427–436. doi: 10.1080/02757540.2013.774379. [DOI] [Google Scholar]
- Shivaji S, Suresh K, Chaturvedi P, Dubeand S, Sengupta S. Bacillus arsenicus sp. nov., an arsenic-resistant bacterium isolated from a siderite concretion in West Bengal, India. Int J Syst Evol Microbiol. 2005;55:1123–1127. doi: 10.1099/ijs.0.63476-0. [DOI] [PubMed] [Google Scholar]
- Simeonova DD, Lievremont D, Lagarde F, Muller DA, Groudeva VI, Lett MC. Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol Lett. 2004;237:249–253. doi: 10.1111/j.1574-6968.2004.tb09703.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tessier A, Campbell PGC, Bisson M. Sequential extraction procedure for the speciation of particulate trace metals. Anal Chem. 1979;51:844–851. doi: 10.1021/ac50043a017. [DOI] [Google Scholar]
- Tsai SL, Singh S, Chen W. Arsenic metabolism by microbes in nature and the impact on arsenic remediation. Curr Opin Biotechnol. 2009;20:659–667. doi: 10.1016/j.copbio.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Voth-Beach LM, Shrader DE. Reduction of interferences in the determination of arsenic and selenium by hydride generation. Spectroscopy. 1985;1:60–65. [Google Scholar]
- Zhang C, Wang K, Tan S, Niu X, Su P. Evaluation and remediation of organics, nutrients and heavy metals in landfill leachate- a case study in Beijing. Chem Ecol. 2013;29:668–675. doi: 10.1080/02757540.2013.841897. [DOI] [Google Scholar]
- Zhang SY, Zhao FJ, Sun GX, Su JQ, Yang XR, Li H, Zhu YG. Diversity and abundance of arsenic biotransformation genes in paddy soils from southern China. Environ Sci Technol. 2015;49:4138–4146. doi: 10.1021/acs.est.5b00028. [DOI] [PubMed] [Google Scholar]