Abstract
Currently, study of the inter and the intra-population genetic disparity was done by use of the 200 Olea europaea L. which is found growing naturally in the nation of Iran, and this study was carried out by AFLP and IRAP markers. The fingerprints that were similar to the AFLP and the IRAP markers were evidence of high concentrations of heterozygosity and this shows that O. europaea L. is primarily the out crossing species. The average percentage of polymorphism is as shown below:
87.15 and 87.38% of the information used in regard to the AFLP and the IRAP, respectively.
The gene disparity numerals on the population researched were 1.087 for HT and 0.871 for HS in regard to AFLP. For the IRAP it was 1.084 for HT and 0.860 for HS.
The general values for genetic variations that are found in the O. europaea L. germplasm in the nation of Iran were then assessed through putting together the AFLP and the IRAP information so as to cover a larger genome. Arguing from the AFLP and the IRAP studies, it can be concluded that there are more levels of genetic variation at inter and the intra-population level for the O. europaea.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0669-x) contains supplementary material, which is available to authorized users.
Keywords: AFLP, IRAP, Population structure, Olea europaea L., Genetic diversity
Introduction
Olea europaea, commonly known as Olive, is one of the most popular tree crops in the agricultural Mediterranean basin. This may be attributed to the high social and financial importance that comes along with it. The wealth of the grown olive germplasm is not very common in plants due to the life span of the trees and the lack of turnover with the invented breeding genotypes (Barranco et al. 2005; Bartolini et al. 2005; Baldoni and Belaj 2009). Some of the factors that lower their genetic diversity in comparison to their wild counterparts are their inability to withstand the extravagance of the developed germplasm (Lumaret et al. 2004; Breton et al. 2006; Belaj et al. 2010), and this is evidence that the last ones may develop the genetic basis of the grown material.
Truly, majority of the studies have concentrated on looking at the difference between the developed and the naturally growing olives (Baldoni et al. 2006; Breton et al. 2006; Belaj et al. 2007; Erre et al. 2010). They are also focusing on coming up with the generic associations in the different O. europaea subspecies that may be found outside of the Mediterranean region (Besnard et al. 2001; García-Verdugo et al. 2010). Due to the late growth in DNA inventions, a majority of these studies have been done by molecular markers and the SSRs are the most commonly used. Even so, ignoring the disadvantages of the regular morphological features, for instance natural effects, the requirement for a wider view of grown plants, SSR markers, and the combined usage of the morpho-agronomical features may provide the opportunity to abuse the essence of these two strategies in studying the genetic diversity of naturally growing olive trees (Karp et al. 1997; Sorkheh and Khaleghi 2016).
It has been largely known that the bias on olive cultivar founded on morphological representations cannot be relied on (Belaj et al. 2002, 2007, 2010, 2011) and in this regard the DNA molecular markers, precisely microsatellites, that is, the SSRs, are very commonly used today so as to add to the morphological studies and to explicitly acknowledge the increases held in collections (Bracci et al. 2011; Noormohammadi et al. 2014; Sorkheh and Khaleghi 2016).
There have been various accounts on genetic variation in the olive clone’s studies that have been done with molecular markers. The clones were acknowledged by use of RADP and ISSR (Gemas et al. 2004; Gomes et al. 2008; Martins-Lopes et al. 2009) also with AFLP (Frane et al. 2010) and last with microsatellites (Lopes et al. 2004; Muzzalupo et al. 2010; Zaher et al. 2011; Albertini et al. 2011; Ipek et al. 2012; Marra et al. 2013; Noormohammadi et al. 2014; Caruso et al. 2014; Abdessemed et al. 2015).
Even though there are ongoing studies to come up with dependable means of differentiating the mutation in genes, acknowledging a clone is still reliant on the study of phenotypic features that are associated with molecular studies. Breeders are coming up with genetic relationships genotypes and phenotypic information (Klocke et al. 2002). Iran happens to be one of the nations that are very rich in olive germplasm (Shahriari et al. 2008; Sorkheh and Khaleghi 2016) and as a result explaining and sorting this germplasm is a fundamental step in the picking and reproduction of olive populations. Due to this, effective use of genetic resources in plant growing ventures needs more knowledge in genetic diversity.
In the current research, AFLP and IRAP studies were used to evaluate the genetic disparity of O. europaea populations with the aim of using them in breeding projects and also in the preservation administration of the germplasm in Iran.
Materials and methods
Plant material
Here, 200 trees in the 20 groups of grown olives were studied and they were acquired from four countries, namely Abosatl (Syria), T5 (unknown), Dezfoli (Iran), Mishen (USA), Masabei (Syria), Conservolia (Europe), Kaeisei (Europe), Kaylit (Europe), T21 (Europe), Khoseari (Syria), Zard (Iran), Roghani (Iran), T7 (Europe), Manzanila (Europe), Kavi (Syria), T2 (Europe), Balidi (Syria), Mari (Syria), Foji (Europe), and Koroneiki (Europe). Ten of these individual represented each nation. The entire samples were taken from olives which were 5–10 years old (for more details see Table S1).
AFLP analysis
The amplification of PCR as well as the product electrophoresis AFLP assays was done as the procedure described by Zabeau (1993) and Vos et al. (1995). But there were some small improvements in regard to olive acquirements as stipulated by Sorkheh et al. (2007). The entire 20 primers were synthesized by use of MWG (Germany) (Supplementary Table S2). About 6 µl of 50 ng genomic DNA was digested with MseI and PstI in 10X buffer where as an MseI and PstI adapter were subsequently ligated to the digested DNA fragments. The arrangement of the adapters and the adjoining limited sites played the role of binding sites so as to amplify the picked fragments.
A certain population of fragments was amplified from the reaction mixture and this was done by adding nucleotides to the 3′ ends of the primers in two steps as stipulated by Sorkheh et al. (2007). In the first phase, one nucleotide was added in the initial amplification while three of them were added for the second amplification. DNA was PCR-amplified for 26 cycles using 5 µl of template DNA and +1 primer (MseI +1 and PstI +1). As for the PCR condition according to Soorni et al. (2013) and Sorkheh et al. (2016b) were as follows:
A first denaturation step was done at 94 °C for 2 min. 26 cycles were run at 94 °C for 60 s, 65 °C for 60 s, 72 °C for 60 s, and one last extension step of 5 min at 72 °C.
The second amplification step used +3 primers (MseI +3 and PstI +3). The DNA template for the second amplification step was the PCR product that was done by the initial phase. DNA was amplified for one cycle at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 60 s, then for 12 cycles with a 0.7 °C annealing temperature decrease per cycle, and last for 23 cycles at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s.
The products of the PCR after the second denaturing were mixed and denatured using an same volume of loading buffer which comprised of 98% formamide, 10 mM EDTA, 0.05% Xylene cyanol, 0.05% Bromophenol blue. It was then warmed for 5 min in 94 °C, frozen on ice then 6 µl of denatured preparation were put on per warmed (50 °C) polyacrylamide sequencing gels (Gibco BRL, model S2) which had 7 M urea in 1 urea in 1X TBE buffer run for 1.5–2 h at 100 W up to a point where the forward running dye (Bromophenol blue) reached the end of the gel.
As it had been earlier stated by Bassam and Caetano-Anollés (1993), the bands of DNA were seen by use of a silver staining. The AFLP bands which were in varying lengths of 67–501 bases were noted as present using (1) or absent using (0).
IRAP analysis
28 of the IRAP primers were evaluated for formal amplification as shown in supplementary Table S4. This amplification was done inside in thin-walled microcentrifuge tubes using thermocycler (iCycler, Bio Rad Co., USA). Reaction volume of 20 µl was composed of 2 µl 10X buffer (100 mmol L−1 Tris–HCl pH 9, 500 mmol L−1 KCl, 20 mmol L−1 MgCl2 and 0.1% Triton X-100), 1 unit Taq polymerase, 200 mmol L−1 of each dNTP, 30 nmol L−1 primer, and 20 ng DNA template. Each reaction mixture was overlaid with 10 µL mineral oil. A negative control reaction, whereby the DNA template was skipped, was then put in all the PCR run so as to make sure that there was no instance of self amplification and even DNA pollution. This amplification of DNA was done following the steps stated by Sorkheh et al. (2016a) and they were as follows: 4 min at 94 °C, 1 min at 36 °C, and 2 min at 72 °C for one cycle; 1 min at 94 °C, 1 min at 36 °C, and 2 min at 72 °C for 40 cycles; and 1 min at 94 °C, 1 min at 36 °C, and 10 min at 72 °C for one cycle. Amplified PCR products were electrophoresed through 1.5% agarose (Roche Co., Germany) stained with Safe stain (SinaClone, Iran) in TBE buffer and photographed under UV light, by a Gel Doc system (UVP, Bio Doc Co., USA).
Data analysis
In all the AFLP and the IRAP that were used in form of a character the presence or absence of the allele was scored in binary code. In the genetic association research, only independent, reproducible, and properly resolved AFLP and IRAP were noted as either present or absent and from the band scores that were noted, an information matrix was done.
GS was then evaluated in the entire groups of accessions by use of scorable fragments of AFLP and IRAP markers. By clustering the information then UPGMA (unweighted pair group method with arithmetic average), a dendrogram of genetic association was produced. After this, the Cophenetic relationship was evaluated, and the Mantel test (Mantel 1967) was applied to determine the suitability of the cluster evaluation to the similarity matrix where it was employed.
Simple matching similarity (SM) was employed for more comparison with the results that have been stated by previous studies. All this was done using the help of the NTSYS-pc 2.02 software package (Rohlf 1998). A bootstrap study using 1000 replicates was conducted to evaluate the relative support for the various groups and the steadiness of the dendrogram by use of the TREECON program verison 1.3 as stated by Van de Peer and De Wachter (1994).
The data content of each single marker was noted as PICi = 1 − ∑p i 2, where p i is the frequency of the ith band. The average PIC (polymorphic information content) was then evaluated for the IRAP and the AFLP markers in all assay units and this was done by using the formula stated above. This formula was invented by Powell et al. (1996).
Each single DNA fragment that was seen in the gel was taken to be a distinct prevailing locus. The polymorphic bands that had a high intensity were scored. Singly, a marker was recognized using the primer combination and the band number as suffix. The data matrix did excluded markers in that had a molecular weight of less than 100 bp thus they were removed from it. The polymorphism information content (PIC) was the one that was used to evaluate the discrimination power of each AFLP and IRAP. Within-accession diversity (HS) and total gene diversity (HT) (Nei 1973) were calculated within the species and major groups using the POPGENE software. The markers, monomorphic and polymorphic, were used to make the calculations. In conclusion, analysis of molecular variance (AMOVA) was used to differentiate total genetic variation among and in between population following Excoffier et al. (1992). The percent of polymorphism, the observed (Na) and effective number (Ne) of alleles, and Shannon’s information desk (SI) were calculated. Population differences were then found out per statistics using the tests made by the students. Those that looked alike genetically within populations were approximated by Nei’s pair-wise genetic distance (D) and genetic identity (I) (Nei 1987); these calculations were made using the GeneAlex ver 6.5 using 1000 permutations (Peakall and Smouse 2006).
There was a tool used for population genetic analysis, (Pritchard et al. 2000, 2010) called the Markov chain Monte Carlo (MCMC), that was used to analyze the population structure., following the steps that were provided and described for the dominant markers by Falush et al. (2007) when used both admixture and no admixture models and go to the optimal number of cluster (K) thought determination of posterior log likelihood.[In (K)] and second order rate of change in the likelihood function (ΔK) as suggested by Evanno et al.(2005). The structure program was then run at preferred values with a burn in period of 100,000 and a MCMC model with repetitions.
Results
AFLP analysis
The level of polymorphism in the 23 AFLP primers was evaluated in the 200 O. europaea L. accessions. The methods of variability were studied as shown in Table S3. 1090 amplification fragments were seen and roughly 47.39 fragments/AFLP and also fragment sizes that had sizes ranging from 80 to 126 and sizes of 65–101 bp. These finding are in line with other studies that have been carried out earlier (Sorkheh et al. 2007, 2016b; Russi et al. 2009; Rahimmalek et al. 2009).
In the current research, 940 bands were polymorphic with an average polymorphism of 87.15% with 40.86 bands/primer combinations and this was more than the study conducted by Russi et al. (2009) and Rahimmalek et al. (2009) in Echinacea and Achillea, respectively. The notable disparity in this research may be attributed to accessions, the various locations, and the used primer combinations. These findings were evidence that the AFLP method can yield results and it is also economical and joins the reliability of RFLP and the power of PCR (Vos et al. 1995).
The amplified band ranges were as follows: from 24 (for M-CCT + P-GGA primer combination) to 78 (for M-G + P-AAC primer combination). M-CAC + P-GCA, M-CAG + P-GTA, M-GAG + P-ACC, M-GCA + P-AAC, M-GTC + P-AAC, M-CCT + P-GGA, and M-CGA + P-GTT primer combination showed the highest 44, 55, 35, 43, 24, and 42 polymorphic bands with 100% polymorphism. Polymorphism information content (PIC) varied from 0.68 (for M-GCA + P-AAC primer combination) to 0.94 (for M-GTC + P-AGC primer combination) with an average of 0.81.
The pair-wise genetic resemblance that was noted in regard to the proportion of shared fragments from 0.06 to 0.70 shows that there was quite some distance and disparity in and between the populations. As it had been stated earlier, these high genetic distances are characteristics AFLP in the sense that they are able to come up with high levels of diversity. This can be confirmed more so among close associates of inter-breeding species where there is a high likelihood of a reticulate evolution occurrence. When the right statistical tools are employed (Yeh and Boyle 1997) AFLPs can also serve the purpose of assessing the genetic structure and the differentiation in and between the species and populations (Sorkheh and Khaleghi 2016; Sorkheh et al. 2016a, b).
The UPGMA cluster study showed the genetic associations in and between the populations. The CCC (cophenetic correlation coefficient) showed a high correlation of 0.94 linking the similarity matrix and the cophenetic matrix (CM). All these were acquired from the UPGMA dendrogram, and it is evidence that there is a proper representation of the molecular associations between the genotypes.
The CCC is regarded as a very ideal representation of the information matrix in the dendrogram, that is, if it is beyond 0.90 (Romesburg 1990). The dendrogram clearly distinguished all the accessions into three key clusters as shown in Fig. 1. Here, the bootstrap values fully back the three clusters. The first key cluster has the accessions from Syria which is group 1. This key cluster that was supported in majority of the cases with values exceeding 78% was evidence that T5 with an unrecognized location could be from Syria. Group 2 had accessions from Europe and the third group had accessions from Iran and the USA. This third group had a bootstrap value exceeding 67%. In general, the accessions were divided in regard to the populations and the sites that they were collected from. Those with similar features were grouped together. Due to this, the similarity matrix and the cluster study showed a high genetic variation on each population that had distinct geographical locations.
Fig. 1.

Dendrogram obtained with the simple matching (SM) similarity coefficient pair group method with arithmetical average clustering algorithm from 1090 AFLP markers for 200 olive accessions. The value on the dendrogram gives the stability of nodes estimated with a bootstrap procedure (no number indicates support less than 10%). Bar graph showing genetic diversity structures for the accessions of O. europaea L. as assessed using STRUCTURE software. Each group is represented by a different color (blue, red and green)
There was a very high genetic diversity from the populations acquired from Iran and the USA by use of the Shannon’s Information Index. The populations from Iran were (H = 0.2730.20, I = 0.401) and those from Syria which were the lowest too were (H = 0.24, I = 0.37) (Table 1). When the AMOVA procedure was carried out, the within-population molecular variation showed a high genetic variation of 58% and the within populations was 42% such that the accessions acquired from Iran had the most variable intra-population diversity as shown in Table 2.
Table 1.
Information on number of observed alleles (Na), number of effective alleles (Ne), Nei’s gene diversity (h), and Shannon’s information index (SI), for AFLP and IRAP markers 200 O. europaea L. accessions utilized within this study
| Population | Naa | Nea | Ha | SIa | Groupc | HT | HS | Gst | Nmb |
|---|---|---|---|---|---|---|---|---|---|
| AFLP | |||||||||
| Iran | 1.95 ± 0.38 | 1.55 ± 0.38 | 0.273 ± 0.18 | 0.401 ± 0.24 | Group I | 0.348 ± 0.014 | 0.271 ± 0.009 | 0.222 | 1.75 |
| Syria | 1.69 ± 0.44 | 1.49 ± 0.36 | 0.24 ± 0.19 | 0.37 ± 0.26 | Group II | 0.382 ± 0.013 | 0.289 ± 0.011 | 0.259 | 1.42 |
| USA | 1.82 ± 0.22 | 1.53 ± 0.21 | 0.36 ± 0.14 | 0.42 ± 0.19 | Group III | 0.341 ± 0.016 | 0.311 ± 0.014 | 0.087 | 5.20 |
| Europe | 1.87 ± 0.35 | 1.42 ± 0.28 | 0.31 ± 0.16 | 0.56 ± 0.22 | |||||
| Total | 1.071 ± 0.043 | 0.871 ± 0.034 | |||||||
| IRAP | |||||||||
| Iran | 1.87 ± 0.32 | 1.45 ± 0.37 | 0.235 ± 0.15 | 0.503 ± 0.25 | Group I | 0.354 ± 0.013 | 0.286 ± 0.006 | 0.234 | 1.64 |
| Syria | 1.56 ± 0.32 | 1.39 ± 0.45 | 0.25 ± 0.13 | 0.43 ± 0.32 | Group II | 0.376 ± 0.015 | 0.276 ± 0.012 | 0.214 | 1.84 |
| USA | 1.74 ± 0.23 | 1.55 ± 0.32 | 0.22 ± 0.17 | 0.36 ± 0.15 | Group III | 0.354 ± 0.017 | 0.298 ± 0.016 | 0.067 | 6.96 |
| Europe | 1.65 ± 0.36 | 1.34 ± 0.28 | 0.29 ± 0.16 | 0.24 ± 0.14 | |||||
| Total | 1.084 ± 0.045 | 0.860 ± 0.034 |
Table 2.
List of the IRAP primers used in the study and Level of polymorphism and informativeness obtained in 200 accessions of O. europaea L.
| IRAP primers | Total bands | Polymorphic bands | Polymorphism (%) | PIC | MI |
|---|---|---|---|---|---|
3 LTR-3 LTR-3 LTR LTR |
15 | 12 | 80.0 | 0.93 | 74.40 |
3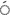 LTR-LTR6150 LTR-LTR6150 |
12 | 10 | 83.3 | 0.62 | 51.67 |
| LTR6150-Sukkula | 11 | 8 | 72.7 | 0.82 | 59.64 |
| Sukkula–Sukkula | 24 | 22 | 91.7 | 0.89 | 81.58 |
| Sukkula-Nikita | 13 | 13 | 100.0 | 0.91 | 91.00 |
Sukkula-3 LTR LTR |
16 | 16 | 100.0 | 0.93 | 93.00 |
5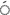 LTR2-Sukkula LTR2-Sukkula |
15 | 13 | 86.7 | 0.97 | 84.07 |
| LTR6150-LTR6150 | 13 | 11 | 84.6 | 0.91 | 77.00 |
| Average | 14.88 | 13.13 | 87.38 | 0.87 | 76.54 |
| Total | 119.00 | 105.00 | 699.01 | 6.98 | 612.35 |
IRAP analysis
From the prescreening assays with two O. europaea L. accessions by use of 28 IRAP primers, 8 primers brought about bright polymorphic products that were incorporated in further studies. Table 2 shows the findings of the IRAP fingerprinting of the 200 researched accessions by use of eight primers. These eight primers are responsible for the amplified polymorphic products and represented polymorphism in all the accessions and the quantity of bands detected was 119, but that number was varying from 11 (LTR6150-Sukkula) to 24 (Sukkula–Sukkula) with an average of 14.88 bands per primer with size of 200–800 bp. The total of separable bands were 105 and ranged from 8 (LTR6150-Sukkula) to 22 (Sukkula–Sukkula) with an average of 13.13 per primer.
In the current research, it was seen that the average polymorphism was 87.38% and that two primers, Sukkula-Nikita and Sukkula-3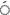 LTR, represented 100% polymorphism. In the germplasm that was studied the average PIC value was 0.87 starting from 0.62 (30 LTR-LTR6150) to 0.97 (5
LTR, represented 100% polymorphism. In the germplasm that was studied the average PIC value was 0.87 starting from 0.62 (30 LTR-LTR6150) to 0.97 (5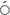 LTR2-Sukkula primer), and the average value for MI was 76.54 starting from 51.67 (3
LTR2-Sukkula primer), and the average value for MI was 76.54 starting from 51.67 (3 LTR-LTR6150) to 93.00 (Sukkula-3
LTR-LTR6150) to 93.00 (Sukkula-3 LTR). Out of the eight primers, seven of them had a PIC value higher than 0.74. Due to the high polymorphic value of the studied band, it was possible to differentiate the genetic variation among and between populations.
LTR). Out of the eight primers, seven of them had a PIC value higher than 0.74. Due to the high polymorphic value of the studied band, it was possible to differentiate the genetic variation among and between populations.
Relying on the findings that were found by the IRAP markers, a similarity matrix was employed to come up with a UPGMA dendrogram as shown in Fig. 2. The CCC between the initial similarity matrix and the CM that was acquired from the UPGMA dendrogram was found to be very high at about 0.96, and this is evidence that there is a good fit of the similarity matrix and the dendrogram. There were three key clusters that were shown by the dendrogram in the group 1 of Syria, group 2 of Europe that had 100% bootstrap, and also group 3 that was inclusive of Iran and the USA. The separation of the accession by the locations in which they were acquired was similar to the diagram acquired from the AFLP markers. More so, the accessions were separated according to their populations and the locations sites then put in the same subgroup.
Fig. 2.
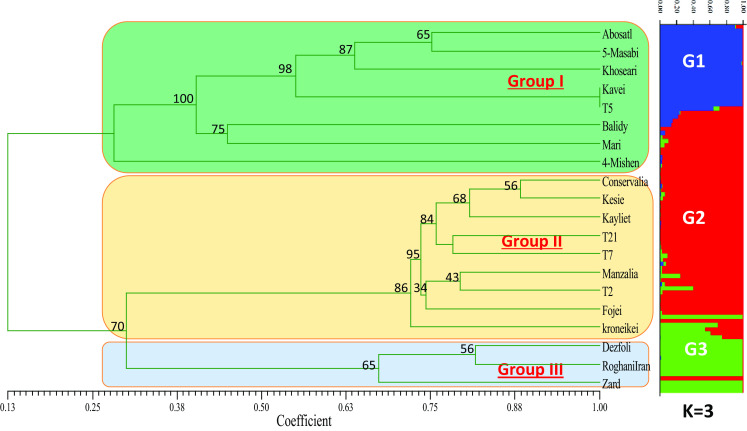
Dendrogram obtained with the simple matching (SM) coefficient pair group method with arithmetical average clustering algorithm from 119 IRAP markers for 200 olive accessions. The value on the dendrogram gives the stability of nodes estimated with a bootstrap procedure (no number indicates support less than 10%). Bar graph showing genetic diversity structures for the accessions of O. europaea L. as assessed using STRUCTURE software. Each group is represented by a different color (blue, red and green)
The genetic disparity in the populations by the use of Shannon’s Information index (I) and the Nei’s gene diversity index (H) produced the greatest genetic diversity from the accessions acquired from Syria (H = 0.25, I = 0.43) and the minimal from those in the USA (H = 0.29, I = 0.24 (Table 3). The AMOVA represented the genetic variation among the populations as 49% but the difference in the populations was at 51% only as represented in Table 3.
Table 3.
Analysis of molecular variance (AMOVA) for 200 olive accessions based on 1090 AFLP and 119 IRAP markers
| Source | AFLP | IRAP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| df | Sum of square | Mean square | Variation (%) | df | Sum of square | Mean square | Variation (%) | Significanceb | |
| Amonga pops | 4 | 1453.545 | 363.38 | 58 | 4 | 673.911 | 168.48 | 49 | P < 0.001 |
| Within pops | 195 | 1751.879 | 8.98 | 42 | 195 | 457.536 | 2.35 | 51 | P < 0.001 |
| Total | 199 | 100 | 199 | 100 | |||||
aThe olive accessions from four origins including Iran, Syria, USA, and Europe with 10 individuals representing each region. All the samples were collected from young orchards composed of 5- to 10-year-old olive trees (see Table 1)
bSignificance of variance component expressed as the probability of obtaining a more extreme random value computed from nonparametric procedures (1000 data permutation)
Discussion
The principal discussion behind the study of the O. europaea L. in this research was that despite the variety of this crop being high in Iran, there was very minimal information about it. The current research brought to surface a very high concentration of the O. europaea L. in regard to the AFLP and the IRAP marker. In total, 940 and 105 polymorphic bands with 87.15 and 87.38% were differentiated by the AFLP and IRAP in the researched species. Yu et al. (2009) detailed 65 polymorphic bands among the accessions of Leonurus japonicus by the AFLP. In addition, Chen et al. (2009) stated that 117 polymorphic bands among L. japonicus populations by ISSR marker, confirming that there is indeed a broad genetic difference in our germplasm. Also, the findings of the study can be depended on with previous discoveries showing the broad genetic bias of olive germplasm (Zohary and Spiegel Roy 1975). It has also been confirmed that got with isozymes (Pontikis et al. 1980; Ouazzani et al. 1993, 1995; Trujillo et al. 1995), with RAPDs (Fabbri et al. 1995) and with AFLP markers (Angiolillo et al. 1999).
Grouping relying on these two markers separated the accessions into three key groups such that the data set by these two markers were similar and it is as if that the two genetic markers could separate promotions at similar genetic levels. UPGMA dendrograms show that the accessions from Iran are the most distinct from the rest in regard to molecular information. The explanation of this maybe its location despite the fact that it has certain morphological characteristics when likened to other populations, for instance, leaf shape, leaf width, leaf length, stone width, stone length, stone shape index, fruit width, fruit length, fruit shape index (Sorkheh and Khaleghi 2016). The dendrogram shows that there is indeed a close association between the researched populations.
The estimated correlation between matrices of AFLP and IRAP data (r = 0.83) showed a good relationship between these information. These markers show certain regions of the nuclear genome and study polymorphism in these certain locations. By this, it can be seen that the recorded areas by AFLP markers has coordinate correspondence with data presented by IRAP. There are some researches of crops with a good relationship between molecular information data (Landry et al. 1994; Shimada et al. 1999; Horvath et al. 2008).
The dendrogram that relies on incorporating the information of the two markers acknowledges the considered populations in reference to the results that were acquired from each. The advantage of these joined datasets was a comprehensive taxonomic picture due to the fact that each is a representative of a distinct level of taxonomic separation and the confirmed view as seen by Sneller et al. (1997) in this particular situation. Last, due to the transfer and the polymorphism of majority of these markers, the authors suggested that the AFLP and the IRAP primers to be enough in the O. europaea L. populations and any associated species. In addition, this result will be an important resource for the future of breeding and the methodologies for operation, for example, foundation and study of seed plantation, evaluation of controlled crosses, and clonal spread and dispersal.
Conclusion
The study of AFLP and IRAP profiles in the group of 200 accessions of O. europaea L. germplasm showed a broad genetic difference in the Iranian olive germplasm and a clear structure of genetic variability can be seen between those cultivars. This new information is very essential for opening extensive horizons and the growth of new kinds with developed chemical and horticultural features.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by Shahid Chamran University of Ahvaz, Iran. We thank the anonymous reviewer for his/her most helpful comments. We are grateful to Mis Kh. Chenaneh-Hanoni for kind help in undertaking this study. This study has been supported by the Project SCH_AGC_No. 1231_1395, entitled “Fingerprinting of olive trees collection” of Shahid Chamran University of Ahvaz and Ataturk University.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
All authors contributed equally to this work.
Change history
8/1/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s13205-022-03266-4
References
- Abdessemed S, Muzzalupo L, Benbouz H. Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. Sci Horti. 2015;192:10–20. [Google Scholar]
- Albertini E, Torricelli R, Bitocchi E, Raggi L, Marconi G, Pollastri L, Di Minco G, Battistini A, Papa R, Veronesi F. Structure of genetic diversity in Olea europaea L. cultivars from central Italy. Mol Breed. 2011;27:533–547. [Google Scholar]
- Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphism. Theor Appl Genet. 1999;98:411–421. [Google Scholar]
- Baldoni L, Belaj A (2009) Oil crops: olive. In: Vollmann, J., Rajcan, I. (Eds.), Handbook of Plant Breeding. Springer Series, doi:10.1007/978-0-387-77594-4 13
- Baldoni L, Tosti N, Ricciolini C, Belaj A, Arcioni S, Pannelli G, Germana MA, Mulas M, Porceddu A. Genetic structure of wild and cultivated olives in the Central Mediterranean Basin. Ann Bot. 2006;98:935–942. doi: 10.1093/aob/mcl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco D, Trujillo I, Rallo L. Elaiografía Hispánica. In: Rallo L, Barranco D, Caballero JM, del Rio C, Martín A, Tous J, Trujillo I, editors. Variedades de olivo en Espa˜na. Madrid: Junta de Andalucía, MAPA-Ediciones Mundi-Prensa; 2005. pp. 48–76. [Google Scholar]
- Bartolini G, Prevost G, Messeri C, Carignani C (2005) Olive Germplasm: Cultivars and World-wide Collections. FAO/Plant Production and Protection, Rome, Available at: http://www.apps3.fao.org/wiews/olive/oliv.jsp
- Bassam BJ, Caetano-Anollés G. Silver staining of DNA in polyacrylamide gels. Appl Biochem Biotechnol. 1993;42(2):181–188. [Google Scholar]
- Belaj A, Satovic Z, Rallo L, Trujillo I. Genetic diversity and relationships in olive (Olea europaea L.) germplasm collections as determined by randomly amplified polymorphic DNA. Theor Appl Genet. 2002;105:638–644. doi: 10.1007/s00122-002-0981-6. [DOI] [PubMed] [Google Scholar]
- Belaj A, Munoz-Diez C, Baldoni L, Porceddu A, Barranco D, Satovic Z. Genetic diversity and population structure of wild olives from North-Western Mediterranean assessed by SSR markers. Ann Bot. 2007;100:449–458. doi: 10.1093/aob/mcm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Munoz-Diez C, Baldoni L, Satovic Z, Barranco D. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Sci Hortic. 2010;124:323–330. [Google Scholar]
- Belaj A, León L, Satovic Rosa R. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agro-morphological traits and SSR markers. Sci Horti. 2011;129:561–569. [Google Scholar]
- Besnard G, Baradat P, Bervillé A. Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theor Appl Genet. 2001;102:251–258. [Google Scholar]
- Bracci T, Busconi M, Fogher C, Sebastiani L. Molecular studies in olive (Olea europaea L.): overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep. 2011;30:449–462. doi: 10.1007/s00299-010-0991-9. [DOI] [PubMed] [Google Scholar]
- Breton C, Tersac M, Bervillé A. Genetic diversity and gene flow between the wild olive (Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J Biogeogr. 2006;33:1916–1928. [Google Scholar]
- Caruso T, Marra FP, Costa F, Campisi G, Macaluso L, Marchese A. Genetic diversity and clonal variation within the main Sicilian olive cultivars based on morphological traits and microsatellite markers. Sci Horti. 2014;180:130–138. [Google Scholar]
- Chen L, Zhao L, Bai Y, Hu R, Si J. Genetic relationship analysis of different provenances of Leonurus japonicus by ISSR marker. Zhongguo Zhong Yao Za Zhi. 2009;34(11):1343–1345. [PubMed] [Google Scholar]
- Erre P, Chessa I, Munoz-Diez C, Belaj A, Rallo L, Trujillo I. Genetic diversity and relationships between wild and cultivated olives in Sardinia as assessed by SSR markers. Genet Res Crop Evol. 2010;57:41–54. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri A, Hormaza JI, Polito VS. Random amplified polymorphic DNA analysis of olive (Olea europaea L.) J Amer Soc Hort Sci. 1995;120:538–542. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null allele. Mol Ecol. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frane S, Dunjia BM, Slavko P, Zlatko C, Zlatko S, Branka J. Genetic variation within the olive (Olea europaea L.) cultivar Oblica detected using amplified fragment length polymorphism (AFLP) markers. Afr J Biot. 2010;9:2880–2883. [Google Scholar]
- García-Verdugo C, Forrest AD, Balaguer L, Fay MC, Vargas P. Parallel evolution of insular Olea europaea subspecies based on geographical structuring of plastid DNA variation and phenotypic similarity in leaf traits. Bot J Linn Soc. 2010;162:54–63. [Google Scholar]
- Gemas VJV, Almadanim MC, Tenreiro R, Martins A, Fevereiro P. Genetic diversity in the olive tree (Olea europaea L. subsp. europaea) cultivated in Portugal revealed by RAPD and ISSR markers. Genet Res Evol. 2004;51:501–511. [Google Scholar]
- Gomes S, Martins-Lopes P, Lima-Brito J, Meirinhos J, Lopes J, Martins A, Guedes-Pinto H. Evidence of clonal variation in olive ‘Verdeal-Transmontana’ cultivar using RAPD, ISSR and SSR markers. J Hortic Sci Biotechnol. 2008;83:395–400. [Google Scholar]
- Horvath A, Christmann H, Laigret F. Genetic diversity and relationships among Prunus cerasifera (cherry plum) clones. Botany. 2008;86:1311–1318. [Google Scholar]
- Ipek A, Barut E, Gulen H, Ipek M. Assessment of inter- and intra-cultivar variations in olive using SSR markers. Sci Agric. 2012;69:327–335. [Google Scholar]
- Karp A, Kresovich S, Bhat KV, Ayad WG, Hodgkin T (1997) Molecular tools in plant genetic resources conservation: a guide to the technologies. IPGRI technical bulletin No. 2. International Plant Genetic Resources Institute, Rome, Italy
- Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocke E, Langbehn J, Grewe C, Pank F. DNA fingerprinting by RAPD on Origanum majorana. J Herbs Spices Med Plants. 2002;9:171–176. [Google Scholar]
- Landry BS, Li RQ, Cheung WY, Granger RL. Phylogeny analysis of 25 apple rootstocks using RAPD markers and tactical gene tagging. Theor Appl Genet. 1994;89:847–852. doi: 10.1007/BF00224507. [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1972) The apportionment of human diversity. In: Dobzhansky T, Hecht M, Steere WC (eds.) Springer book archive, evolutionary biology pp 381–398. doi:10.1007/978-1-4684-9063-3_14
- Lopes MS, Mendonc AD, Sefc MK, Gil FS, Da Camara Machado A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. Hort Sci. 2004;39:1562–1565. [Google Scholar]
- Lumaret R, Ouazzani N, Michaud H, Vivier G, Deguilloux MF, Di Giusto F. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean basin. Heredity. 2004;92:343–351. doi: 10.1038/sj.hdy.6800430. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Marra FP, Caruso T, Costa F, Di Vaio C, Mafrica R, Marchese A. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. subsp. europaea) cultivated in Southern Italy revealed by SSR markers. Tree Gene Genom. 2013;9:961–973. [Google Scholar]
- Martins-Lopes P, Gomes S, Lima-Brito J, Lopes J, Guedes-Pinto H. Assessment of clonal genetic variability in Olea europaea L. ‘Cobrancosa’ by molecularmarkers. Sci Hortic. 2009;123:82–89. [Google Scholar]
- McDermott JM, McDonald BA. Gene flow in plant pathosystems. Ann Rev Phytopath. 1993;31:353–373. doi: 10.1146/annurev.py.31.090193.002033. [DOI] [Google Scholar]
- Muzzalupo I, Chiappetta A, Benincasa C, Perri E. Intra-cultivar variability of three major olive cultivars grown in different areas of central-southern Italy and studied using microsatellite markers. Sci Hortic. 2010;126:324–329. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci (USA) 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- Noormohammadi Z, Trujillo I, Belaj A, Ataei A, Hosseini-Mazinan M. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. Sci Horti. 2014;178:175–183. [Google Scholar]
- Ouazzani N, Lumaret R, Villemur P, Di Giusto F. Leaf allozyme variation in cultivated and wild olive trees (Olea europaea L.) J Hered. 1993;84:34–42. doi: 10.1038/sj.hdy.6800430. [DOI] [PubMed] [Google Scholar]
- Ouazzani N, Lumaret R, Villemur P. Apport du polymorphisme alloenzymatique a` l’identification vari_etale de l’olivier (Olea europaea L) Agronomie. 1995;15:31–37. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontikis CA, Loukas M, Kousounis C. The use of biochemical markers to distinguish olive cultivars. J Hort Sci. 1980;55(4):333–343. [Google Scholar]
- Powell W, Morgante M, Andre C, Hanafey Mm Vogel J, Tingey S, Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225–238. [Google Scholar]
- Pritchard JK, Stevens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D (2010) Documentation for structure software: Version 2.3. http://pritch.bsd.uchicago.edu/software/structure_v.2.3.1/documentation.pdf. (accessed 15 Sept. 2012)
- Rahimmalek M, Ebrahim B, Arzani A, Etemadi N. Assessment of genetic diversity among and within Achillea species using amplified fragment length polymorphism (AFLP) Biochem Systemat Ecol. 2009;37:354–361. [Google Scholar]
- Rohlf FJ (1998) NYSYS-pc. Numerical taxonomy and Multivariate Analysis System, Version 2.02 Exeter Software, Setauket, NY
- Romesburg HC. Cluster analysis for researchers. Malabar: Krieger Publishing; 1990. [Google Scholar]
- Russi L, Moretti C, Raggi L, Albertini E, Falistocco E (2009) Identifying commercially relevant Echinacea species by AFLP molecular markers, vol. 52. NRC Research Press, pp 912–918 [DOI] [PubMed]
- Shahriari M, Omrani A, Falahati-Anbaran A, Ghareyazei B, Nankali A. Identification of Iranian olive cultivars by using RAPD and microsatellite markers. Acta Horti. 2008;791:109–115. [Google Scholar]
- Shimada T, Hayama H, Haji T, Yamaguchi M, Yoshida M. Genetic diversity of plums characterized by random amplified polymorphic DNA (RAPD) analysis. Euphytica. 1999;109:143–147. [Google Scholar]
- Sneller CH, Miles JW, Hoyt JM. Agronomic performance of soybean plant introductions and their genetic similarity to elite lines. Crop Sci. 1997;37:1595–1600. [Google Scholar]
- Soorni A, Nazeri V, Fattahi R, Khadivi-Khub A. DNA fingerprinting of Leonurus cardiaca L. germplasm in Iran using amplified fragment length polymorphism and interretrotransposon amplified polymorphism. Bioch Syst Ecolo. 2013;50:438–447. [Google Scholar]
- Sorkheh K, Khaleghi E. Molecular characterization of genetic variability and structure of olive (Olea europaea L.) germplasm collection analyzed by agromorphological traits and microsatellite markers. Turk J Agric For. 2016;40:583–596. [Google Scholar]
- Sorkheh K, Shiran B, Gradziel TM, Epperson BK, Martinez-Gomez P, Asadi E. Amplified fragment length polymorphism as a tool for molecular characterization of almond germplasm: genetic diversity among cultivated genotypes and related wild species of almond, and its relationships with agronomic traits. Euphytica. 2007;156:237–344. [Google Scholar]
- Sorkheh K, Amirbakhtiar N, Ercislie S. Potential starts codon targeted (SCoT) and interretrotransposon amplified polymorphism (IRAP) markers for evaluation of genetic diversity and conservation of wild Pistacia species population. Bioch Genet. 2016;54:368–387. doi: 10.1007/s10528-016-9725-1. [DOI] [PubMed] [Google Scholar]
- Sorkheh K, Masaeli M, Hosseini Chaleshtori M, Adugna A, Ercisli S. AFLP-based analysis of genetic diversity, population structure, and relationships with agronomic traits in rice germplasm from north region of IRAN and world core germplasm set. Biochem Genet. 2016;54:177–193. doi: 10.1007/s10528-016-9711-7. [DOI] [PubMed] [Google Scholar]
- Trujillo I, Rallo L, Arus P. Identifying olive cultivars by isozyme analysis. J Amer Soc Hort Sci. 1995;120(2):318–324. [Google Scholar]
- Van de Peer Y, De Wachter R. TREECON for Windows: as software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucl Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Boyle TJB. Population genetic analysis of codominant and dominant markers and quantitative traits. Belg J Bot. 1997;129:157. [Google Scholar]
- Yu Q, Shen X, Shen Y, Chen J, Shi C, Wang Z. AFLP analysis of genetic diversity of Leonurus japonicus germplasm resources. Zhongcaoyao. 2009;40:1296–1299. [Google Scholar]
- Zabeau M (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application No. 0-534-858-A1
- Zaher H, Boulouha B, Baaziz M, Sikaoui L, Gaboun F, Sripada M. Morpho-logical and genetic diversity in olive (Olea europaea subsp. europaea L.) clones and varieties. POJ. 2011;4:370–376. [Google Scholar]
- Zohary H, Spiegel Roy P. Beginnings of fruits growing in the old world. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


