Abstract
Coptis herbs, which are important herbal medicinal materials, are the dried rhizomes of various plants. In China’s herbal market, drying herbs can severely influence morphological markers such as shape, color, and odor, which make consumers difficult to precisely identify the herbs and effectively evaluate the quality. Here, we present the phylogenetic analysis of four Coptis herbal medicinal materials based on internal transcribed spacer sequences. C. chinensis, C. omeiensis, C. deltoidea and C. teeta constituted a monophyletic group. In this group, C. omeiensis and C. deltoidea cluster together, and they form the sister group with C. teeta, and C. chinensis locate the outermost of C. omeiensis, C. deltoidea and C. teeta. At the same time, the HPLC–DAD method was applied to simultaneously analyze main alkaloids from seventeen different samples. On the basis of the robust phylogenetic topology, the parsimony reconstructions of six effective medicinal constituents were implemented to elucidate the evolutionary history in Coptis herbs, and thus evaluate the herbal quality. The results showed that C. chinensis had been considered to be the best herb, not only the high content in single constituent but also in total alkaloids. In addition, all the samples from seventeen habitats were considered as qualified herbs, and they could meet the requirements of national quality standards for Coptis herbs in China.
Keywords: Coptis herbs, Internal transcribed spacer (ITS), Evolution, Alkaloid, Herbal quality
Introduction
Coptis herbs, as famous Traditional Chinese Medicine, have been used for prevention and treatment of human diseases, such as cancer and inflammation for centuries (He et al. 2015). Coptis herbs can clear away heat and drying dampness in the treatment of diarrhea, dysentery, jaundice, hematemesis, carbuncles, and abscesses. They also purge the sthenic fire and clear away toxic material in cases of seasonal febrile disease, carbuncles, sore throats, etc. (The Pharmacopoeia of the People’s Republic of China 2015; Liu et al. 2006). According to the Pharmacopoeia of the People’s Republic of China (2015 edition), Coptis herbs are the dried rhizomes of ranunculaceous plants including Coptis chinensis Franch., Coptis deltoidea C.Y. Cheng et Hsiao, and Coptis teeta Wall (Qiao et al. 2009), and these three species are all distributed and endemic in Southwest China (Teng and Choi 2012), especially in Sichuan, Chongqing, Yunnan, and Hubei. Concurrently, the Coptis omeiensis C.Y. Cheng was always used as the substitute because of their similar medicinal constituents. Traditional means of herbal species classification or taxonomy mainly rely on the inspection of morphological markers such as shape, color, texture, and odor (Heinrich 2007). However, the morphological markers were destroyed by dry herbs, and thus, it became difficult to classify the different species and evaluate the quality even for the specialists (Song et al. 2009). So it is imperative to find new ways and/or methods to classify the Coptis herbs from different sources and to evaluate the herbal quality.
A number of research groups have used molecular systematics methods to reconstruct plant systematics and solve systematic problems, as it is a difficult problem in previous taxonomic studies (Hörandl et al. 2005; Lane et al. 2007; Yassin et al. 2010). To date, the molecular phylogeny of genus Coptis has been reported (He et al. 2014; Xiang et al. 2016), but too little attention was paid to the medicinal plants of genus Coptis. In Southwest China, various climatic zones and different soil types provided a wide range of environmental conditions for the growth with profoundly different vegetation types (Zhang et al. 2004), and environmental factors possibly produced conservation of genetic mutations (He et al. 2014). So, molecular phylogenetic methods could be used to fully resolve the species taxonomy of Coptis herbs.
Phytochemical and pharmacological studies on medicinal plants revealed that Coptis herbs contained a number of alkaloids, such as berberine, palmatine, jatrorrhizine, coptisine, columbamine, and epiberberine (Kuang 2000; Qiao et al. 2009). Since application of Coptis herbs is growing steadily, development of a useful and suitable method for their quality control was urgently required (Kong et al. 2009). Berberine had been usually used as a marker to monitor the drug quality in the official Chinese Pharmacopoeia. Comparing with a single berberine, the assay for a set of main alkaloids is an effective way to evaluate the quality of botanical drugs. Thus, the qualitative and quantitative analyses of several high-content constituents in Coptis herbal extracts will help us better evaluate and regulate the drug quality.
Methods
Medicinal plant materials and sequencing
Samples were collected from seventeen different habitats in Southwest China (Fig. 1). Leave and rhizome samples were separated from different plant sources. Microquantities of DNA were isolated directly from dried specimens using a modified CTAB method (Doyle and Doyle 1987). Amplifications of the nuclear ITS regions were carried out using the universal primer set, forward primer (5′-AACAAGGTTTCCGTAGGTGA-3′), and reverse primer (5′-TATGCTTAAA YTCAGCGGGT-3′) (Nickrent et al. 1994). The PCR was conducted as follows: denaturation at 94 °C for 4 min, 30 cycles at 94 °C for 30 s, 45 °C for 30 s, and 72 °C for 2 min, and then 72 °C for 5 min (Schuettpelz et al. 2002). PCR products were sent to Sangon Biotech (Shanghai, China) Co., Ltd., for sequencing. And the sequencing results were submitted to NCBI (Accession numbers: KX983995-KX984011).
Fig. 1.
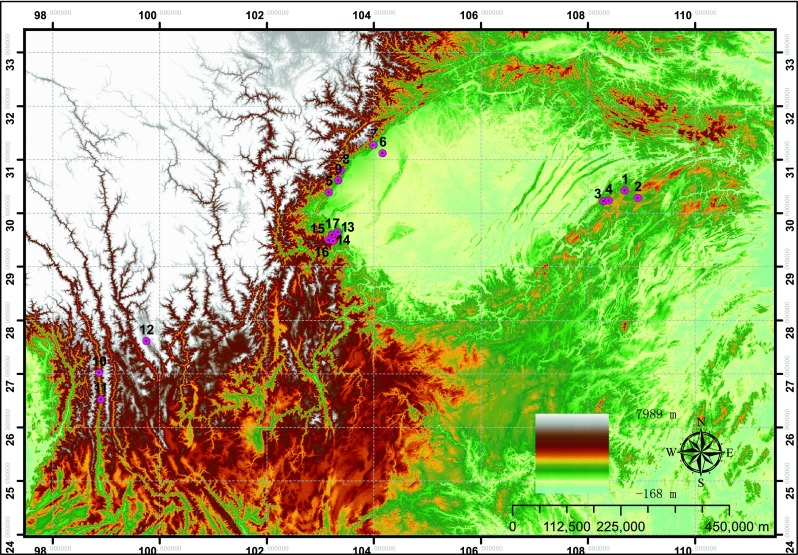
Sampling of the four Coptis species. Map showing the sampling sites of the here included specimens
Preparation of sample solution
All the tested samples were crushed into powder and dried at 50 °C for 24 h before use. The dried powders (approximately 100 mg) were accurately weighed into a clean Erlenmeyer flask, and extracted with 50 mL of hydrochloric acid–methanol solution (1:100, v/v) by ultrasonication for 30 min. After filtration, the filtrate was adjusted to the original weight with the extraction solvent. The sample solutions were filtered through a 0.45-μm membrane prior to an injection into the HPLC system. The concentrations of berberine, palmatine, coptisine, epiberberine, columbamine, and jatrorrhizine in each sample were determined using the established calibration curves.
Apparatus and chromatographic conditions
HPLC analysis was performed on an Agilent 1200 Series LC system (Agilent Technologies, Santa Clara, CA, USA) equipped with an auto-sampler, a quaternary pump, a diode array detector (DAD), and a column thermostat. The chromatographic separation was carried out on a Xtimate C18 HPLC column (4.6 mm × 250 mm, 5 μm), and maintained at 30 °C. The detection wavelength was set at 270 nm. The mobile phase was acetonitrile (A) and 30 mmol/L ammonium bicarbonate buffer containing 0.7% (v/v) ammoniae aqua and 0.1% (v/v) triethylamine (B) with a gradient elution program of 10–25% A at 0–15 min, 25–30% A at 15–25 min, and 30–45% A at 25–40 min. All injection sample volumes were 10 μL.
Phylogenetic analysis
The successfully sequenced genes were initiated a complete alignment by Clustal X 1.81 (Thompson et al. 1997) with default parameters, and then the result was repeatedly checked by eye to ensure its accuracy. After the nucleotide substitution model within each gene was tested by software jModeltest 2.1.4 (Darriba et al. 2012), the best evolutionary model was selected by Akaike information criterion (AIC).
According to the best evolutionary model selection, the whole sequence was, respectively, constructed with maximum likelihood (ML) and Bayesian inference (BI). ML with RAxML 8.0.17 (Stamatakis 2014) was performed using a GTR+G model of sequence evolution for the final likelihood search and switched to the per-site rate category model during the fast bootstrapping with 1000 runs. BI was conducted using MrBayes 3.2.2 (Ronquist and Huelsenbeck 2003), while the evolutionary model was used according to the result of jModeltest, and we ran four concurrent Markov chain Monte Carlo (MCMC) calculations of five million generations while sampling once every 100 generations. The first 25% of sampled trees were discarded as burnin and we used the remaining samples to generate a majority-rule consensus tree. We repeated all MCMC runs twice to confirm the consistent approximation of the posterior parameter distributions.
For understanding the evolutionary changes of effective constituents in our selected four species, the parsimony reconstruction was used to estimate the tendency of variation, which was implemented in Mesquite 3.0.4 (Maddison and Maddison 2009).
Results
Geographic character of Coptis herbs
The seventeen samples were successfully collected from the main habitats. Figure 1 indicates that the Coptis herbs were mainly distributed at the edge of Sichuan Basin and Yunnan Province. The Coptis herbs were mainly grown in 500–2000 m above sea level in the mountain forest or in the valley shade. Because of similar environmental conditions for the growth, the Coptis herbs have the similar quality in different species. Moreover, they are all distributed and endemic in Southwest China, and almost all wild plants of Coptis herbs are endangered medicinal herbs.
Sequencing and molecular evolution analyses
Seventeen ITS sequences were successfully obtained from the genomic DNA of different samples by the specific primer set. The efficiency of ITS amplification from C. chinensis and C. omeiensis reached up to 100%; however, C. deltoidea and C. teeta were 92 and 90%, respectively.
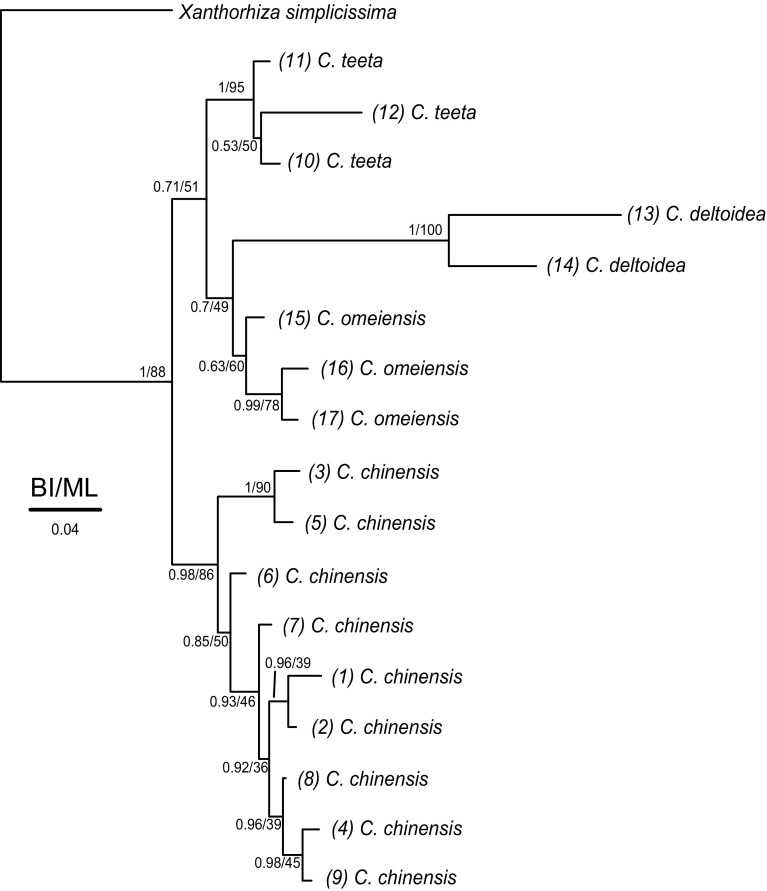
After alignment, the seventeen ITS sequences produced about 700 nucleotide sites. The best evolutionary model was selected as GTR+G model according to the Akaike’s information criterion (AIC) (Akaike 1973). The topology produced from maximum likelihood (ML) and Bayesian inference (BI) was coincident, and all currently recognized species were monophyletic. In general, C. chinensis, C. omeiensis, C. deltoidea and C. teeta constituted a monophyletic group. In this group, C. omeiensis and C. deltoidea cluster together, and they form the sister group with C. teeta, and C. chinensis locate the outermost of C. omeiensis, C. deltoidea and C. teeta (Fig. 2). The Coptis herbs in Southwest China can be successfully classified by the ITS sequences.
Fig. 2.
Bayesian inference tree derived from the part of ITS genes. Numbers above the lines or besides the nodes are Bayesian posterior probabilities and maximum likelihood analyses (1000 replicates)
Chemical analysis of main alkaloids
The proposed HPLC–DAD method was applied to simultaneously analyze main alkaloids from seventeen different samples. The main six peaks were identified, i.e., berberine, palmatine, coptisine, epiberberine, columbamine, and jatrorrhizine, and subsequently evaluated by comparison with authentic standards (Table 1). C. chinensis, as the most important species for daily use in clinical medication, contain the obviously different contents not only in total alkaloids but also in six main alkaloids compared with C. teeta, C. omeiensis and C. deltoidea. From Table 1, we can find that the content of jatrorrhizine in C. deltoidea and C. teeta is marginally higher than that in C. chinensis or C. omeiensis. Especially in epiberberine, C. teeta, C. omeiensis, and C. deltoidea have lower contents than C. chinensis, but it was not detected in C. omeiensis. When it comes to other contents, C. chinensis has far higher than that in C. teeta, C. omeiensis, or C. deltoidea.
Table 1.
Contents of six main alkaloids of four Coptis herbal medicinal materials from 17 different habits (mg/g)
| No. | Species | Source | Jatrorrhizine | Columbamine | Epiberberine | Coptisine | Palmatine | Berberine | Total |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Coptis chinensis Franch | Moudao village, Lichuan city, Hubei province | 4.66 ± 0.42 | 4.37 ± 0.34 | 15.46 ± 0.86 | 25.34 ± 0.54 | 16.76 ± 0.75 | 80.34 ± 0.95 | 146.89 ± 3.54 |
| 2 | Yulong village, Lichuan city, Hubei province | 5.23 ± 0.27 | 6.76 ± 0.15 | 16.87 ± 1.54 | 26.34 ± 1.21 | 20.36 ± 1.44 | 81.22 ± 1.11 | 156.78 ± 2.63 | |
| 3 | Yuelai village, Shizhu county, Chongqing city | 3.43 ± 0.36 | 5.34 ± 0.21 | 13.44 ± 0.67 | 27.53 ± 0.98 | 16.64 ± 0.82 | 75.46 ± 1.36 | 141.84 ± 2.35 | |
| 4 | Huangshui village, Shizhu county, Chongqing city | 4.22 ± 0.26 | 4.74 ± 0.17 | 15.71 ± 0.76 | 25.67 ± 0.84 | 16.46 ± 0.72 | 76.81 ± 1.26 | 143.63 ± 3.63 | |
| 5 | Nanbao village, Qionglai city, Sichuan province | 5.45 ± 0.64 | 6.12 ± 0.24 | 16.76 ± 1.27 | 23.23 ± 0.63 | 21.76 ± 1.26 | 73.87 ± 0.88 | 147.19 ± 4.23 | |
| 6 | Wazhai village,Shifang city, Sichuan province | 4.65 ± 0.25 | 3.86 ± 0.07 | 15.87 ± 0.65 | 26.36 ± 1.47 | 18.76 ± 1.14 | 85.23 ± 1.36 | 154.73 ± 3.26 | |
| 7 | Bajiao village, Shifang city, Sichuan province | 4.07 ± 0.22 | 4.14 ± 0.08 | 11.60 ± 0.58 | 19.83 ± 0.75 | 18.48 ± 1.43 | 66.10 ± 1.11 | 124.23 ± 2.87 | |
| 8 | Lanniba village, Chongzhou city, Sichuan province | 3.34 ± 0.28 | 3.76 ± 0.42 | 12.35 ± 1.22 | 26.67 ± 0.56 | 16.88 ± 0.85 | 75.35 ± 0.95 | 138.35 ± 3.83 | |
| 9 | Xieyuan village, Dayi country, Sichuan province | 6.45 ± 0.15 | 8.78 ± 0.57 | 17.50 ± 0.37 | 27.35 ± 1.55 | 26.54 ± 0.45 | 88.86 ± 1.82 | 175.48 ± 2.64 | |
| 10 | Coptis teeta Wall | Lumadeng village, Fugong county, Yunnan province | 6.38 ± 0.21 | 1.58 ± 0.65 | 0.46 ± 0.10 | 15.07 ± 0.78 | 5.23 ± 0.75 | 83.67 ± 0.97 | 112.42 ± 3.13 |
| 11 | Pihe village, Fugong county, Yunnan province | 6.07 ± 0.09 | 1.73 ± 0.23 | 0.36 ± 0.07 | 14.93 ± 0.37 | 4.61 ± 0.58 | 78.99 ± 1.58 | 106.71 ± 3.03 | |
| 12 | Zhongdian country, Diqing city, Yunnan province | 7.76 ± 0.28 | 1.58 ± 0.37 | 0.66 ± 0.11 | 17.81 ± 1.61 | 5.24 ± 0.98 | 84.85 ± 1.74 | 117.93 ± 3.95 | |
| 13 | Coptis deltoidea C. Y. Chenag et Hsiao | Heishan village, Hongya county, Sichuan province | 7.27 ± 0.16 | 2.15 ± 0.17 | 2.73 ± 0.15 | 11.82 ± 0.71 | 5.88 ± 0.55 | 47.06 ± 0.66 | 76.93 ± 2.75 |
| 14 | Heilin village, Hongya county, Sichuan province | 8.81 ± 0.21 | 1.80 ± 0.14 | 1.90 ± 0.22 | 12.18 ± 1.25 | 6.77 ± 0.77 | 48.37 ± 1.13 | 79.85 ± 3.64 | |
| 15 | Coptis omeiensis C.Y. Cheng | Heishan village, Hongya county, Sichuan province | 4.43 ± 0.17 | 3.17 ± 0.06 | 0.00 | 21.23 ± 0.66 | 9.14 ± 0.52 | 63.29 ± 1.76 | 101.22 ± 2.27 |
| 16 | Heilin village, Hongya county, Sichuan province | 4.72 ± 0.28 | 2.85 ± 0.31 | 0.00 | 16.68 ± 0.42 | 9.43 ± 0.96 | 64.72 ± 0.47 | 98.40 ± 2.26 | |
| 17 | Gaomiao village, Hongya county, Sichuan province | 4.97 ± 0.13 | 2.30 ± 0.75 | 0.00 | 17.39 ± 1.36 | 8.50 ± 0.86 | 56.39 ± 1.23 | 89.55 ± 3.56 |
The quality evaluation of Coptis herbs
The six indexes of effective medicinal constituents (jatrorrhizine, columbamine, epiberberine, coptisine, palmatine, berberine) were successfully achieved corresponding to each sequenced specimen. According to the confirmed phylogenetic topology from ML and BI, the parsimony reconstructions about the six effective medicinal constituents were implemented. The evolutionary changes of effective medicinal constituents are shown in Fig. 3. From the ancestor node, we can find that the content of each effective medicinal constituent is different, the content in C. chinensis is almost higher, and other three species are lower than that of ancestor node. At the same time, Fig. 3 shows that there were some differences in the content in regard to the different intraspecific habitats. These results provided us with the possibility to evaluate the quality of Coptis herbs.
Fig. 3.
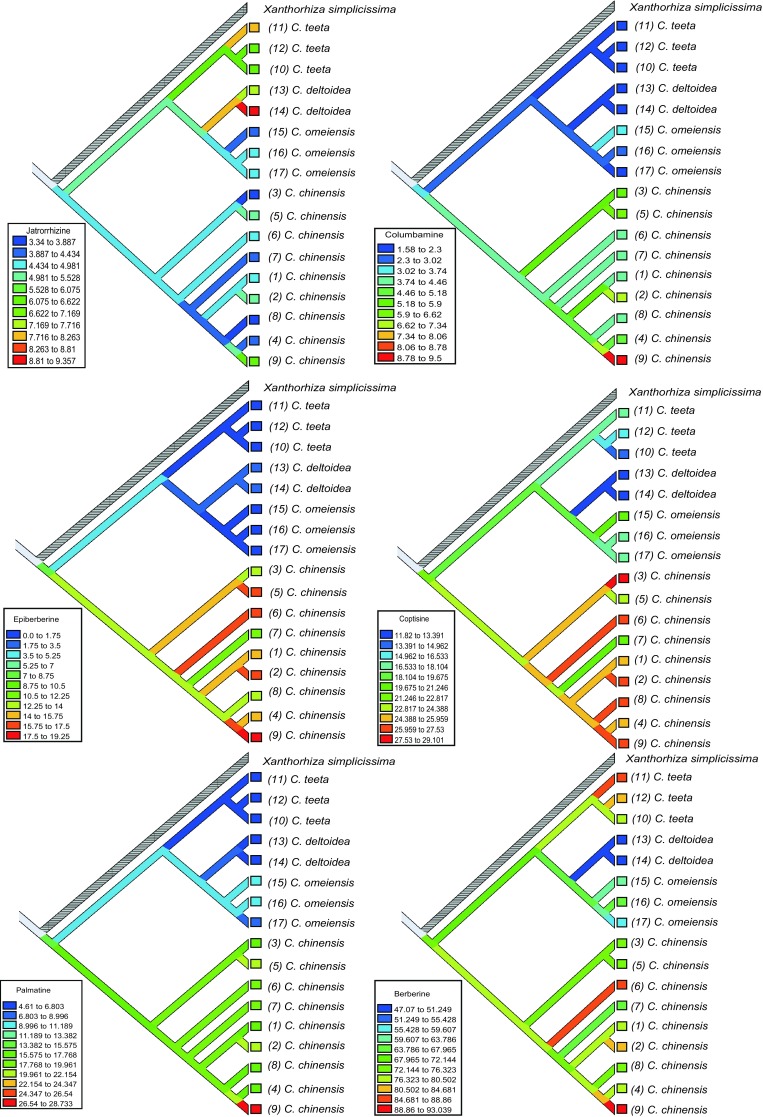
Parsimony reconstruction of six selected effective components (Jatrorrhizine, Columbamine, Epiberberine, Coptisine, Palmatine, Berberine). The topology of phylogenetic tree was used from the result of Bayesian inference
Discussion
The distribution and evolution of Coptis herbs
Seventeen samples of Coptis herbs were collected to reconstruct the phylogeny for species evolution by ITS sequences. Originating from the common ancestor, as a result of the differences in growth environment, late evolution and herbal quality may result in large differences in Coptis herbs. As shown in Fig. 2, C. teeta, C. deltoidea, and C. omeiensis cluster as one group, and this group is sister to C. chinensis. The results indicated that the Coptis herbs from different species can be successfully classified by phylogenetic analyses.
Previous studies have shown that C. deltoidea and C. omeiensis have the most close genetic distance in the medicinal material in Coptis (He et al. 2014; Xiang et al. 2016), also as shown in Fig. 2. This result may be attributed to their closely geographic intercept (Fig. 1), and the two species are mainly distributed on both sides of Mount Emei in Sichuan province (Hu et al. 2011); the uplift of Mount Emei may cause the biotic geographic isolation, leading to the speciation of C. deltoidea and C. omeiensis. The C. teeta is grown only in Yunnan province, and it has more special growth environment than other three species. The C. chinensis has the most extensive distribution and growth quantity in R. coptidis, and we collected the samples from Sichuan, Chongqing, and Hubei (Wagner et al. 2011). Different growth circumstances caused differential genetic relationship in C. chinensis and gave rise to difference in constituents and contents of alkaloids, thereby resulting in different attributes in drug quality.
Chemical analysis of Coptis herbs
Coptis herbs are commonly used herbal drugs in TCM, and the detection of chemical constituents has been reported by many researchers (Chen et al. 2008; Li et al. 2009). The main active constituents of Coptis herbs are protoberberine alkaloids, which are also important marker constituents in the quality evaluation of Coptis herbs (Kuang 2000; Kong et al. 2009). Therefore, in this study, the six main protoberberine alkaloids (i.e., berberine, coptisine, palmatine, epiberberine, columbamine, and jatrorrhizine) were determined simultaneously in Coptis herbs by conventional HPLC method. It was found that the six alkaloids were detected in almost all samples of Coptis herbs with the exception of epiberberine, which was absent in three Coptis omeiensis samples. Compared to other constituents, the berberine content always showed the highest in all the samples, as previously reported (Qiao et al. 2009). On the other hand, the content of total alkaloids ranged from 76.93 to 175.48 mg/g, showing a variation of about twofold in the four species, which indicated that the contents of the six active alkaloids were quite different according to their origins. Moreover, C. chinensis has the higher content of total alkaloids than those in C. teeta, C. omeiensis and C. deltoidea. This is also the reason why the C. chinensis can be acted as one of the main Coptis herbs for cultivation and application.
The evolution and quality evaluation of Coptis herbs
In order to elucidate the evolutionary relationship of Coptis herbs, especially derived from the content of alkaloids, more attention should be paid. Parsimony reconstructions of the six selected effective constituents (jatrorrhizine, columbamine, epiberberine, coptisine, palmatine, and berberine) herein were implemented and no previous studies had been focused on this aspect. The topology of phylogenetic tree was used from the result of Bayesian inference. From these six constituents, they might have the same evolutionary trends and the ancestor node that is located between the content of C. chinensis and other three species (i.e., C. teeta, C. omeiensis, and C. Deltoidea) has the same evolutionary direction, but C. chinensis has the opposite direction for evolution. The inconsistency in the direction of evolution caused differences not only in the content of effective constituents but also in the herbal quality.
In the official Chinese Pharmacopoeia, berberine alone was assigned as the marker composition for the quality evaluation of Coptis herbs (The Pharmacopoeia of the People’s Republic of China 2015). However, in general practices, assessment of multiple constituents is objective and reasonable than a constituent in quality evaluation of Coptis herbs (Kong et al. 2009). Moreover, simultaneous determination of major bioactive constituents is also necessary in the practice of quality control. In this work, C. chinensis has been considered to be the best herb among Coptis herbs, because it has the high content not only in single constituent but also in total alkaloids. All the samples from seventeen habitats were recognized as qualified herbs, and they could meet the requirements of national quality standards in China. Due to different growth environments, different habitats of C. chinensis have different herbal qualities. The samples from Dayi have the highest quality, which has the highest content in single constituent and high content of total alkaloids in all samples, compared to other samples.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (Grant No. 81303168), and the Science and technology special fund of Sichuan Provincial Administration of traditional Chinese Medicine (Grant No. 2016Q060).
Compliance with ethical standards
Conflict of interest
The author declares no conflict of interest.
References
- Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F (eds) Second international symposium on information theory. Akademiai Kiado, Budapest, pp 267–281
- Chen J, Zhao H, Wang X, Lee FSC, Yang H, Zheng L. Analysis of major alkaloids in Rhizoma coptidis by capillary electrophoresis-electrospray-time of flight mass spectrometry with different background electrolytes. Electrophoresis. 2008;29:2135–2147. doi: 10.1002/elps.200700797. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- He Y, Hou P, Fan G, Arain S, Peng C. Comprehensive analyses of molecular phylogeny and main alkaloids for Coptis (Ranunculaceae) species identification. Biochem Syst Ecol. 2014;56:88–94. doi: 10.1016/j.bse.2014.05.002. [DOI] [Google Scholar]
- He S, Liu X, Zhang W, Xie W, Zhang H, Fu W, Liu H, Liu X, Xu Y, Yang D, Gao Y. Discrimination of the Coptis chinensis geographic origins with surface enhanced Raman scattering spectroscopy. Chemometr Intell Lab. 2015;146:472–477. doi: 10.1016/j.chemolab.2015.07.002. [DOI] [Google Scholar]
- Heinrich M (2007) The identification of medicinal plants. A handbook of the morphology of botanicals in commerce, W. Applequist. American Botanical Council, Austin, TX, USA and University of Missouri Botanical Garden Press, St. Louis, 2006
- Hörandl E, Paun O, Johansson JT, Lehnebach C, Armstrong T, Chen L, Lockhart P. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Mol Phylogenet Evol. 2005;36:305–327. doi: 10.1016/j.ympev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Hu J, Shao A, Song L, Yuan Q, Huang L (2011) Development and characterization of microsatellite markers for the endangered medicinal plant Coptis omeiensis (Ranunculaceae). Mol Ecol Resour 1–5
- Kong WJ, Zhao YL, Xiao XH, Jin C, Li ZL. Quantitative and chemical fingerprint analysis for quality control of Rhizoma Coptidis chinensis based on UPLCePAD combined with chemometrics methods. Phytomedicine. 2009;16:950–959. doi: 10.1016/j.phymed.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Kuang HX (2000) Chinese medicine chemistry. China Press of Traditional Chinese Medicine
- Lane CE, Lindstrom SC, Saunders GW. A molecular assessment of northeast Pacific Alaria species (Laminariales, Phaeophyceae) with reference to the utility of DNA barcoding. Mol Phylogenet Evol. 2007;44:634–648. doi: 10.1016/j.ympev.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Li CY, Tsai SI, Damu AG, Wu TS. A rapid and simple determination of protoberberine alkaloids in Rhizoma Coptidis by 1H NMR and its application for quality control of commercial prescriptions. J Pharmaceut Biomed Anal. 2009;49:1272–1276. doi: 10.1016/j.jpba.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Liu B, Li WJ, Chang YL, Dong WH, Ni L. Extraction of berberine from rhizome of Coptis chinensis Franch using supercritical fluid extraction. J Pharmaceut Biomed Anal. 2006;41:1056–1060. doi: 10.1016/j.jpba.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (2009) Mesquite: a modular system for evolutionary analysis
- Nickrent DL, Schuette KP, Star EM. A molecular phylogeny of Arceuthobium based upon rDNA internal transcribed spacer sequences. Am J Bot. 1994;81:1149–1160. doi: 10.2307/2445477. [DOI] [Google Scholar]
- Qiao YL, Shen YX, Wang LQ, Zhang JL. Development of a rapid resolution liquid chromatographic method for simultaneous analysis of four alkaloids in Rhizoma Coptidis under different cultivation conditions. J AOAC Int. 2009;92:663–671. [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schuettpelz E, Hoot SB, Samuel R, Ehrendorfer F. Multiple origins of Southern Hemisphere Anemone (Ranunculaceae) based on plastid and nuclear sequence data. Plant Syst Evol. 2002;231:143–151. doi: 10.1007/s006060200016. [DOI] [Google Scholar]
- Song JY, Yao H, Li Y, Li XW, Lin YL, Liu C, Han JP, Xie CX, Chen SL. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol. 2009;124:434–439. doi: 10.1016/j.jep.2009.05.042. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng H, Choi YH. Optimization of extraction of total alkaloid content from Rhizome Coptidis (Coptis chinensis Franch) using response surface methodology. J Korean Soc Appl Biol Chem. 2012;55:303–309. doi: 10.1007/s13765-012-1148-z. [DOI] [PubMed] [Google Scholar]
- The Pharmacopoeia of the People’s Republic of China . The Pharmacopoeia Commission of PRC. Beijing: Chemical Industry Publishing House; 2015. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H, Bauer R, Melchart D, Xiao PG, Staudinger A. Rhizoma Coptidis—Huanglian. Chromatographic fingerprint analysis of herbal medicines. 2. New York: Springer; 2011. pp. 301–309. [Google Scholar]
- Xiang KL, Wu SD, Yu SX, Liu Y, Jabbour F, Erst AS, Zhao L, Wang W, Chen ZD. The first comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS ONE. 2016;11(4):e0153127. doi: 10.1371/journal.pone.0153127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A, Markow TA, Narechania A, O’Grady PM, DeSalle R. The genus Drosophila as a model for testing tree- and character-based methods of species identification using DNA barcoding. Mol Phylogenet Evol. 2010;57:509–517. doi: 10.1016/j.ympev.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang J, Yang Z. Molecular phylogeny of eastern Asian species of Amanita (Agaricales, Basidiomycota): taxonomic and biogeographic implications. Fungal Divers. 2004;17:219–238. [Google Scholar]