Abstract
Metabolomics is a rapidly growing field with potential applications in various disciplines. In particular, metabolomics has received special attention in the discovery of biomarkers and diagnostics. This is largely due to the fact that metabolomics provides critical information related to the downstream products of many cellular and metabolic processes which could provide a snapshot of the health/disease status of a particular tissue or organ. Many of these cellular products eventually find their way to urine; hence, analysis of urine via metabolomics has the potential to yield useful diagnostic and prognostic information. Although there are a number of analytical platforms that can be used for this purpose, this review article will focus on nuclear magnetic resonance–based metabolomics. Furthermore, although there have been many studies addressing different diseases and metabolic disorders, the focus of this review article will be in the following specific applications: urinary tract infection, kidney transplant rejection, diabetes, some types of cancer, and inborn errors of metabolism. A number of methodological considerations that need to be taken into account for the development of a clinically useful optimal test are discussed briefly.
Keywords: Diagnosis, metabolomics, NMR spectroscopy, urine
Introduction
The number of disciplines ending with the suffix “omics” is constantly increasing. However, the major ones remain to be genomics, transcriptomics, proteomics, and metabolomics. The genome is known as the entry point into the sciences of the “omics,” whereas the metabolome is considered to be the end point of the pyramid.1 Genomics is the study of the genome, with its complete set of genes, whereas proteomics focuses on the proteome, with its entire set of proteins produced or modified by an organism. Transcriptomics deals with the study of messenger RNA. Metabolomics, a relatively newer member of the “omics” family, consists of the complete set of the metabolites found in a biological fluid or matrix. It is the comprehensive and systematic profiling of metabolite concentrations and their systematic response to different factors.2 Through multiple analyses, a metabolic profile of a biological sample can be acquired, and that can be used to identify groups of diseases and to determine a comprehensive mechanism of the pathology. Metabolic changes due to certain diseases can be detected in biological fluids before the clinical symptoms develop, generating useful fingerprints. Further along, these characteristic fingerprints of the pathology may then serve as metabolic biomarkers, in which various diseases can be detected through the analysis of tissues or biofluids.3–5 This lends way to the importance of biomarkers in medicine.
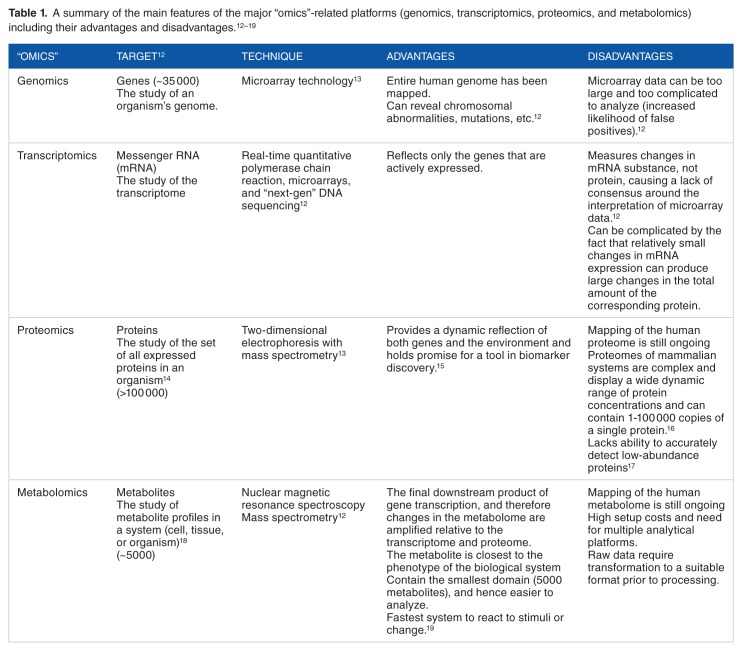
Unlike metabolomics, genomics and proteomics lack the information needed for an understanding of the cellular function in living systems, because both sciences do not provide information on the dynamic metabolic status of the organism.6 Metabolomics has an advantage over the other “omics” technologies as it is the most predictive of the phenotypic properties of a biological system.7,8 The metabolites are the downstream products of numerous biochemical interactions and can be a very sensitive measure of an organism’s phenotype, making metabolomics very useful in the perturbations and interactions of genetic and environmental factors.3,9,10 In addition, metabolomics elucidates the nature and identity of the processes themselves rather than just examining the compounds, and the relatively small number of metabolites makes it easier to analyze the data.11 Table 1 shows the main features of the major “omics”-related platforms.
Table 1.
The 2 major components of a biomarker identification strategy are the analytical technique and the statistical analysis.20 Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry are suitable techniques for metabolomics. Both have different analytical strengths and weaknesses but give complementary information.2,3 In both techniques, robust statistical data analysis is necessary due to the complex nature of the multivariate data sets. Mass spectrometry can provide metabolite identification with high sensitivity. The analysis of nonvolatile metabolites has mainly been utilized for metabolic profiling in which multiple classes of metabolites are determined in one analysis. However, matrix effects, ionization suppression and enhancement, can be affected by the presence of other chemical species and cause inconsistent results in the analysis.21 Sample preparation is also extensive, and the sample is destroyed in the process. In contrast, NMR does not require preselection of analysis conditions, and sample preparation is more straightforward.21,22 Furthermore, NMR is nondestructive, thereby allowing further analysis of the same sample, if needed.23 In addition, each separate resonance observed in an NMR spectrum is specifically assigned to an individual compound, while simultaneously providing comprehensive structural information. The information gathered via NMR lends insight to the mechanisms of biochemical disease processes and drug metabolism.24 It is paramount that the analysis has high reproducibility, confirming that the metabolic effects detected by the instrument are much higher than the analytical variability. Nuclear magnetic resonance–based metabolite profiling allows a “snapshot” of the metabolite’s molecular dynamics and mobility, simultaneously detecting the wide range of metabolites in the sample.2
Advancement in metabolomics has included biomarker determination indicative of disease and clinical diagnostics.2 In addition, NMR-based metabolomics has also shown the potential for monitoring the progression of toxicological effects and the identification of biomarkers of toxicity.24 Pioneer biomarkers such as creatinine, glucose, and cholesterol have been utilized to assess kidney function, diabetes, and lipid metabolism.20 A human metabolome database, with detailed information on small-molecule metabolites, has already been established and is available to the public at no cost, and it contains 41 993 metabolite entries, with both water and lipid soluble metabolites.25
Several studies have utilized hydrogen-1 NMR (1H NMR) spectroscopy to analyze various body fluids such as plasma, blood, and urine. Blood samples are composed of a complex mixture of high- and low-molecular-weight metabolites, ranging from fatty acids/lipoproteins to amino acids. This wide range of metabolites can compromise spectral quality and often requires special NMR pulse sequence or multiple sample extractions. In contrast, under normal conditions, urine requires minimal sample preparation and contains very low concentration of macromolecules. The most common metabolites identified in urine are those associated with major endogenous pathways along with their intermediates, such as citrate, succinate, -oxaloacetate, and α-ketoglutarate.23 Moreover, metabolic phenotyping has provided the development of patient stratification and personalized medicine.26 Urine samples are also more convenient as they can be readily obtained noninvasively from clinical patients. For disease characterization, urine is often the preferred sample because of its role as the major secretion of xenobiotics20 and its metabolite-rich nature.27 In addition, urine contains the metabolic signatures of many biochemical pathways.28
Hydrogen-1 NMR spectroscopy of urine samples has proven to be an eligible analytical and diagnostic tool.3,29 Early detection of various diseases is crucial and can sometimes only be analyzed at the molecular level by investigating the chemical interactions of metabolites. The advancement in urinary metabolomics has paved the way for potentially early detection and disease characterization, especially in the studies of urinary tract infections (UTIs), kidney transplant rejection, diabetes, malignancies, and inborn errors of metabolism.30 These applications will be discussed in detail below, following the section on methodological considerations.
Methodological Considerations
Sample preparation
For logistical reasons, urine samples are generally frozen immediately after collection and analyzed later. According to a study by Rist et al, the freezing procedure plays a major role in sample preparation, due to its effects on the variation of the spectral data. The study recommended a freezing temperature of −20° C and storage at lower temperatures within 1 week. Samples frozen on dry ice, in comparison with liquid nitrogen, showed the largest deviations. This was proven to be dependent on pH differences introduced by the range of CO2 concentrations brought on by the freezing procedure used.31 Urinary pH has proven to cause spectral variability between similar samples, particularly in the chemical shift of citrate.32 Similarly, according to Lauridsen et al, urine samples should be stored at or below −25°C. They have shown that samples stored at this temperature for up to 26 weeks show no significant change in their 1H NMR spectra. At about 4°C, formation of acetate can be observed due to microbial contamination.33 After a 4-week period, the degree of change in metabolite concentrations was influenced by the various methods of sample preparation and storage used. The significant deviations of metabolite concentrations that were observed were largely due to bacterial contamination, which significantly altered the metabolic profile of urine over time.
The sample preparation for 1H NMR analysis is fast and straightforward. However, one has to be careful regarding the pH of the samples. The pH of normal human urine samples generally falls in the range of 5.5 to 6.5. Under physiological stress, the pH of urine further varies, falling in the range of 4.6 to 8.0. To minimize the pH-related variation in NMR chemical shifts, buffer solutions are added to urine.33 Frozen urine samples are thawed on ice and are vortexed for 30 seconds before use. Aliquots of 500 µL of the samples are then transferred into Eppendorf tubes and treated with 250 µL of 0.33 M phosphate buffer prepared in D2O (pH = 7.4). A buffer concentration of 0.33 M is sufficient to minimize pH-related variations in the NMR chemical shifts for most urine samples. Lauridsen et al,33 suggested using a 1.0 M phosphate buffer solution for more concentrated urine samples. The deuterated solvent (D2O) allows for deuterium locking, in which the magnetic field is better controlled. For sample analysis using internal standard solutions (such as Chenomx ISTD, Chenomx Inc., Edmonton, Canada), the samples are vortexed and centrifuged at 1200g for 15 minutes after adding approximately 70 µL of the Chenomx ISTD solution to 630 µL of urine samples into the Eppendorf tubes. A volume of 600 µL of the supernatant is then transferred into a 5-mm NMR tube for analysis.33–35 Similar methodology can be adopted with the use of other more common internal standards such as Trimethylsilylpropanoic acid (TSP) or 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). DSS is less sensitive than TSP to variations in pH of the sample. However, given its more hydrophobic nature, its resonance can be broadened due to binding with macromolecular structures such as proteins in the sample.36
Nowadays, given the availability of automatic sample preparation and robotic liquid handling technologies, large-scale studies can be carried out for high-throughput applications.37 Moreover, with the combined use of flow injection (FI) probes and liquid handling systems, it is possible to perform “tubeless” NMR using 96-well plates. Samples can be identified by unique barcode, prepared for analysis by a robot handling 96-well plates, and finally transferred to the FI probe for NMR analysis. Such large-scale metabolomics studies have already been reported in the analysis of urine samples, which allowed the sampling and 1H NMR data collection of approximately 100 samples per day, with an acquisition period of 5 minutes per sample.2
Water suppression
Sample collection for urine metabolomics is a very convenient and noninvasive method. However, the large concentration of water in urine makes it difficult to see metabolites that are present at a very low concentration, resulting in a distorted spectrum. Water suppression has become necessary to address this setback. Water presaturation is often applied to samples to mitigate the effects of excess water. In this technique, a transmitter is set to the water frequency, and a selective pulse is used to saturate the water resonance. However, solvent suppression can sometimes have unintended effect on the spectrum by causing a damping effect on the peaks of the neighboring clusters, which may result in artificially lower concentrations.34 The water suppression techniques commonly used in metabolomics are briefly discussed in the following section.
NMR data acquisition and analysis
For urinalysis by NMR, 1-dimensional (1D) NMR techniques with presaturation of water signal are mostly widely used. There are several water suppression techniques available for metabolomics applications.38 The experimental details of these techniques have been discussed in detail in a recent review article.39 One dimensional nuclear Overhauser enhancement spectroscopy pulse sequence with water presaturation is the most recommended technique. One drawback of using presaturation would be in the quantification of urea. The pulse train in a typical hard pulse–based water suppression by gradient tailored excitation (WATERGATE) may negatively impact the urea signal due to lack of sufficient selectivity. In a study by Liu et al,40 the authors used water suppression enhanced through t1 effects (WET) for water suppression and chose its pulses to be long enough so that it had sufficient selectivity to exert minimal impact on the urea signal. The pulse duration was also kept short to make sure that no significant proton exchange takes place between water and urea.40
Other techniques such as excitation sculpting would entirely eliminate the water signal from the spectrum, but the intensity of the signals resonating adjacent to water signal is greatly affected.38 The next step is to identify metabolites in urine samples, which can be achieved by the NMR spectral analysis. Some of the common metabolites, eg, lactate, alanine, acetate, citrate, creatine, creatinine, trimethylamine N-oxide (TMAO), hippurate, and formate, can be identified by looking at their chemical shift values. However, the identification of other metabolites, which are in low concentration and are overlapping with other high-concentration metabolites, is particularly challenging. The identification process mainly involves comparison of the 1D and 2-dimensional (2D) spectral data of the metabolites with that of NMR metabolites repositories such as the Human Metabolome Database,25 the Biological Magnetic Resonance Data Bank (BMRB),41 and the Birmingham Metabolite Library (BML).42 The 2D J-resolved spectroscopy (JRES), correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and heteronuclear multiple bond correlation (HMBC) data of reference compounds in these databases will be valuable in the detailed comparison and identification of metabolites. A detailed methodology on the metabolite identification using 1D (1H, 13C) and 2D (JRES, COSY, TOCSY, and HMBC) has been discussed in recent review articles by Everett and colleagues.43–46
The issue of normalization is something that needs to be given a serious consideration given the dilution issues faced when analyzing urine specimens. Normalization is routinely used to account for the dilution effect of each sample as well as variation from different batch of measurement.47 This is especially important in urine samples given the different water intake of individuals. Normally, each metabolite peak integral is normalized to the creatinine level based on the assumption that the creatinine level is an indicator of the metabolite concentrations in urine.47 However, creatinine normalization has been questioned because factors such as muscle mass, age, and gender were found to affect the normal urine creatinine level.47 Another commonly used method is the total area/integral normalization, which assumes the total integral of all metabolites in a sample to be a constant throughout all samples. However, this method is not applicable if drug metabolites also appeared in the sample. A different approach, named probabilistic quotient normalization, has been proposed recently.48 This method calculates a most probable dilution factor (eg, the median of the quotients) among all the metabolite variables, which is then utilized to perform normalization of all variables in the sample.
Given the complex nature of urine samples, the 1H NMR spectrum of urine shows well-resolved peaks only for a few metabolites. Such metabolites can be quantified by manual integration. However, metabolites that are found in low concentration are generally overlapping with other metabolites that are present in high concentration. As a result, other more robust quantification methodologies such as targeted profiling49 and/or Bayesian deconvolution50 have been developed for such purposes. Targeted profiling relies on database of compounds modeled to behave like the pure spectra of the individual compounds under comparable experimental conditions of pH and ionic strength. A Lorentzian peak shape model of each reference compound is generated from the database information and superimposed on the actual spectrum. The linear combination of all modeled metabolites gives rise to the total spectral fit, which gives an estimate of peak areas of experimental spectral peak. Using this information, concentrations of various metabolites can be estimated. Analysis of samples using target profiling allows both identification and quantification of individual compounds. Chenomx NMR Suite is a commercial software that provides a comprehensive database of metabolites (>350 compounds) making use of targeted profiling51 and has been used in many metabolomics studies for the analysis of biofluids including urine.52 On the contrary, the Bayesian model makes extensive use of prior information on the characteristic spectral pattern of each metabolite. It also accounts for shifts in the position of peaks commonly seen in NMR spectra of biological samples. It is an “R”-based public domain software package, “Bayesian AuTomated Metabolite Analyzer for NMR spectra (BATMAN),” which deconvolutes peaks from 1D NMR spectra and automatically assigns them to specific metabolites from a target list, obtaining concentration estimates. It applies a Markov chain Monte Carlo algorithm to sample from a joint posterior distribution of the model parameters and obtains concentration estimates with reduced error compared with conventional numerical integration and comparable with that of manual deconvolution.50–52
Data analysis in metabolomics is usually performed via unsupervised or supervised analysis. Unsupervised analysis involves the application of statistical models without prior knowledge of the sample identity or classification assignment. This is often times done to see if the data separate into certain patterns or clusters. This is usually the first step in data pattern exploration. Principal component analysis (PCA) is representative of the unsupervised method and is commonly applied to reduce the dimensionality and examine the structure of the data set.53–55 Scores plot is generated to assess the clustering of different samples, with the corresponding loadings plot demonstrating the variables accounting for the most variation in the specified principal component. In supervised analysis, information of sample class labels (eg, disease and control) is utilized in building the statistical models. One commonly used supervised analysis is partial least squares discriminant analysis (PLS-DA) which maximizes the covariance between predictor variables (eg, metabolite intensities from NMR measurements) and the response variables (eg, the classes of each sample).53 SIMCA (soft independent modelling of class analogies) forming the basis of the readily available software SIMCA-P is commonly used in metabolomics analysis.56 The strategy of SIMCA-P is to rely almost exclusively on PCA, and for classification, on its supervised versions, partial least squares (PLS) or principal component regression (PCR). (For 2-class problems, PLS and PCR are equivalent.)54,55
One of the important steps in metabolomics data analysis is identifying which of the original features in the spectra are relevant and meaningful in the final outcome. A genetic algorithm–based optimal region selection algorithm has been developed specifically for such feature extraction.54 An important advantage of this is that it retains spectral identity: the new features being functions (typically the averages) of adjacent spectral data points and hence readily interpretable. Such an algorithm has been part of a statistical classification strategy used in several studies involving NMR data.57–62 A schematic diagram of the metabolomics workflow is depicted in Figure 1.
Figure 1.
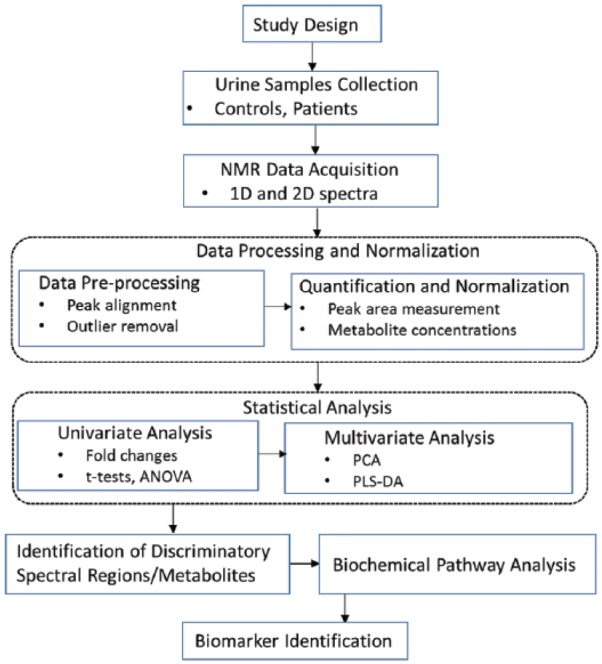
Schematic diagram of metabolomics workflow in the urinalysis.43,60,63
1D indicates 1-dimensional; 2D, 2-dimensional; ANOVA, analysis of variance; NMR, nuclear magnetic resonance; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis.
Other special considerations
There are also some other special considerations that need to be taken into account with this technique. High-protein diets such as fish have proven to cause increases in creatinine concentrations, which can cause distortion in some 1H NMR spectra.64 A study by Lenz et al65 has suggested that endogenous urinary profiles are affected by cultural and severe dietary influences, emphasizing that variation in profiles can occur between different populations. The study compared the urinary metabolic profile of the Swedish and the British population, in which they found that high levels of TMAO were observed in Swedish population, due to their fish diet. This effect was also consistent in a study by Dumas et al,66 in which a similar trend was observed in Japanese population, where fish diet is also dominant. This variation is an important factor that needs to be considered when interpreting NMR spectral profiles for diagnostic purposes. Similarly, Zuppi et al67 tested subjects living in different parts of Europe and found that a diet rich in carbohydrates resulted in increased excretion of citrate, lactate, and glycine.
Slupsky et al68 investigated the effects of diurnal variation, gender, and age in urinary metabolomic profiles. Their results showed that gender and age affected metabolites related to energy metabolism, whereas diurnal variation affected metabolites and dietary components associated with circadian rhythms. In a different study by Psihogios et al,69 they determined the most influential metabolites responsible for the differences in gender groups, which included citrate, creatinine, TMAO, and an unidentified compound. Overall, Rasmussen et al suggest it is important to perform diet standardization before dietary intervention in metabolomics study. This will help reduce intrasubject and intersubject variability.70
Applications
Urinary tract infections
Urinary tract infections are the second most common type of infection in the human body. Due to anatomical reasons, the chance for contracting this infection in women is greater than 50%. Urinary tract infection is commonly diagnosed through the examination of a patient’s cultured urine sample. This traditional method is labor-intensive and time-consuming, taking approximately 24 hours for culturing and additional time for identification. The dipstick methods have shown to relay false-negative/false-positive results.
In multiple studies by Gupta et al,71 1H NMR spectroscopy has been utilized for identifying and quantifying the common uropathogens of UTI (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Proteus mirabilis). This metabolic approach has been successfully demonstrated as a viable tool in diagnosing UTI pathology. In a study of 617 urine samples from suspected patients and 50 samples from healthy volunteers, UTI was detected from the quantification of 6-hydroxynicotinic acid (6-OHNA), which is metabolized from nicotinic acid by the bacterium, P aeruginosa. Similarly, the production of 1,3-propanediol from glycerol metabolism was also used to detect UTI caused by K pneumoniae, along with the production of 4-methylthio-2-oxobutyric acid (MOBA) from methionine metabolism, found in P mirabilis. The most common UTI-causing bacterium, E coli, metabolizes lactose to lactate, which can also be detected through 1H NMR, as is evident in the spectrum in Figure 2. The results demonstrated the power of 1H NMR for developing quick identification of microorganisms in chronic and severe UTI. Bacterial identification from 1H NMR was found to have very high specificity (97%) relative to the conventional culture method.71 In a similar pilot study by Bezabeh et al,72 33 urine samples were collected from children (1–16 years old) with and without UTI and analyzed by NMR. The results showed 9 patients to have elevated levels of TMAO and 8 patients with elevated levels of creatine. In another study by Wan Lam et al,73 88 samples from UTI patients and 61 samples from controls were analyzed by 1H NMR, and the results showed urine acetic acid/creatinine level was the most discriminatory marker for bacterial UTI.
Figure 2.
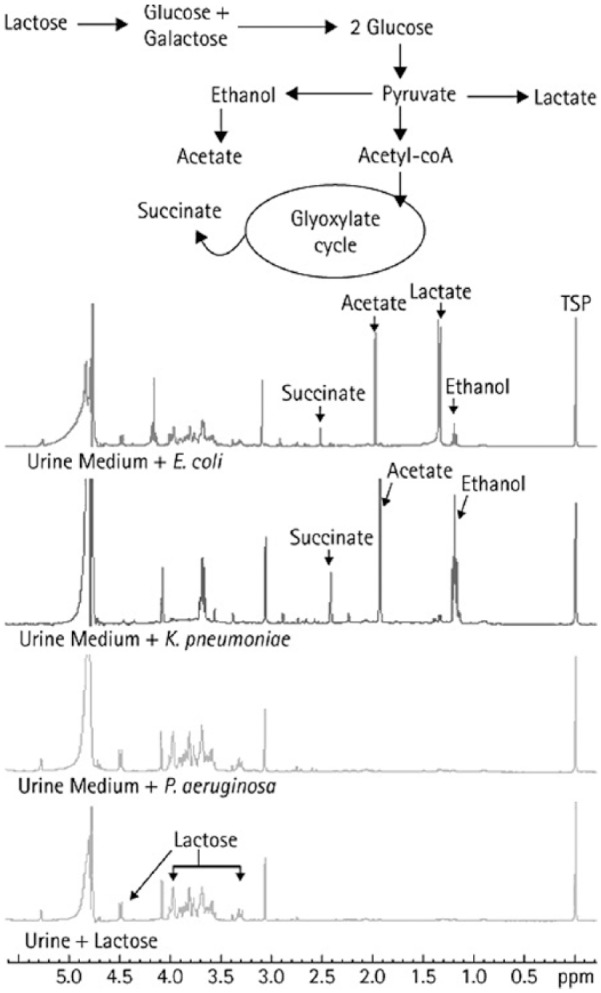
Nuclear magnetic resonance spectra of lactose metabolism in urine.
Escherichia coli produces lactate, acetate, succinate, and ethanol through lactose metabolism. Lactate is the specific product in lactose metabolism by E coli; other bacilli (Pseudomonas aeruginosa, K pneumoniae, etc) do not produce lactate in urine.71
Kidney transplant rejection
In renal transplants, symptoms are often not detected until there is already a marked impairment in renal function. Acute rejection has been reported to cause a 20% reduction in 1-year survival of the allograft.74 Frequent biopsies are necessary to test for kidney transplant rejections. These procedures are often expensive and are inconvenient for the patient. Through the advancement of metabolomics, 1H NMR is able to detect organ rejection by analyzing urine samples through noninvasive means.
In 2 pilot studies by Rush et al,75 significant differences were observed in the spectra between patients with apparent graft dysfunction and those with relatively normal allograft function. Patients with graft dysfunctions due to transplant rejection had increased TMAO and dimethylamine (DMA) levels in their urine. Detecting subclinical rejection using 1H NMR could pave the way for early treatment.
In a similar study by Le Moyec et al, urine and plasma samples from 39 renal transplant patients were collected and analyzed with 1H NMR spectroscopy. When compared with the levels of creatinine, the relevant resonances for identifying renal function were from TMAO, citrate, alanine, and lactate. The urine spectra from those who required hemodialysis showed an ischemic pattern of elevated TMAO, lactate, and alanine. The results showed that a combination of measured metabolites can be used in the follow-up of transplant patients and management of cyclosporine A dosage.76
In a study by Foxall et al,77 urine samples were collected from 33 renal allograft transplantation patients for 14 consecutive days following transplant and were analyzed using 1H NMR spectroscopy. As shown in Figure 3, their results revealed that during the early phase of post-transplantation, the high levels of urinary excretion of the low-molecular-weight metabolites (TMAO, DMA, lactate, acetate, succinate, glycine, and alanine) seemingly correlated with occurrences of graft dysfunction. More specifically, the urinary concentration of TMAO was statistically significantly higher in patients with graft dysfunctions than the patients with good graft function. This was suggested to be due to the effects of graft dysfunctions on the renal medulla, which resulted in its higher secretion of TMAO from the damaged cells. Their goal was to see if there was an increase in TMAO excretion in the individuals with graft dysfunction. The results revealed that during the early phase of post-transplantation, the high levels of urinary excretion of the low-molecular-weight metabolites seemingly correlated with occurrences of graft dysfunction. Early diagnosis is vital for early intervention therapy, thereby improving graft outcome. This study highlights the importance of NMR spectroscopy in the investigation of metabolic perturbations posttransplantation.77
Figure 3.
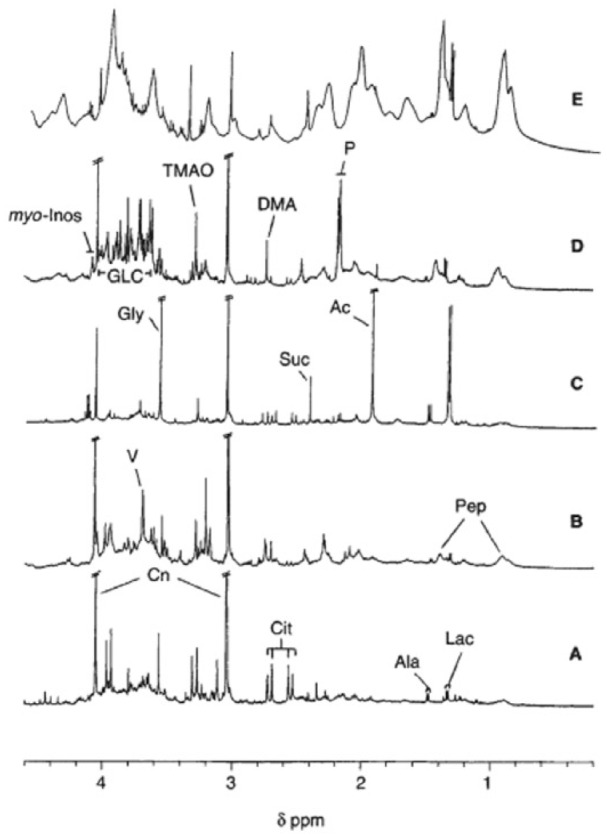
Partial 500-MHz single-pulse hydrogen-1 nuclear magnetic resonance spectra of normal human urine (A) and urine collected from 4 patients on day 3 postrenal transplantation showing immediate functioning graft (B), urinary tract infection (C), renal tubular ischemia, (D) and nonfunctioning graft (E).77
DMA indicates dimethylamine; GLC glucose; TMAO, trimethylamine N-oxide.
Diabetes
Twenty-nine million people in the United States are plagued with diabetes. Among those 29 million, 8 million are undiagnosed.78 In a study by Messana et al,29 33 urine samples from patients with type 2 diabetes mellitus (T2DM) and 20 control subjects were examined by 1H NMR spectroscopy. The results showed significantly higher levels of lactate, citrate, glycine, alanine, hippurate, TMAO, and DMA in diabetic patients.
In a related study by Doorn et al, the effect of thiazolidinediones, a medication that lowers insulin resistance in muscles and fat, was investigated in patients with type 2 diabetes and healthy volunteers. The traditional method for identifying type 2 diabetes is through the measurement of plasma concentrations of glucose and hemoglobin A1c (HbA1c). However, significant downstream metabolic effects are overlooked. Nuclear magnetic resonance–based metabolic profiling provided a more comprehensive “snapshot” of the metabolic changes induced by thiazolidinediones. The study identified putative disease and gender-specific metabolites in urine. The findings agreed with those of Messana et al, with an increase in citrate and hippurate concentrations in the urine of T2DM patients.79 In a different study by Nicolescu et al, NMR data from nonbuffered urine samples of 72 controls and 94 patients with type II diabetes were subjected to a customized statistical classifier. Their results achieved 83% sensitivity, with 83.6% specificity and an overall accuracy of 83.2%. Their results were based on the nonglucose regions of the spectra (Figure 4). This protocol has the potential for the automated and clinical diagnosis of diabetes.80
Figure 4.
Hydrogen-1 nuclear magnetic resonance spectrum (400 MHz) of a urine sample from a patient with type 2 diabetes showing the 2 input subregions and the 4 discriminatory regions identified by the optimal region selection algorithm.80
Although most of the NMR work to date has focused on type 2 diabetes, there have been some studies dealing with type 1 diabetes. In a study by Balderas et al, urine samples from children with type 1 diabetes were investigated. Their findings showed that the diabetic group excreted larger amounts of carboxyethylarginine and fructosamine in their urine, which are glycation end products.81 In another study by Deja et al,82 the relationship between metabolite concentration in urine of patients with type 1 diabetes and their level of HbA1c was investigated. They collected urine samples from 30 children and teenagers between the ages of 4 and 19 years with type 1 diabetes, while using 12 samples collected from healthy 9-year-old children as controls. Their results proved that targeted analysis of low-molecular-weight compounds in urine can serve as a way to monitor the changes in diagnosed patients. The comparison between patients with low and high levels of HbA1c revealed the concentrations of the metabolites—alanine, valine, acetate, pyruvate, and citrate—were significantly higher in the latter group. The elevation of these metabolites suggests that in patients with high HbA1c, the tricarboxylic acid cycle is the pathway that is commonly disturbed. Their results also revealed that patients with normal levels of HbA1c still had differences in their urine compound composition in comparison with the nondiabetic children. In retrospect, their study allows for an additional methodology to monitor the progression in patients with type 1 diabetes.82
Cancer
The 4 most common cancers occurring worldwide are lung, female breast, bowel, and prostate cancer (PCa). These 4 cancers account for about 4 in 10 of all cancers diagnosed worldwide. Noninvasive diagnosis can be done through the utilization of modalities such as computed tomography, magnetic resonance imaging, or positron emission tomography. However, definitive diagnosis relies on biopsies, which is invasive in nature. Currently, the general screening methods are not ideal, and early-stage tumors are often not detected due to their lack of symptoms. The diagnosis of patients frequently occurs during the late stages of cancer development, resulting in very poor prognosis. Urinary metabolomics has been applied to lung, gastric adenocarcinoma (GC), prostate, and bladder cancer (BCa). Based on the accumulated analyses of cancer marker metabolites, it has been suggested that various cancers have common, yet distinct metabolic phenotypes that correspond to their perturbed biochemical pathway.83
Various studies have explored the applications of 1H NMR-based metabolomics as a powerful early diagnostic tool. In a study by Carrola et al, 1H NMR-based metabolomics has been applied for the first time to investigate lung cancer metabolic signatures in urine. Their goal was to acquire information on lung cancer metabolism and its systemic effects. Seventy-one urine samples from cancer patients and 54 from a control group were collected and analyzed by 1H NMR. The spectral profiles were analyzed with PCA, PLS-DA, and orthogonal partial least squares discriminant analysis (OPLS-DA), and their results showed a significant discrimination between the 2 groups, as shown in Figure 5. The most intense signals originated from the metabolites: creatinine, TMAO/betaine, hippurate, citrate, α-ketoglutarate, and glycine. Hippurate and trigonelline were reduced and β-hydroxyisovalerate, α-hydroxyisobutyrate, N-acetylglutamine, and creatinine were elevated in the cancer patients. The PLS-DA model showed a 93% sensitivity and 94% specificity. The study also tested for factors such as gender and age since metabolic composition of urine was modulated. The gender variation test showed negative Q2 values, which indicated poor predictive ability. In addition, when the classifier was based on age, the PLS-DA model showed poor predictive power (negative Q2), a low classification rate, specificity, and sensitivity. When the presence of the disease was used as the classifier for the PLS-DA modeling, the overall classification rate was 84%, indicating good separation.84
Figure 5.
The 500-MHz hydrogen-1 nuclear magnetic resonance spectra of urine from (A) a healthy (control) subject, and (B) a lung cancer patient. Signal assignment: 1, α-hydroxybutyrate; 2, valine; 3, isobutyrate; 4, β-aminoisobutyrate; 5, methyl-β-hydroxybutyrate; 6, β-hydroxyisovalerate; 7, lactic acid and threonine; 8, α-hydroxyisobutyrate; 9, alanine; 10, N-acetylglutamine; 11, pyruvate; 12, succinate; 13, α-ketoglutarate; 14, citrate; 15, dimethylamine; 16, creatinine; 17, trimethylamine N-oxide and betaine; 18, scyllo-inositol; 19, glycine; 20, hippurate; 21, trigonelline; 22, p-hydroxyphenylacetate; 23, phenylacetylglycine; 24, histidine; 25, 3-methylhisitidine; 26, formate; 27, trigonellinamide.84
In a study by Chan et al, GC, the third most deadly cancer worldwide, was analyzed through 1H NMR urinary metabolomics. Seventy-seven metabolite concentrations were detected and pairwise comparisons between GC patients, healthy subjects, and patients with benign gastric disease were performed followed by the Benjamini and Hochberg correction methods. The results showed elevated levels of alanine in healthy subjects compared with GC patients. Alanine was thus suggested to be a potential biomarker for GC.85 However, further studies with larger sample sizes are necessary to confirm a definitive biomarker for this disease.
Major advances have been made in both NMR and mass spectrometry in identifying metabolic changes in PCa that can serve as potential biomarkers.86 In PCa, the prostate-specific antigen (PSA) blood test is the most frequently used tool for PCa detection. However, it has its limitations. The PSA test has low specificity. Results can often lead to false-negative and overdiagnosis. Currently, researchers are looking at 1H NMR-based metabolomics of urine samples as a potential diagnostic tool.
In a study by Zaragoza et al, 113 urine samples were collected from patients and were tested for PCa. The model accurately classified 14 of 14 samples as controls and 36 of 50 as patients with PCa in the validation set (n = 64). They concluded that the presence of PCa could not be associated with a unique analyte, but rather it is most likely linked to the presence or changes in concentration of multiple metabolites. This set of metabolites is composed of phosphocholine, myo-inositol, spermine, glutamine, citrate, alanine, lactate, OH-butyrate, valine, and leucine. This study confirms the potential of 1H NMR spectroscopy as a noninvasive diagnostic tool for the detection of PCa.87 In addition, NMR-metabolomics has also shown promise in monitoring the progression of PCa. In a study by Sreekumar et al,88 in which they analyzed 110 urine samples, the results showed increased levels of sarcosine, an N-methyl derivative of glycine, during PCa progression to metastasis.
Bladder cancer, a common type of cancer, has been known to have a very high mortality rate, and early detection has been proven a vital component in survival. In a study by Shen et al, urine samples from 23 early-stage BCa and 21 healthy controls were prepared and analyzed with 1H NMR. Their results identified 3 upregulated metabolites (nicotinuric acid, trehalose, and AspAspGlyTrp) and 3 downregulated metabolites (inosinic acid, ureidosuccinic acid, and GlyCysAlaLys) for BCa. Their results showed that these metabolites revert back to normal levels after tumor removal, confirming that they are a good representation of metabolomics features associated with BCa. This study showed a high diagnostic performance for detecting BCa with area under curve (AUC) values of 0.919 and 0.934.89
In a study by Slupsky et al, urinary metabolic profiling showed changes in metabolite concentrations that were specifically correlated with ovarian and breast cancer.90 They collected urine samples from early- and late-stage breast and ovarian cancer patients and randomly from females with no known cancer. Their results revealed that numerous metabolites decreased in concentration among those patients with ovarian or breast cancer when compared with the healthy group. They compared 67 metabolite concentrations from healthy subjects (n = 62), breast cancer patients (n = 38), and ovarian cancer patients (n = 40), and the results revealed significant differences. However, the extent of the change differed for the breast and ovarian cancer patients. The unknown singlet found at 3.35 ppm, suggested to be methanol, was ranked as the most important metabolite for distinguishing the ovarian patients with 65% decrease in concentration relative to normal subjects, whereas in breast cancer patients, formate was ranked first with a decreased metabolite concentration change of 43%. The potential of this technique as an effective screening tool was reaffirmed with its almost zero false negatives (98% and 100% sensitivity) and a few false positives (99% and 93% specificity) for ovarian and breast cancer, respectively. In comparison with mammography, which produces numerous false positives and false negatives, this technique is faster, less costly, noninvasive, and more efficient for early cancer screening.90
Inborn errors of metabolism
Inborn errors of metabolism are rare genetic disorders in which the body is not capable of converting food into energy. The current screening tests are not able to detect all inborn errors of metabolism and may yield false-positive results. Hydrogen-1 NMR–based metabolomics has been investigated as a potential diagnostic tool for this disorder. In a study by Constantinou et al, 47 urine samples from healthy newborns, 9 from newborns with phenylketonuria, and 1 from a child with maple syrup urine disease were tested from a Greek population. The spectra of the samples showed variation in phenylalanine, leucine, valine, and isoleucine resonances. Principal component analysis and PLS-DA were performed to create accurate models for the discrimination between the samples.91 This method is quick and noninvasive, and further research can help solidify it as an ideal mass screening tool.
In a similar study by Wevers et al, 1H NMR spectroscopy was used to identify inborn errors of purine and pyrimidine metabolism. For the inborn errors of pyrimidine deficiency, patients with dihydropyrimidine dehydrogenase deficiency showed elevated levels of uracil and thymine, in comparison with healthy urine samples in which thymine was not detected and uracil was only observed in trace amounts. For the inborn errors of purine metabolism, patients with a deficiency of hypoxanthine-guanine phosphoribosyltransferase had increased levels of hypoxanthine and xanthine. In most cases, hypoxanthine was elevated. For the patients with a deficiency in purine nucleoside phosphorylase, increased concentrations of inosine, guanosine, and their deoxy forms were observed in the spectra.92
In a related study by Aygen et al,93 989 urine samples from newborns were collected and analyzed using 1H NMR spectroscopy. Forty-five pathological metabolites were discovered that were not present in the healthy samples. The study demonstrated that 1H NMR can detect numerous metabolites in urine along with the enzyme defects that cause the inborn errors of metabolism. Based on the findings of this study, a statistical model of normality in the healthy population of newborns in Turkey was established, resulting in known distributions of metabolites to help identify inborn errors.93 Future studies will focus on unknown “pathological peaks” in the spectra to explore the progression of metabolic phenotypes with time.
Conclusions
Although metabolomics can be considered to be still in its infancy (compared with other “omics” platforms such as genomics), it has seen significant progress over the past few years. However, many challenges still remain. Although an effort can be made to the extent possible to impose restrictions on the samples collected for a study (eg, diet), the more practical approach would be to have a standardized protocol whereby samples are collected under similar conditions. The standardization should be applied to every stage of the process, including patient recruitment, sample collection, transport, storage, preparation, data acquisition, and analysis.2,36,94 Genetic factors, ethnicity, gender, age, and diet all do affect the metabolome, and hence any classifier developed for diagnostic purposes has to be made robust by the inclusion of a diverse patient and control population.27,36,67,93 Moreover, sample sizes in such studies have to be significantly large to ensure the necessary statistical power and avoid any bias. In this regard, multicenter and multinational studies that will facilitate access to patients having genetically and ethnically diverse samples should be encouraged.
As shown in the above applications, metabolomics of urine has a potential to play a significant role in diagnostics and biomarker discovery. The metabolic profile of urine obtained by NMR spectroscopy could provide useful information related to the different processes and pathways that may not be functioning properly in the human body. Given the recent focus on noninvasive and personalized medicine, urinary metabolomics could play an important role in that regard. Over the past few years, there has been significant development in NMR technology related to both hardware and software pushing urinary metabolomics further into the clinical arena. On the hardware side, there have been considerable improvements in sensitivity of the technique, progress in making it highly automated and user-friendly. On the software side, there has been a significant development in the way data are acquired, analyzed, and presented. With robust, fast, and user-friendly methods of data analysis generating diagnostic output that both practitioners and patients can easily understand, urinary metabolomics is finding its way in the mainstream of clinical diagnostics.
Footnotes
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1589 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) received no financial support for the research, authorship, and/or publication of this article.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
Conceived the idea of writing the review article and drafted an outline: TB. Performed extensive literature search: AC, TB. Wrote the first draft of the manuscript: AC, TB. Made critical revisions: OI, AC, TB. Final editing of the manuscript: AC, OI, TB. Approved final version: AC, OI, TB.
REFERENCES
- 1.Dettmer K, Hammock B. Metabolomics—a new exciting field within the “omics” sciences. Environ Health Perspect. 2004;112:A396–A397. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckonert O, Keun H, Ebbels T, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 3.Roessner U, Bowne J. What is metabolomics all about? Biotechniques. 46:363–365. doi: 10.2144/000113133. [DOI] [PubMed] [Google Scholar]
- 4.Van QN, Veenstra TD, Issaq HJ. Metabolic profiling for the detection of bladder cancer. Curr Urol Rep. 2011;12:34–40. doi: 10.1007/s11934-010-0151-3. [DOI] [PubMed] [Google Scholar]
- 5.Duarte LF, Gil AM. Metabolic signatures of cancer unveiled by NMR spectroscopy of human biofluids. Prog Nucl Magn Reson Spectrosc. 2012;62:51–74. doi: 10.1016/j.pnmrs.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson JK, Lindon JC, Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 7.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 8.Weckweth W. Metabolomics in systems biology. Annu Rev Plant Biol. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- 9.Duarte L, Diaz S, Gil A. NMR metabolomics of human blood and urine in disease research. J Pharm Biomed Anal. 2014;93:17–26. doi: 10.1016/j.jpba.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Bouatra S, Aziat F, Guo AC, et al. The human urine metabolome. PLoS ONE. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Aronov P, Zakharkin S. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horgan RP, Kenny LC. SAC review “omic” technologies: genomics, transcriptomics, proteomics and metabolomics. SAC Rev. 2011;12:189–195. [Google Scholar]
- 13.Cunningham MJ. Genomics and proteomics: the new millennium of drug discovery and development. J Pharmacol Toxicol Methods. 2000;44:291–300. doi: 10.1016/s1056-8719(00)00111-8. [DOI] [PubMed] [Google Scholar]
- 14.Theodorescu D, Mischak H. Mass spectrometry based proteomics in urine biomarker discovery. World J Urol. 2007;25:435–443. doi: 10.1007/s00345-007-0206-3. [DOI] [PubMed] [Google Scholar]
- 15.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 16.Celis JE, Gromov P. 2D protein electrophoresis: can it be perfected? Curr Opin Biotechnol. 1999;10:16–21. doi: 10.1016/s0958-1669(99)80004-4. [DOI] [PubMed] [Google Scholar]
- 17.Chandramouli K, Quian P. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009:239204. doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Urbanczyk-Wochniak E, Luedemann A, Kopka J, et al. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep. 2003;4:989–993. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 21.Dunn W, Bailey N, Johnson H. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 22.Psihogios N, Gazi I, Elisaf M, Seferiadis K, Bairaktari E. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR Biomed. 2008;21:195–207. doi: 10.1002/nbm.1176. [DOI] [PubMed] [Google Scholar]
- 23.Lehman-McKeeman LD, Car B. Metabonomics: application in predictive and mechanistic toxicology. Toxicol Pathol. 2004;32:94–96. [Google Scholar]
- 24.Neild GH, Foxall PJ, Lindon JC, Holmes EC, Nicholson JK. Uroscopy in the 21st century: high-field NMR spectroscopy. Nephrol Dial Transplant. 1997;12:404–417. doi: 10.1093/ndt/12.3.404. [DOI] [PubMed] [Google Scholar]
- 25.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindon J. The emergent role of metabolic phenotyping in dynamic patient stratification. Expert Opin Drug Metab Toxicol. 2014;10:915–919. doi: 10.1517/17425255.2014.922954. [DOI] [PubMed] [Google Scholar]
- 27.Saude EJ, Sykes BD. Urine stability for metabolomic studies: effects of preparation and storage. Metabolomics. 2007;3:19–27. [Google Scholar]
- 28.Kind T, Tolstikov V, Fiehn O, Weiss R. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 29.Messana I, Fomi F, Ferrari F, Rossi C, Giardina B, Zuppi C. Proton nuclear magnetic resonance spectral profiles of urine in type II diabetic patients. Clin Chem. 1998;44:1529–1534. [PubMed] [Google Scholar]
- 30.Smolinska A, Blanchet L, Buydens LMC, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 31.Rist MJ, Muhle-Goll C, Gorling B, et al. Influence of freezing and storage procedure on human urine samples in NMR-based metabolomics. Metabolites. 2012;3:243–258. doi: 10.3390/metabo3020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyataka H, Ozaki T, Himeno S. Effect of pH on 1H-NMR spectroscopy of mouse urine. Biol Pharm Bull. 2007;30:667–670. doi: 10.1248/bpb.30.667. [DOI] [PubMed] [Google Scholar]
- 33.Lauridsen M, Hansen S, Jaroszewski J, Cornett C. Human urine as test material in 1H NMR-based metabonomics: recommendations for sample preparation and storage. Anal Chem. 2007;79:1181–1186. doi: 10.1021/ac061354x. [DOI] [PubMed] [Google Scholar]
- 34.Vitols C, Fu H. Targeted Profiling of Common Metabolites in Urine. Edmonton, AB: Chenomx Inc; 2006. [Google Scholar]
- 35.Rosewell R, Vitols C. Identifying Metabolites in Biofluids. Edmonton, AB: Chenomx Inc; 2006. [Google Scholar]
- 36.Emwas AH, Luchinat C, Turano P, et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics. 2015;11:872–894. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Da Silva L, Godejohann M, Martin FP, et al. High-resolution quantitative metabolome analysis of urine by automated flow injection NMR. Anal Chem. 2013;84:5801–5809. doi: 10.1021/ac4004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraudeau P, Silvestre V, Akoka S. Optimizing water suppression for quantitative NMR-based metabolomics: a tutorial review. Metabolomics. 2015;11:1041–1055. [Google Scholar]
- 39.Emwas AH, Roy R, McKay RT, et al. Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. J Proteome Res. 2016;15:360–373. doi: 10.1021/acs.jproteome.5b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, Mo H, Wei S, Raftery D. Quantitative analysis of urea in human urine and serum by 1H nuclear magnetic resonance. Analyst. 2012;137:595–600. doi: 10.1039/c2an15780b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulrich EL, Akutsu H, Doreleijers JF, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig C, Easton JM, Lodi A, et al. Birmingham Metabolite Library: a publicly accessible database of 1-D 1H and 2-D 1H J-resolved NMR spectra of authentic metabolite standards (BML-NMR) Metabolomics. 2012;8:8–18. [Google Scholar]
- 43.Dona AC, Kyriakides M, Scott F, et al. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput Struct Biotechnol. 2016;12:135–153. doi: 10.1016/j.csbj.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Everett JR. A new paradigm for known metabolite identification in metabonomics/metabolomics: metabolite identification efficiency. Comput Struct Biotechnol. 2015;13:131–144. doi: 10.1016/j.csbj.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oman T, Tessem MB, Bathen TF, et al. Identification of metabolites from 2D (1) H-(13)C HSQC NMR using peak correlation plots. BMC Bioinformatics. 2014;15:413. doi: 10.1186/s12859-014-0413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadal-Desbarats L, Aidoud N, Emond P, et al. Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders. Analyst. 2014;139:3460–3468. doi: 10.1039/c4an00552j. [DOI] [PubMed] [Google Scholar]
- 47.Torgrip RJO, Aberg KM, Alm E, Schuppe-Koistinen I, Lindberg J. A note on normalization of biofluid 1D H-1NMR data. Metabolomics. 2008;4:114–121. [Google Scholar]
- 48.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 49.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 50.Hao J, Liebeke M, Astle W, De lorio M, Bundy JG, Ebbels TM. Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN. Nat Protoc. 2015;9:1416–1427. doi: 10.1038/nprot.2014.090. [DOI] [PubMed] [Google Scholar]
- 51.Chenomx NMR Suite http://www.chenomx.com/software/software.php.
- 52.Friedrich N, Budde K, Suhre K, et al. Sex differences in urine metabolites related with risk of diabetes using NMR spectroscopy: results of the study of health in Pomerania. Metabolomics. 2015;11:1405–1415. [Google Scholar]
- 53.Barker M, Ravens W. Partial least squares for discrimination. J Chemometr. 2003;3:166–173. [Google Scholar]
- 54.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi- and Megavariate Data Analysis: Principles and Applications. Umeå, Sweden: Umetrics Academy; 2001. [Google Scholar]
- 55.Jackson JE. A User’s Guide to Principal Components. New York, NY: Wiley; 1991. [Google Scholar]
- 56.Nikulin AE, Dolenko B, Bezabeh T, Somoriaj RL. Near-optimal region selection for feature space reduction: novel preprocessing methods for classifying MR spectra. NMR Biomed. 1998;11:209–216. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<209::aid-nbm510>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 57.Somorjai RL. Creating robust, reliable, clinically relevant classifiers from spectroscopic data. Biophys Rev. 2009;1:201–211. doi: 10.1007/s12551-009-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lean CL, Somorjai RL, Smith ICP, Russell P, Mountford CE. Accurate diagnosis and prognosis of human cancers by proton MRS and a three-stage classification strategy. Annu Rep NMR Spectrosc. 2002;48:71–111. [Google Scholar]
- 59.Bezabeh T, Somorjai RL, Dolenko B, et al. Detecting colorectal cancer by 1H magnetic resonance spectroscopy of fecal extracts. NMR Biomed. 2009;22:593–600. doi: 10.1002/nbm.1372. [DOI] [PubMed] [Google Scholar]
- 60.Bezabeh T, Ijare OB, NIkulin AE, Somoriai R, Smith ICP. MRS-based metabolomics in cancer research. Magn Reson Insights. 2014;7:1–14. doi: 10.4137/MRI.S13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somorjai RL, Nikulin AE, Pizzi N, et al. Computerized consensus diagnosis: a classification strategy for the robust analysis of MR spectra. I. Application to 1H spectra of thyroid neoplasms. Magn Reson Med. 1995;33:257–263. doi: 10.1002/mrm.1910330217. [DOI] [PubMed] [Google Scholar]
- 62.Somorjai RL, Dolenko B, Nikulin AK, et al. Classification of 1H MR spectra of human brain neoplasms: the influence of preprocessing computerized consensus diagnosis on classification accuracy. J Magn Reson Imaging. 1996;6:437–444. doi: 10.1002/jmri.1880060305. [DOI] [PubMed] [Google Scholar]
- 63.Xiao J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using metaboanalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 64.Rasmussen LG, Winning H, Savorani F, et al. Assessment of the effect of high or low protein diet on the human urine metabolome as measured by NMR. Nutrients. 2012;4:112–131. doi: 10.3390/nu4020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lenz EM, Bright J, Wilson I, et al. Metabonomics, dietary influences and cultural differences: a 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. J Pharm Biomed Anal. 2004;36:841–849. doi: 10.1016/j.jpba.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Dumas ME, Maibaum EC, Teague C, et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP study. Anal Chem. 2006;78:2199–2208. doi: 10.1021/ac0517085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuppi C, Messana I, Forni F, Ferrari F, Rossi C, Giardina B. Influence of feeding on metabolite excretion evidenced by urine 1H NMR spectral profiles: a comparison between subjects living in Rome and subjects living at arctic latitudes (Svaldbard) Clin Chim Acta. 1998;278:75–79. doi: 10.1016/s0009-8981(98)00132-6. [DOI] [PubMed] [Google Scholar]
- 68.Slupsky CM, Rankin KN, Wagner J, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 69.Psihogios N, Gazi I, Elisaf M, Seferiadis K, Bairaktari E. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR Biomed. 2008;21:195–207. doi: 10.1002/nbm.1176. [DOI] [PubMed] [Google Scholar]
- 70.Rasmussen LG, Savorani F, Larsen TM, Dragsted LO, Astrup A, Engelsen SB. Standardization of factors that influence human urine in metabolomics. Metabolomics. 2011;7:71–83. [Google Scholar]
- 71.Gupta A, Dwivedi M, Ali Mahdi A, Nagana Gowda G, Lal Khetrapal C, Bhandari M. 1H-nuclear magnetic resonance spectroscopy for identifying and quantifying common uropathogens: a metabolic approach to the urinary tract infection. BJU Int. 2009;104:236–244. doi: 10.1111/j.1464-410X.2009.08448.x. [DOI] [PubMed] [Google Scholar]
- 72.Bezabeh T, Ijare OB, Reed M, Blydt-Hansen T, Smith IC. NMR spectroscopy of urine for the detection of urinary tract infection (UTI) in children. Proc Intl Soc Mag Reson Med. 2010:3367. [Google Scholar]
- 73.Lam CW, Law CY, To KK, et al. NMR-based metabolomic urinalysis: a rapid screening test for urinary tract infection. Clin Chim Acta. 2014;436:217–223. doi: 10.1016/j.cca.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 74.Clarke W, Silverman B, Zhang Z, Chan D, Klein A, Molmenti E. Characterization of renal allograft rejection by urinary proteomic analysis. Ann Surg. 2003;237:660–665. doi: 10.1097/01.SLA.0000064293.57770.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rush D, Somorjai R, Deslauriers R, Shaw A, Jeffery J, Nickerson P. Subclinical rejection—a potential surrogate marker for chronic rejection—may be diagnosed by protocol biopsy or urine spectroscopy. Ann Transplant. 2000;5:44–49. [PubMed] [Google Scholar]
- 76.Le Moyec L, Pruna A, Eugene M, et al. Proton nuclear magnetic resonance spectroscopy of urine and plasma in renal transplantation follow-up. Nephron. 1993;65:433–439. doi: 10.1159/000187525. [DOI] [PubMed] [Google Scholar]
- 77.Foxall P, Mellotte G, Bending M, Lindon J, Nicholson J. NMR spectroscopy as a novel approach to the monitoring of renal transplant function. Kidney Int. 1993;43:234–245. doi: 10.1038/ki.1993.37. [DOI] [PubMed] [Google Scholar]
- 78.National Diabetes Statistics Report. http://www.cdc.gov/diabetes. Published 2014.
- 79.Doorn M, Vogels J, Tas A, et al. Evaluation of metabolite profiles as biomarkers for the pharmacological effects of thiazolidinediones in type 2 diabetes mellitus patients and healthy volunteers. Br J Clin Pharmacol. 2007;63:562–574. doi: 10.1111/j.1365-2125.2006.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicolescu A, Dolenko B, Bezabeh T, et al. Diagnosis of type II diabetes based on non-glucose regions of 1H NMR spectra of urine. Rev Chem. 2011;62:1150–1153. [Google Scholar]
- 81.Balderas C, Ruperez FJ, Ibanez E, et al. Plasma and urine metabolic fingerprinting of type 1 diabetic children. Electrophoresis. 2013;34:2882–2890. doi: 10.1002/elps.201300062. [DOI] [PubMed] [Google Scholar]
- 82.Deja S, Barg E, Mlynarz P, Basiak A, Willac-Janc W. 1H NMR-based metabolomics studies of urine reveal differences between type 1 diabetic patients with high and low HbAc1 values. J Pharm Biomed Anal. 2013;83:43–48. doi: 10.1016/j.jpba.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 83.Ng DJY, Paskinati KK, Chan ECY. Trend analysis of metabonomics and systematic review of metabonomics-derived cancer marker metabolites. Metabolomics. 2011;7:155–178. [Google Scholar]
- 84.Carrola J, Rocha C, Barrios A, et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 85.Chan A, Mercier P, Schiller D, et al. H-NMR urinary metabolomic profiling for diagnosis of gastric cancer. BR J Cancer. 2016;114:59–62. doi: 10.1038/bjc.2015.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trock B. Applications of metabolomics to prostate cancer. Urol Oncol. 2011;29:572–581. doi: 10.1016/j.urolonc.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaragoza P, Ruiz-Cerda JL, Quintas G, et al. Towards the potential use of (1)H NMR spectroscopy in urine samples for prostate cancer detection. Analyst. 2014;139:3875–3878. doi: 10.1039/c4an00690a. [DOI] [PubMed] [Google Scholar]
- 88.Sreekumar A, Poisson LM, Rejendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 89.Shen C, Sun Z, Chen D, et al. Developing urinary metabolomic signatures as early bladder cancer diagnostic markers. OMICS. 2015;19:1–11. doi: 10.1089/omi.2014.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Slupsky CM, Steed H, Wells TF, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. 2010;16:5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 91.Constantinou MA, Papakonstantinou E, Spraul M, et al. 1H NMR-based metabonomics for the diagnosis of inborn errors of metabolism in urine. Anal Chim Acta. 2005;542:169–177. [Google Scholar]
- 92.Wevers RA, Engelke UF, Moolenaar SH, et al. 1H-NMR spectroscopy of body fluids: inborn errors of purine and pyrimidine metabolism. Clin Chem. 1999;45:539–548. [PubMed] [Google Scholar]
- 93.Aygen S, Durr U, Hegele P, et al. NMR-based screening for inborn errors of metabolism: initial results from a study on Turkish neonates. JIMD Rep. 2014;16:101–111. doi: 10.1007/8904_2014_326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keun HC, Ebbels TM, Antti H, Bollard ME, Beckonert O, Scholtterbeck G. Analytical reproducibility in (1)H NMR-based metabonomic urinalysis. Chem Res Toxicol. 2002;15:1380–1386. doi: 10.1021/tx0255774. [DOI] [PubMed] [Google Scholar]