Abstract
Herein, we investigated the effects of new cofactors and inhibitors on an oxidative cleavage of DNA catalysis, known as a pistol-like DNAzyme (PLDz), to discuss its catalytic mechanism. PLDz performed its catalytic activity in the presence of ascorbic acid (AA), in which Cu2+ promoted, whereas Fe2+ significantly inhibited the catalytic function. Since Fe2+/AA-generated hydroxyl radicals are efficient on DNA damage, implying that oxidative cleavage of PLDz had no relation with hydroxyl radical. Subsequently, we used Fe2+/H2O2 and Cu2+/H2O2 to identify the role of hydroxyl radicals in PLDz catalysis. Data showed that PLDz lost its activity with Fe2+/H2O2, but exhibited significant cleavage with Cu2+/H2O2. Because Fe2+/H2O2 and Cu2+/H2O2 are popular reagents to generate hydroxyl radicals and the latter also produces superoxide anions, we excluded the possibility that hydroxyl radical participated in oxidative cleavage and confirmed that superoxide anion was involved in PLDz catalysis. Moreover, pyrogallol, riboflavin and hypoxanthine/xanthine oxidase with superoxide anion and hydrogen peroxide generation also induced self-cleavage of PLDz, where catalase inhibited but superoxide dismutase promoted the catalysis, suggesting that hydrogen peroxide played an essential role in PLDz catalysis. Therefore, we proposed a catalytic mechanism of PLDz in which superoxide anion and hydrogen peroxide mediated an oxidative cleavage process.
Introduction
In living systems, reactive oxygen species (ROS) include superoxide anion (O2 •−), hydrogen peroxide (H2O2), hydroxyl radical (OH•), organic (lipid, alkyl, or short chain) hydroperoxides and hydroperoxide radicals (ROOH, ROO•), hypochlorous acid (HOCl), singlet oxygen (1O2), and ozone (O3)1, 2. Nowadays, it is widely accepted that ROS play a dual physiological role, not only in various diseases, but also in cellular homeostasis3. Among reactive oxygen species, hydroxyl radical is extremely reactive and able to attack many biomolecules, such as nucleic acids, proteins and lipids.
In biological studies, the major source of hydroxyl radicals comes from Fenton reaction of Fe2+ with H2O2. The Fenton reaction initiates the Equation 1, which is then followed by the indicated in Equations 2 and 3. The reaction termination is caused by the Equation 4. The Equations 2 and 3 were commonly known as the Haber-Weiss cycle4.
| 1 |
| 2 |
| 3 |
| 4 |
Fenton reagents have been expanded from the original Fe2+/H2O2 system to the Fe2+/H2O2/AA (Fe2+/AA) system by introducing ascorbic acid (AA)5, 6. Meantime, it was reported that Cu2+ could also induce Fenton-like reactions into the Cu2+/AA (Cu2+/H2O2) system. In some studies, researchers have found that the Cu2+/AA (Cu2+/H2O2) system leads to DNA cleavage and damage7, 8. The cause has been attributed to the production of hydroxyl radicals by Fenton reaction (Eqs 5–8) to attack the deoxyribose DNA backbone and bases.
| 5 |
| 6 |
| 7 |
| 8 |
In the mid-1990s, Carmi et al. using Cu2+/AA (or Cu2+) as cofactors obtained a series of oxidative cleavage DNA catalysis by in vitro selection, in which a pistol-like DNAzyme (PLDz) was the most active structure9–11. Based on its cofactors and structure, PLDz has been developed into Cu2+-, AA-, and glucose-biosensors, DNA molecular logic gates and a dual-catalytic allosteric DNAzyme12–18. However, little is known about the catalytic mechanism of PLDz except the oxidative cleavage of DNA. In this study, we found new cofactors for PLDz catalysis and further investigated their effects on the catalytic activity of PLDz. Our experimental data excluded that PLDz catalyzed a hydroxyl radical-mediated cleavage reaction and supported that superoxide anion and hydrogen peroxide might play a critical role in the oxidative cleavage process.
Results
General cofactors requirements for PLDz function
A 56-nucleotide version of cis pistol-like DNAzyme (PLDz) was shown in Fig. 1. PLDz composes of a 15-nucleotide active core surrounded by a triple helix in the left arm and a double stranded helix in the right arm. The addition of GAGA at 5′ end allows separation of cleavage fragments by denaturing gel electrophoresis.
Figure 1.
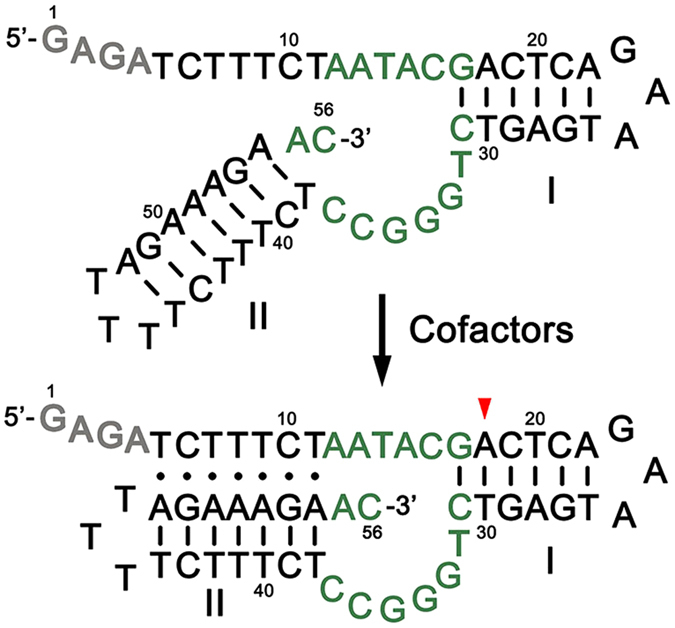
The sequence and secondary structure of a cis pistol-like DNAzyme. The green letters correspond to the conserved nucleotides of catalytic core. The red arrowhead indicates the major site of DNA cleavage. I and II designate stem-loop structures, where lines indicate Watson-Crick base pairs and dots represent triplex interactions.
In previous studies, PLDz was generally labeled at the 5′ end with [γ-32P] ATP by T4 polynucleotide kinase9–11, 18. Using 5′ end-labeled PLDz for analysis, only 5′ cleavage fragments can be observed by autoradiography, while 3′ and middle cleavage fragments can′t be identified. In here, we used label-free PLDz for analysis. All cleavage products including 5′, 3′ and middle cleavage fragments can be separated by gel electrophoresis and detected with GelRed staining. As shown in Fig. 2A, catalysis by PLDz with Cu2+ or H2O2 alone has only been observed with low efficiency, which is consistent with previously reported data10, 11, 18. Surprisingly, AA as the sole cofactor was able to efficiently support cleavage of PLDz. In addition, PLDz showed slightly better catalytic activity in the Cu2+/AA system as compared to the Cu2+/H2O2 system. And addition of H2O2 did not improve cleavage efficiency compared with the corresponding Cu2+/AA. In Fig. 2A, self-cleavage of PLDz can generate multiple fragments, indicating that there exist several cleavage sites. These multiple cleavage sites could be predicted by comparison of DNA fragments lengths with 5′ and 3′ marker DNAs (Supplementary Figure 1). The a1 and a2 bands corresponded to the 5′ and 3′ cleavage fragments occurred at the cleavage site A18, which has been reported as the major cleavage site10, 11. The b1–3 and b4–7 bands could lead to the 5′ and 3′ cleavage fragments occurred at the minor cleavage sites. The c1–3 bands could be small cleavage fragments cleaving simultaneously at both major and minor cleavage sites. As a result of b1–7 and c1–3 bands, the minor cleavage sites in PLDz could be concluded as T32, G33 and G34. In Supplementary Figure 1, DNA cleavage fragments migrated differently with the same length of DNA markers, due to deoxynucleotide moieties bearing at their 3′ and 5′ ends. The newly generated 3-terminal end can be phosphoglycolate11, 19, 20 and might be further converted into 3′ phosphate21, while the 5′ end has a terminal phosphate group (Fig. 2B)21. In order to assess the influence of cofactors on PLDz activity, we conducted a series of DNAzyme assays using five groups of cofactors in a concentration series from 10 μM to 10 mM. As shown in Fig. 2C, PLDz showed a relation of linear dependence on H2O2 concentration, but exhibited bell-shaped dependence on other cofactors (Cu2+, AA, AA/Cu2+ and H2O2/Cu2+). Since a higher concentration of PLDz (1 μM) was used in our study, the optimal concentration of Cu2+ is 100 μM, ten times higher than concentration in previous reports11, 18.
Figure 2.
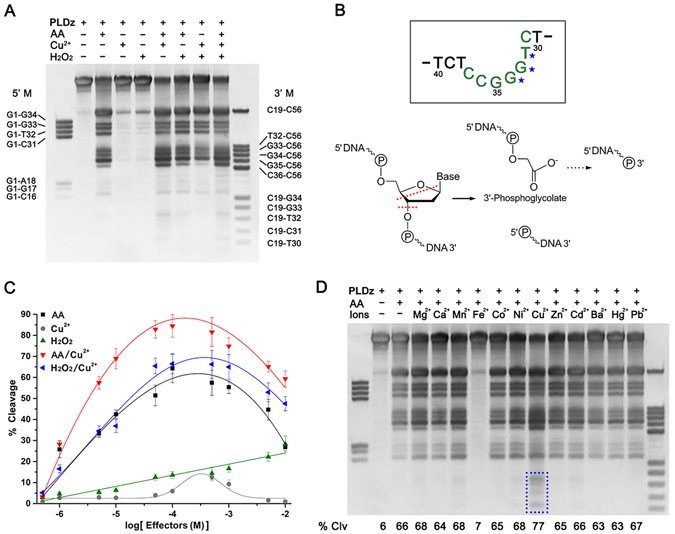
PLDz catalysis in the presence of different cofactors. (A) Effects of general cofactors on PLDz-catalyzed cleavage. PLDz (1 μM) was incubated with 100 μM cofactors at 23 °C for 2 hr in a mixture containing 50 mM Tris-HCl (7.0) and 300 mM NaCl. Lanes designated 5′ M and 3′ M were synthetic DNAs of different lengths as indicated, each with a sequence that corresponds to the 5′ and 3′ terminus (including middle fragments) of the PLDz, respectively (Supplementary Table 1). Letter-numbers indicate the lengths of these marker DNAs. Reaction products were separated by denaturing (7 M urea) 20% PAGE and were visualized by GelRed dye staining. (B) Oxidative cleavage of the target site deoxynucleotide. The DNA in the grey square represents the part structure of PLDz. The blue asterisks indicate the minor sites of DNA cleavage. The red dashed lines depict the possible locations of deoxynucleoside cleavage fragmentation. Dashed arrow indicates the possible conversion of phosphoglycolate into phosphate. (C) Concentration-dependent induction of PLDz function by cofactors. (D) Analysis of the function of PLDz with divalent metal ions in the presence of AA. The dashed box represents small cleavage fragments of PLDz including c19-c31, c19-t32 and c19-g33 (Supplementary Figure 1), which appeared more clearly with increased levels of exposure (Supplementary Figure 2) Reactions were conducted as described in part A. Note that only Fe2+ inhibited PLDz activity. Cropped gels are used in Fig. 2A and D, their full-length gels are presented in Supplementary Figure 3.
In the following, we tested the activities of PLDz in the presence of AA and different metal ions (Mg2+, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Ba2+, Hg2+ and Pd2+) (Fig. 2D). At constant 100 μM AA, the equal concentration of Cu2+ was able to enhance PLDz catalysis. As shown in Fig. 2D, small cleavage fragments were observed upon addition of Cu2+and the cleavage yield was slightly higher than others. Similarly, PLDz only displayed a slight improved DNA cleavage activity in the presence of Cu2+, when incubated with different divalent metal ions alone (Supplementary Figure 4). Additionally, as shown in Fig. 2D, Fe2+ significantly inhibited the cleavage reaction of PLDz with AA as cofactor. PLDz lost its half catalytic activity by addition of 10 μM Fe2+ and almost abolished it at 100 μM Fe2+ (Supplementary Figure 5). In fact, the Fe2+/AA system generating hydroxyl radicals are efficient on DNA damage22–25. Therefore, we inferred that oxidative cleavage of PLDz had nothing to do with hydroxyl radicals.
The role of hydroxyl radicals in the catalytic activity of PLDz
To further investigate the role of hydroxyl radicals in the catalytic activity of PLDz, we first evaluated the effects of different divalent metal species (Mg2+, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Ba2+, Hg2+ and Pd2+) on hydroxyl radical generation in the presence of H2O2 as measured by 3,3′,5,5′-tetramethylbenzidine (TMB) oxidation. Apparently, Fe2+/H2O2 can produce a more amount of hydroxyl radicals comparing with Cu2+/H2O2 (Fig. 3A). In addition, no hydroxyl radical was produced by treatment with other metal ions/H2O2. The hydroxyl radical formation rate from Fe2+/H2O2 was very high and its initial point had already reached its maximum peak (Supplementary Figure 6). In contrast, the hydroxyl radicals produced in Cu2+/H2O2 system grew linearly over the time.
Figure 3.
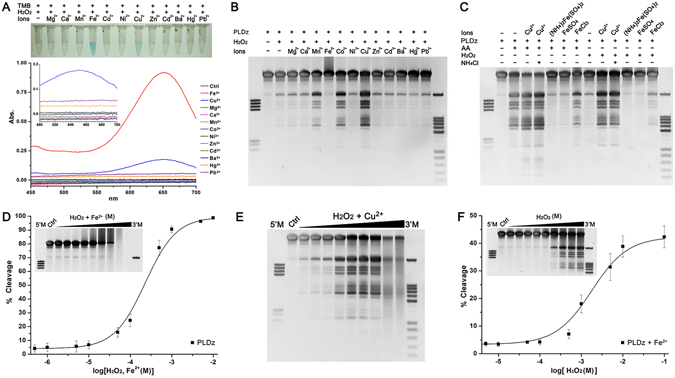
The role of hydroxyl radicals in the catalytic activity of PLDz. (A) Comparison of hydroxyl radical generation by treatment with different metal ions in the presence of H2O2. (B) Effects of divalent metal ions/H2O2 (100 μM) on the cleavage yield of PLDz. (C) Effects of Fe2+ (100 μM), Fe3+ (100 μM) and NH4 + (200 μM) on the cleavage yield of PLDz. (D) Effects of Fenton reagent (Fe2+/H2O2, 1 μM–0.01 M) on the cleavage yield of PLDz. (E) Effect of Cu2+/H2O2 concentration (1 μM–0.01 M) on the cleavage yield of PLDz. (F) Effect of H2O2 concentration (10 μM–0.1 M) on the cleavage yield of PLDz in present of 100 μM Fe2+. Reaction condition: 300 mM NaCl and 50 mM Tris-HCl (pH 7.0) at 23 °C for 2 hr. The error bars represented the standard deviations from three repeated measurements. Cropped gels are used in Fig. 3B–F, their full-length gels are presented in Supplementary Figure 8.
We analyzed the different effects of metal ions/H2O2 on the cleavage reaction of PLDz (Fig. 3B). PLDz exhibited enhanced activities when treated separately with Cu2+, Co2+ and Mn2+ in the presence of H2O2, in the order Cu2+ > Co2+ > Mn2+. Other metal ions (Mg2+, Ca2+, Ni2+, Zn2+, Cd2+, Ba2+, Hg2+ and Pd2+) had no effect and Fe2+ showed a significant inhibitory effect. To further confirm the inhibitory effect on PLDz catalysis is due to Fe2+ rather than NH4 + in (NH4)2Fe(SO4)2 or Fe3+, we also investigated the effects of FeSO4, FeCl3 and NH4Cl on PLDz catalysis. Results showed that (NH4)2Fe(SO4)2 and FeSO4 inhibited the cleavage of PLDz, but FeCl3 and NH4Cl did not lower PLDz catalysis in the presence of AA or H2O2 (Fig. 3C). These results indicated that Fe2+ but not Fe3+ had an inhibitory effect on PLDz catalysis, while NH4 + in (NH4)2Fe(SO4)2 did not interfere with self-cleavage of PLDz.
So far, many papers have reported that hydroxyl radicals produced by Fe2+/H2O2 possess the ability to significantly destroy the DNA structure26–28. Our observation on DNA damage by Fe2+/H2O2 (Fig. 3D) is in agreement with previous reports. At 100 μM Fe2+/H2O2, there was no site-specific cleavage fragment but a smeared DNA observed, indicating that DNA damage occurred. At 1 mM Fe2+/H2O2, obvious smeared bands were displayed on the gel; at 5 mM Fe2+/H2O2, DNA was significantly damaged and could not bind the DNA staining reagent, showing a significant smear on the gel. At 10 mM Fe2+/H2O2, DNA was completely damaged and nothing appeared on the gel. This result provided a proof that hydroxyl radicals did not play a role in catalysis of PLDz. In Fig. 3D, Fe2+ resulted in an upper diffusion band, because a brown precipitate was formed by oxidation of Fe2+ to Fe3+ (Supplementary Figure 7) and decreased DNA electrophoretic mobility.
Both Fe2+ and Cu2+ can generate hydroxyl radicals in the presence of H2O2 (Fig. 3A and Supplementary Figure 6), however, they performed the opposing effects on PLDz catalysis (Fig. 3B and C). The rapid reaction of Fe2+ and H2O2 produces a large amount of hydroxyl radicals (Eq. 1), whereas the Cu2+/H2O2 system generates hydroxyl radicals also superoxide anions (Eq. 9 and 10)29, 30. Data (Fig. 3B and C) showed that Cu2+/H2O2 rather than Fe2+/H2O2 led to self-cleavage of PLDz, which indicated that superoxide anion took part in the cleavage process of PLDz. According to Eq. 10, high level of Cu2+/H2O2 (≥5 mM) can produce a large amount of hydroxyl radicals, which can non-specifically damage PLDz and leave smeared bands in gel, consistent with Fig. 3E. Other metal ions, Mn2+ and Co2+, were found to increase PLDz catalysis in the presence of H2O2 (Fig. 3B), suggesting that the Mn2+/H2O2 and Co2+/H2O2 systems can also generate superoxide anions.
| 9 |
| 10 |
At constant 100 μM Fe2+ and a range of H2O2 concentrations from 10 μM to 100 mM, we found that the activity of PLDz was recovered at the concentrations of H2O2 above 5 mM (Fig. 3F). This result seems inconsistent with previous Fe2+/H2O2-induced unspecific DNA degradation shown in Fig. 3D. But in fact, hydroxyl radicals reacted with excess of H2O2 in Fe2+/H2O2 system via the Haber-Weiss reaction (Eq. 2) to form superoxide anions. Moreover, high level of H2O2 can also promote the cleavage of PLDz, Therefore, it was a clue that superoxide anion and hydrogen peroxide could play an essential role in the cleavage reaction of PLDz.
Effects of pyrogallol and riboflavin on PLDz
Pyrogallol (PG) and riboflavin (RF) are generally utilized to generate superoxide anion and hydrogen peroxide by autoxidation and photo-oxidation, respectively31, 32. We chose both of them for further analysis. Results shown in Fig. 4A displayed that PLDz was self-cleaved when incubated with either pyrogallol or riboflavin with a concentration over 100 μM, suggesting that superoxide anion and hydrogen peroxide participated in PLDz-catalyzed DNA cleavage reaction. Addition of pyrogallol or riboflavin to 100 μM Cu2+ resulted in specific cleavage of PLDz in high yield. It may result from the superoxide anions generated through the reaction of Cu2+ and hydrogen peroxide. Fe2+ still inhibited the stimulatory effects of pyrogallol and riboflavin on PLDz catalysis, because Fe2+ not only participated in redox processes with pyrogallol and riboflavin but also used hydrogen peroxide to generate hydroxyl radical.
Figure 4.
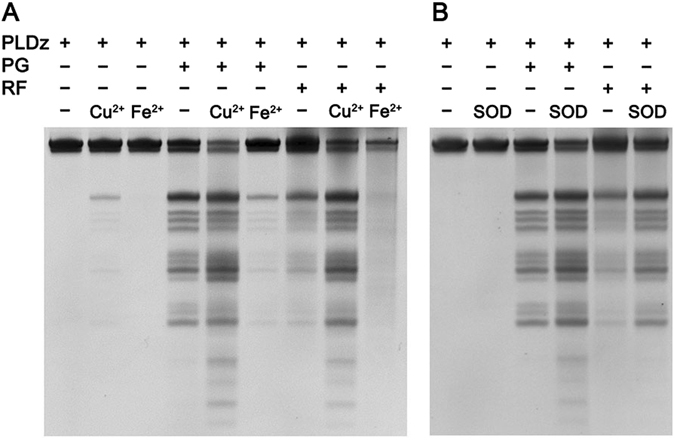
The function of pyrogallol (PG) and riboflavin (RF) on the catalytic activity of PLDz. (A) Effects of autoxidation of PG and photo-oxidation of RF on the cleavage reaction of PLDz. Reaction condition: 0.4 μM PLDz, 100 μM PG (RF), 100 μM Cu2+ (Fe2+), 300 mM NaCl and 50 mM Tris-HCl (pH 7.0) at 23 °C for 2 hr. The reaction system containing RF need to performed under a sunlight lamp. (B) Effects of SOD on the cleavage reaction of PLDz in the presence of H2O2. Reaction condition: 0.4 μM PLDz, 100 μM PG (RF), 0.3 U/μl SOD, 300 mM NaCl and 50 mM Tris-HCl (pH 7.0) at 23 °C for 2 hr. Cropped gels are used in Fig. 4A,B, their full-length gels are presented in Supplementary Figure 9.
It is well known that superoxide dismutase (SOD) can efficiently convert superoxide anion to dioxygen and hydrogen peroxide (Eq. 11). We added SOD into the reaction system of pyrogallol or riboflavin to evaluate the effects of superoxide anion on PLDz catalysis. Results showed that PLDz performed an even stronger cleavage activity with the induction of pyrogallol/SOD (riboflavin/SOD) than pyrogallol (riboflavin) alone (Fig. 4B), indicating that hydrogen peroxide played an important role in pyrogallol- and riboflavin-mediated cleavage of PLDz. Similarly, hydrogen peroxide with the addition of SOD also improved the cleavage yield of PLDz (Supplementary Figure 10). This result is consistent with the earlier result about DNA cleavage mediated by H2O2/SOD by Han et al. 33.
| 11 |
| 12 |
In order to improve detection sensitivity, we designed a fluorescent labeled trans-PLDz (Fig. 5A), in which DNAzyme (PLB) contained 5′-quencher (BHQ2) and the substrate (SBC) was labeled with a Cy3 at the 3′-end and BHQ2 at the 5′-end. When PLB bound SBC forming an enzyme-substrate complex, Cy3 emission was quenched by the nearby BHQ2. In the presence of cofactors, PLB catalyzed SBC cleavage and the cleaved DNA fragments with Cy3 were released, resulting in increased fluorescence. As shown in Fig. 5B, the PLB system emitted fluorescent signal in the presence of pyrogallol. The fluorescent intensity enhanced with the addition of SOD, which is in agreement with Fig. 4B. But it was dramatically reduced with the introduction of catalase (CAT) due to quick decomposition of hydrogen peroxide by CAT (Eq. 12) and inhibition of PLB catalysis. These results suggested that SOD and CAT had opposite effects on PLB and can be used as the positive and negative regulatory factors of PLDz to regulate its enzymatic activity. Besides, similar results in PLB system were observed using riboflavin as cofactor (Supplementary Figure 11).
Figure 5.
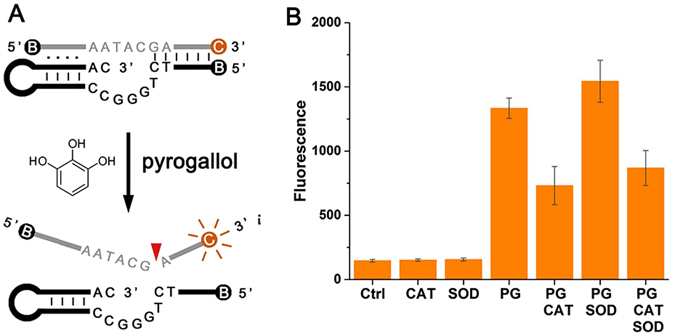
Effect of pyrogallol (PG) on a fluorescent labeled trans-PLDz. (A) Design of a fluorescent labeled trans-PLDz. C indicates Cy3, B indicates BHQ2. (B) Effects of CAT/SOD on the cleavage reaction of PLDz in the presence of PG. Reaction condition: 50 nM PLB, 50 nM SBC, 10 μM PG (50 mU/μl CAT, 5 mU/μl SOD), 300 mM NaCl and 50 mM MES (pH 6.0) at 37 °C for 1 hr.
Effects of hypoxanthine/xanthine oxidase on PLDz
We chose pyrogallol and riboflavin as generators of superoxide anion and hydrogen peroxide in previous text. Since the oxidation process of pyrogallol and riboflavin also produce other byproducts, we further used enzyme reactions to produce superoxide anion and hydrogen peroxide in order to decrease interference. Xanthine oxidase (XO) is generally recognized as a key enzyme in the catabolism of purines, which can catalyze hypoxanthine (HX) to generate uric acid, superoxide anion and hydrogen peroxide34, 35. In here, the hypoxanthine/xanthine oxidase system was utilized to produce superoxide anion and hydrogen peroxide. As shown in Fig. 6, hypoxanthine/xanthine oxidase led PLB to cleave its DNA substrate. Fluorescent signal increased with the addition of SOD and decreased with additional CAT, consistent with the experimental results of pyrogallol and riboflavin.
Figure 6.
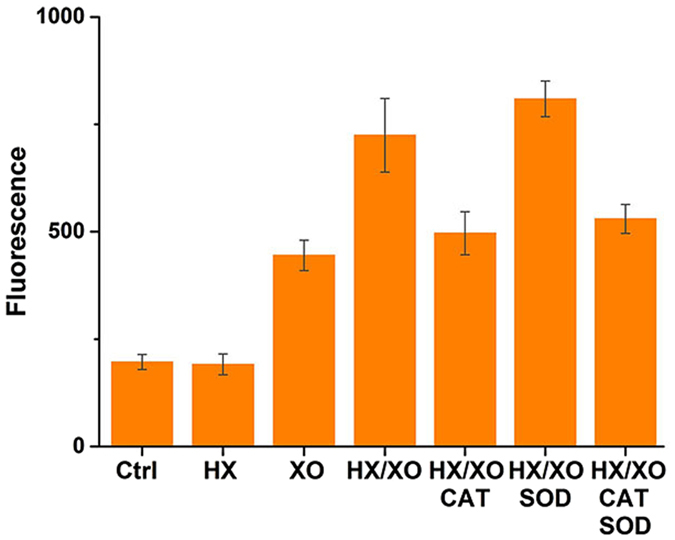
Effects of HX/XO on PLDz catalysis. Reaction condition: 50 nM PLB, 50 nM SBC, 10 μM HX, 1 mU/μl XO (50 mU/μl CAT, 5 mU/μl SOD), 300 mM NaCl and 50 mM MES (pH 6.0) at 37 °C for 1 hr.
Hydrogen peroxide played a key role in the process of PLDz cleavage according to the experimental results of hypoxanthine/xanthine oxidase system. Based on this principle, the oxidases that can generate hydrogen peroxide could also cause PLDz catalysis, such as amino-acid oxidase, glucose oxidase, gulonolactone oxidase, lysyl oxidase, monoamine oxidase, NAD(P)H oxidase, urate oxidase, and so on. In addition, we thought that some compounds similar to pyrogallol and riboflavin in structure can be used as the cofactors of PLDz, such as catechol, phloroglucinol, flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), etc.
Conclusions
Our study indicates that pistol-like DNAzyme promoted an oxidative DNA cleavage using many cofactors and inhibitors. Newly discovered cofactors of PLDz include Mn2+/H2O2, Co2+/H2O2, pyrogallol(/Cu2+), riboflavin(/Cu2+), superoxide dismutase and hypoxanthine/xanthine oxidase, while its inhibitors include Fe2+ and catalase. The results that PLDz catalysis was assisted by Cu2+/H2O2 and inhibited by Fe2+/H2O2 excluded hydroxyl radical-mediated catalytic mechanism of PLDz. Data from the Cu2+/H2O2 system proved that superoxide anion played a critical role in the process of PLDz catalysis. Moreover, experimental results from pyrogallol, riboflavin and hypoxanthine/xanthine oxidase systems supported that hydrogen peroxide played an essential role in PLDz catalysis. Therefore, we proposed a catalytic mechanism of PLDz in which superoxide anion and hydrogen peroxide mediated an oxidative cleavage process.
Methods
Reagents and chemicals
The DNA oligonucleotides were purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and purified by denaturing PAGE or HPLC (Supplementary Table 1). GelRed was purchased from Biotium Inc. Metal ions, cofactors and other reagents were of analytical reagent grade. Deionized and distilled water was used throughout the experiments.
Detection of cleavage fragments of PLDz by polyacrylamide gel electrophoresis
DNA self-cleavage assays were conducted in 100 μl reaction solution containing 1 μM PLDz, 50 mM Tris-HCl (pH 7.0), 300 mM NaCl and cofactors. The reaction mixture was incubated at 37 °C for 2 hr and stopped by adding precipitants (200 μl (2× vol) 100% ethanol, 10 μl (1/10 vol) 3 M NaOAc (pH 5.2), 1 μl 10 mg/ml glycogen) for precipitation. The dried samples were dissolved in 20 μl loading buffer (4 M Urea, 10 mM EDTA, 25 mM Tris-HCl pH 7.5, 0.125‰ xylene cyanol FF, 0.125‰ bromophenol blue) and separated by electrophoresis in denatured 20% polyacrylamide gel. Gel was stained with GelRed dye for 10 min and visualized by UV transillumination. The cleavage yield was determined by the equation: cleavage (%) = all cleaved fragments/(non-cleaved fragments + all cleaved fragments) × %. Note: GelRed is a highly sensitive, low toxicity, fluorescent DNA stain designed to replace the highly toxic ethidium bromide (EtBr) and has similar staining protocol as EtBr. The post-staining protocol is to dilute GelRed 10,000× stock solution ~3300 fold to prepare a 3× working solution in 0.1 M NaCl. For example, 50 ml of the staining solution is composed of 15 μl GelRed 10,000× stock solution, 5 mL of 1 M NaCl, and 45 ml H2O.
Visible spectroscopy detection
A 200 μl reaction mixture containing 200 μM metal ions, 1 mM H2O2, 0.1 mg/ml TMB and 50 mM Tris-HCl (pH 7.0). The reaction mixture was incubated at 23 °C for 3 min and scanned 450–750 nm by UV-2550 (Shimadzu), or measured the absorbance at 652 nm in the range of 0–300 sec.
Fluorescence spectroscopy detection
A 100 μl reaction mixture containing 50 nM PLB, 50 nM SBC, 10 μM pyrogallol (10 μM riboflavin, 10 μM hypoxanthine/5 mU/μl xanthine oxidase), 50 mU/μl catalase, 5 mU/μl superoxide dismutase), 300 mM NaCl and 50 mM MES (pH 6.0) were incubated at 37 °C for 1 hr and scanned by Infinite F200 (Tecan). Parameter setting: Plate: Corning (3925), Excitation Wavelength: 530 nm and Emission Wavelength: 590 nm.
Electronic supplementary material
Acknowledgements
This work was supported by the S&T Development Planning Program of Jilin Province (20111806, 20130101169JC, 20170101157JC), the Fundamental Research Funds for the Central Universities (450060501435, 450060488087 and 450060445666), Graduate Innovation Fund of Jilin University (2015097) and College Students’ Innovative Entrepreneurial Training Plan of Jilin University (2013A34095).
Author Contributions
D. Jiang and J. Xu designed the study. Y. Sun, R. Ma, S. Wang and D. Jiang performed the experiment. D. Jiang, J. Xu, Y. Sun, R. Ma, S. Wang, G. Li, Y. Sheng, H. Rui and J. Zhang analysed the data. J. Xu and D. Jiang wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yanhong Sun and Rulong Ma contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00329-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiacui Xu, Email: jcxu@jlu.edu.cn.
Dazhi Jiang, Email: jiangdz@jlu.edu.cn.
References
- 1.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat. Rev. Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valko M, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Koppenol WH. The Haber-Weiss cycle - 70 years later. Redox Rep. 2001;6:229–234. doi: 10.1179/135100001101536373. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Lee GJ, Jang KJ, Cho TS, Kim SK. Real-time detection of Fe·EDTA/H2O2-induced DNA cleavage by linear dichroism. Nucleic Acids Res. 2008;36:e85. doi: 10.1093/nar/gkn370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duarte TL, Jones GDD. Vitamin C modulation of H2O2-induced damage and iron homeostasis in human cells. Free Radical Biol. Med. 2007;43:1165–1175. doi: 10.1016/j.freeradbiomed.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 7.LaMarr WA, Sandman KM, Reeve JN, Dedon PC. Differential effects of DNA supercoiling on radical-mediated DNA strand breaks. Chem. Res. Toxicol. 1997;10:1118–1122. doi: 10.1021/tx970072c. [DOI] [PubMed] [Google Scholar]
- 8.Duarte TL, Lunec J. When is an antioxidant not an antioxidant? A review of novel actions and reactions of vitamin C. Free Radical Biol. Med. 2005;39:671–686. doi: 10.1080/10715760500104025. [DOI] [PubMed] [Google Scholar]
- 9.Carmi N, Shultz LA, Breaker RR. In vitro selection of self-cleaving DNAs. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/S1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 10.Carmi N, Balkhi SR, Breaker RR. Cleaving DNA with DNA. Proc. Natl. Acad. Sci. USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmi N, Breaker RR. Characterization of a DNA-cleaving deoxyribozyme. Bioorg. Med. Chem. 2001;9:2589–2600. doi: 10.1016/S0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Lu Y. A DNAzyme catalytic beacon sensor for paramagnetic Cu2+ ions in aqueous solution with high sensitivity and selectivity. J. Am. Chem. Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- 13.Miao X, Ling L, Cheng D, Shuai X. A highly sensitive sensor for Cu2+ with unmodified gold nanoparticles and DNAzyme by using the dynamic light scattering technique. Analyst. 2012;137:3064–3069. doi: 10.1039/c2an35217f. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Huang X, Kong D, Shen H, Liu Y. Ultrasensitive, high temperature and ionic strength variation-tolerant Cu2+ fluorescent sensor based on reconstructed Cu2+-dependent DNAzyme/ substrate complex. Biosens. Bioelectron. 2013;42:225–228. doi: 10.1016/j.bios.2012.10.070. [DOI] [PubMed] [Google Scholar]
- 15.Malashikhina N, Pavlov V. DNA-decorated nanoparticles as nanosensors for rapid detection of ascorbic acid. Biosens. Bioelectron. 2012;33:241–246. doi: 10.1016/j.bios.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Liu C, et al. A glucose oxidase-coupled DNAzyme sensor for glucose detection in tears and saliva. Biosens. Bioelectron. 2015;70:455–561. doi: 10.1016/j.bios.2015.03.070. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. Construction of molecular logic gates with a DNA-cleaving deoxyribozyme. Angew. Chem. Int. Ed. 2006;45:1759–1762. doi: 10.1002/anie.200502511. [DOI] [PubMed] [Google Scholar]
- 18.Jiang D, Xu J, Sheng Y, Sun Y, Zhang J. An allosteric DNAzyme with dual RNA-cleaving and DNA-cleaving activities. FEBS J. 2010;277:2543–2549. doi: 10.1111/j.1742-4658.2010.07669.x. [DOI] [PubMed] [Google Scholar]
- 19.Taverna Porro ML, Greenberg MM. DNA double strand cleavage via interstrand hydrogen atom abstraction. J. Am. Chem. Soc. 2013;135:16368–16371. doi: 10.1021/ja409513q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Klauser PC, Brandsen BM, Zhou C, Li X, Silverman SK. DNA-catalyzed DNA cleavage by a radical pathway with well-defined products. J. Am. Chem. Soc. 2017;139:255–261. doi: 10.1021/jacs.6b10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Zhang H, Zhang W, Zhao Y, Yasmeen A, Zhou L, Yu X, Tang Z. In vitro selection of DNA-cleaving deoxyribozyme with site-specific thymidine excision activity. Nucleic Acids Res. 2014;42:9262–9269. doi: 10.1093/nar/gku592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronovitch J, Godinger D, Samuni A, Czapski G. Ascorbic acid oxidation and DNA scission catalyzed by iron and copper chelates. Free Radic. Res. Commun. 1987;2:241–258. doi: 10.3109/10715768709065289. [DOI] [PubMed] [Google Scholar]
- 23.Hu ML, Shih MK. Ascorbic acid inhibits lipid peroxidation but enhances DNA damage in rat liver nuclei incubated with iron ions. Free Radic. Res. 1997;26:585–592. doi: 10.3109/10715769709097828. [DOI] [PubMed] [Google Scholar]
- 24.Rehman A, et al. The effects of iron and vitamin C co-supplementation on oxidative damage to DNA in healthy volunteers. Biochem. Biophys. Res. Commun. 1998;246:293–298. doi: 10.1006/bbrc.1998.8592. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad S, Kitchin KT, Cullen WR. Plasmid DNA damage caused by methylated arsenicals, ascorbic acid and human liver ferritin. Toxicol. Lett. 2002;133:47–57. doi: 10.1016/S0378-4274(02)00079-6. [DOI] [PubMed] [Google Scholar]
- 26.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-H. [DOI] [PubMed] [Google Scholar]
- 27.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 28.Jomova K, Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283:65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Pham AN, Rose AL, Waite TD. Kinetics of Cu(II) reduction by humic substances in natural waters. J. Phys. Chem. A. 2012;116:6590–6599. doi: 10.1021/jp300995h. [DOI] [PubMed] [Google Scholar]
- 30.Pham AN, Xing G, Miller CJ, Waite TD. Fenton-like copper redox chemistry revisited: Hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J. Catal. 2013;301:54–64. doi: 10.1016/j.jcat.2013.01.025. [DOI] [Google Scholar]
- 31.Marklund S, Marklund C. Involvement of the superoxide anion redical in the autoxidation of pyrogallol and a convenient assay of superoxide dismutase. Eur. J. Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 32.Ito K, Inoue S, Yamamoto K, Kawanishi S. 8-hydroxydeoxyguanosine formation at the 5′ site of 5′-GG-3′ sequences in double-stranded DNA by UV radiation with riboflavin. J. Biol. Chem. 1993;268:13221–13227. [PubMed] [Google Scholar]
- 33.Han Y, Shen T, Jiang W, Xia Q, Liu C. DNA cleavage mediated by copper superoxide dismutase via two pathways. J. Inorg. Chem. 2007;101:214–224. doi: 10.1016/j.jinorgbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Bonini MG, Miyamoto S, Mascio PD, Augusto O. Production of the carbonate radical anion during xanthine oxidase turnover in the presence of bicarbonate. J. Biol. Chem. 2004;279:51836–51843. doi: 10.1074/jbc.M406929200. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Rogers SC, Kavdia M. Analysis of kinetics of dihydroethidium fluorescence with superoxide using xanthine oxidase and hypoxanthine assay. Ann. Biomed. Eng. 2012;41:327–337. doi: 10.1007/s10439-012-0653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


