Abstract
Neoseiulus cucumeris is a predatory mite used for biological control of arthropod pests. Mass-reared predators are fed with factitious prey mites such as Tyrophagus putrescentiae. Although some information on certain endosymbionts of N. cucumeris and T. putrescentiae exists, it is unclear whether both species share bacterial communities. The bacterial communities in populations of predator and prey mites, as well as the occurence of potential acaropathogenic bacteria were analyzed. The comparisons were based on the following groups: (i) N. cucumeris mass-production; (ii) N. cucumeris laboratory population with disease symptoms; (iii) T. putrescentiae pure populations and; (iv) T. putrescentiae from rearing units of N. cucumeris. Only 15% of OTUs were present in all samples from predatory and prey mite populations (core OTUs): the intracellular symbionts Wolbachia, Cardinium, plus other Blattabacterium-like, Solitalea-like, and Bartonella-like symbionts. Environmental bacteria were more abundant in predatory mites, while symbiotic bacteria prevailed in prey mites. Relative numbers of certain bacterial taxa were significantly different between the microbiota of prey mites reared with and without N. cucumeris. No significant differences were found in the bacterial communities of healthy N. cucumeris compared to N. cucumeris showing disease symptoms. We did not identify any confirmed acaropathogenic bacteria among microbiota.
Introduction
Phytoseiid mites (Acari: Phytoseiidae) are amongst the most important predators used in plant protection against arthropod pests such as spider mites, whiteflies and thrips1–3. Many species of commercially available phytoseiid mites are mass reared using astigmatid mites (Acari: Astigmata) as factitious prey4,5. Like many other arthropod species, predatory and prey mites are closely associated with symbiotic and pathogenic bacteria that may have variable yet critical impact on several fitness parameters of their arthropod hosts6,7. Diseases and/or reproductive disorders associated with endosymbiotic bacteria can have devastating effects on the mass-rearing of predatory mites8. Similarly, poor quality of prey mites due to infestation with pathogenic bacteria will compromise the production of predatory mites9,10. Until recently, studies on microbiota relied on the use of molecular markers targeting specific endosymbiont species. Recent advances in molecular biology and bioinformatics allow for the rapid screening of the whole microbiome and provide useful insights into the bacterial communities of predatory and prey mites. Given the prominent role of phytoseiid mites as biological control agents, screening the symbiotic and pathogenic bacterial community and establishing an association with phenotypic traits can potentially impact rearing protocols used by the biocontrol industry11. Moreover, the high densities of mites in mass rearing conditions and reports of horizontal bacterial transmission between trophic levels12 suggest these environments offer an ideal setting for the comparative study of the bacterial microbiota of predatory and prey mites.
Previous studies reported the presence of endosymbiotic and pathogenic bacteria in phytoseiid mites8,9. Metaseiulus occidentalis (Nesbitt) is probably the best studied species7. Several bacteria were detected including: the pathogenic Serratia marcescens; two Rickettsia-like bacteria; the gut symbionts Enterobacter and Bacteroidetes; and the endosymbionts Wolbachia and Cardinium7. Enigl and Schausberger13 screened several phytoseiid species for the presence of Wolbachia, Cardinium and Spiroplasma, they found Cardinium in Euseius finlandicus (Oudemans) and Spiroplasma in Neoseiulus californicus (McGregor). Similarly, Wolbachia and Cardinium showed different patterns of infection in the phytoseiid mite N. paspalivorus (De Leon) depending on geographic origin14. Interestingly, the observed postzygotic reproductive incompatibility among populations was associated with the presence of endosymbiotic bacteria. Gols et al.15 discovered the bacterium Acaricomes phytoseiuli in several commercial populations of the phytoseiid mite Phytoseiulus persimilis (Athias-Henriot). The infected predatory mites exhibited lower fecundity and longevity and reduced attraction to plant volatiles induced by spider mites. This was designated as non-responding syndrome and rendered infected P. persimilis populations unsuitable for effective control of spider mites.
Regarding prey astigmatid mites, sequencing of the 16S rRNA gene revealed that the bacterial communities in Acarus siro L., Lepidoglyphus destructor (Schrank) and Tyrophagus putrescentiae (Schrank) were formed by ingested bacteria. These included: Bacillus, Staphylococcus and Kocuria spp.; the gut bacteria Enterobacteriaceae and Bartonella-like bacteria; endosymbiotic bacteria such as Cardinium; and/or entomopathogenic bacteria Xenorhabdus and Photorhabdus16,17.
Neoseiulus cucumeris (Oudemans) is one of the most widely employed predators in augmentative biological control programs against thrips species such as Frankliniella occidentalis (Pergande) and Thrips tabaci Lindeman (Thysanoptera: Thripidae) mostly in protected crops18–20. For the mass-rearing of N. cucumeris, the acaricid mite T. putrescentiae is used as factitious prey21. While some information on certain endosymbionts of N. cucumeris and T. putrescentiae2,3 exists there is no information regarding the intrinsic microbiota of these species, or the effects of predatory-prey mite interactions on the ecology of these microbiota. Although mites are kept at high densities in mass rearing units it is still unclear whether both species share bacterial communities due to horizontal transfer via predation, contact or the feces6. The current study, to our knowledge, is the first direct comparison of bacterial microbiota from Phytoseiidae mites and their factitious prey mites.
Understanding the composition of the bacterial microbiota in both N. cucumeris and T. putrescentiae will allow for detecting pathogenic bacteria in the mass-rearing systems and perhaps offer opportunities for the manipulation of the bacterial community to improve predatory mite health. Both of the above mentioned scenarios can potentially have a substantial impact on biological control. In the present study we use Illumina sequencing of the 16S rRNA gene region and bioinformatics tools to (i) characterize the entire bacterial microbiota (ii) identify potentially pathogenic bacteria and iii) compare bacterial microbiota to examine the effects of predator-prey interactions and different rearing conditions on microbial ecology.
Results
Bacterial microbiota characterization
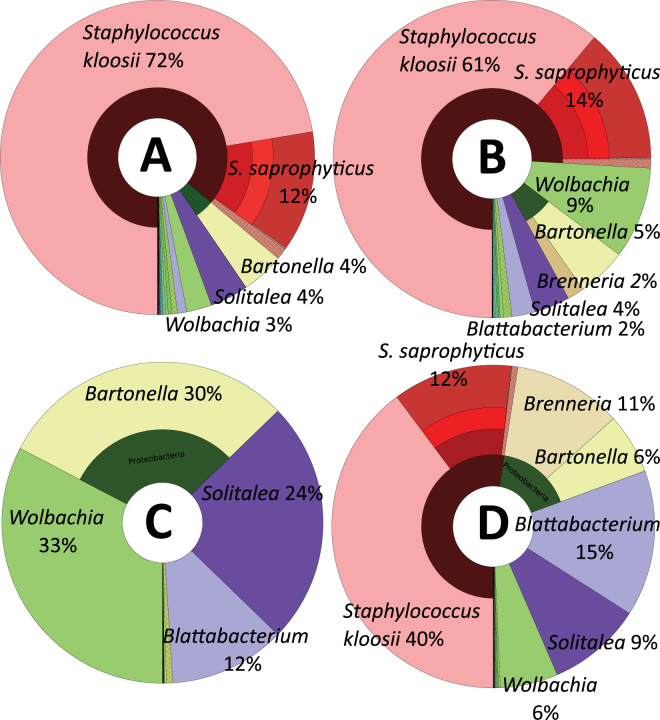
The 16S rRNA gene libraries included 818,413 sequences classified in 75 OTUs. The proportion of bacterial taxa in the groups of predatory and prey mites were visualized in Krona projection (Fig. 1). The minimal number of reads was 17,613 (Fig. 2A). No known acaropathogenic bacteria were identified in the prey or predatory mites either from the mass rearing or the laboratory population with disease symptoms (Table S1).
Figure 1.
The Krona projections of bacterial taxa found in the samples of predator (N. cucumeris) and prey (T. putrescentiae) mites. (A) Predatory mite (N. cucumeris) from mass-production population; (B) predatory mite from a laboratory population with disease symptoms; (C) the prey mite (T. putrescentiae) population from laboratory culture without the presence of predatory mite (Tyro pure), (D) prey mite from culture with the presence of predatory mite in mass rearing.
Figure 2.
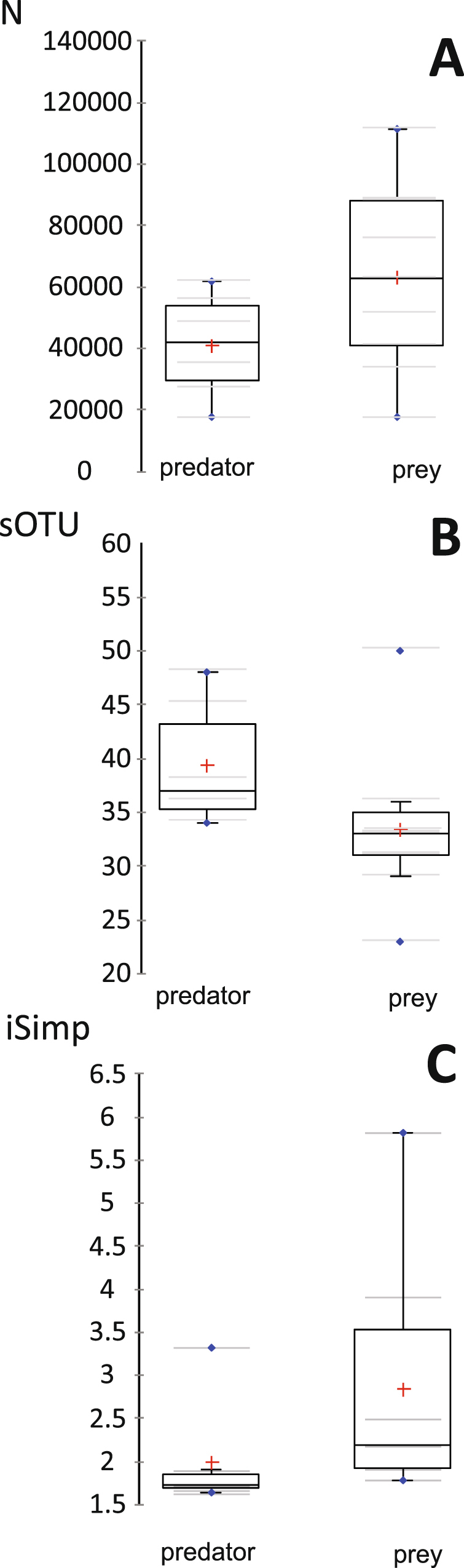
Comparisons of beta-diversity between bacterial microbiota from N. cucumeris compared to T. putrescentiae. (A) Number of sequences analyzed across samples; (B) Species Richness, number of species-level OTUs across samples; (C) Inverse Simpson Diversity Index comparing predatory vs. prey mites (see Table 1 for description of samples).
Bacterial microbiota comparisons: N. cucumeris (predator) vs. T. putrescentiae (factitious prey)
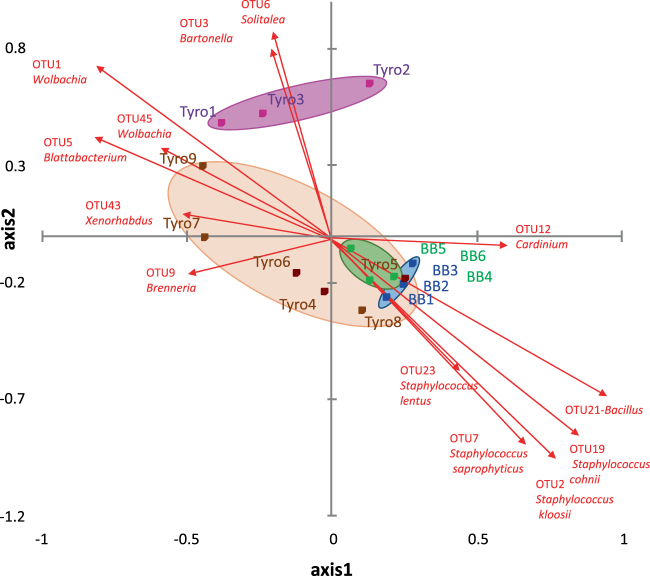
Total numbers of OTUs found in predatory mites compared to prey mites were marginally non-significant (Fig. 2B). The Inverse Simpson index was 1.5 times higher in prey compared to predatory mite samples (U(1,14) = 8, P = 0.029) (Fig. 2C). Bray-Curtis dissimilarity index showed higher dissimilarity in bacterial composition within prey mite samples than within the predatory mite samples. There were significant differences between the bacterial composition of prey mites compared to predator mites (i.e. factor 1, Table S2) (two-way PERMANOVA, F = 0.947, P = 0.011). There was also significant difference between the populations of mites (i.e. factor 2, Table S2) (two-way PERMANOVA, F = 0.66, P = 0.012), but the interaction between mite species and different population factors was not significant (F = −2.19, P = 0.999). When analyzed using Jaccard similarity matrix, the results were in agreement with previous analyses. Species of mite was a significant factor affecting the bacterial diversity of microbiota (F = 0.245, P = 0.045), as was the population that they originated from (F = 0.303, P < 0.001) but their interaction was not significant (F = −1.89, P = 0.972). Sample dissimilarity was visualized by PCoA; the first axis explained 56% and the second axis 15% of variability in the data set (Fig. 3). Microbiota of predator and prey mites formed two distinct clustered when represented by PCoA. Distances between OTUs along the first axis can be attributed to differences between predator and prey microbiota, with the exception of Tyro5 and Tyro8 (T. putrescentiae from N. cucumeris mass rearing units) where the bacterial composition was similar to that of the predatory mites. The following bacterial taxa were associated with prey mites: Solitalea-like (OTU6), Bartonella-like (OTU3), Wolbachia (OTU1 and 45), Blattabacterium-like (OTU5), Brenneria (OTU9) and Xenorhabdus (OTU43). The following taxa were associated with predatory mites: Cardinium (OTU12), Bacillus (OTU21) and Staphylococcus (OTUs2, 7, 19 and 23).
Figure 3.
Principal coordinate analyses (PCoA) of microbiota in the samples of predatory (N. cucumeris) and prey (T. putrescentiae) mites. The microbiome was analyzed using Bray-Curtis dissimilarity matrix; OTUs responsible for significant differences between microbiotas are represented by arrows (calculated via Pearson correlation coefficient). BB1-3 = N. cucumeris (laboratory); BB4-6 = N. cucumeris (mass-reared); Tyro1-3 = T. putrescentiae laboratory culture without the presence of predatory mite (pure) and Tyro4-9 = from N. cucumeris mass-rearing The samples are described in Table 1, the OTUs are identified in Table S1.
Venn diagrams show that 11 OTUs were shared by prey mites and 16 OTUs by predatory mites. Altogether 10 core OTUs (15% of the total OTUs) were present in all samples of both predatory and prey mites (Fig. 4). This group contained: endosymbiotic bacteria such as Wolbachia (OTU1), Cardinium (OTU12), Bartonella-like (OTU3), Blattabacterium-like (OTU5), Solitalea-like (OTU6) and environmental bacteria Brevibacterium (OTU16), Staphylococcus spp. (OTUs2, 7 and 9) and Bacillus cereus (OTU21).
Figure 4.
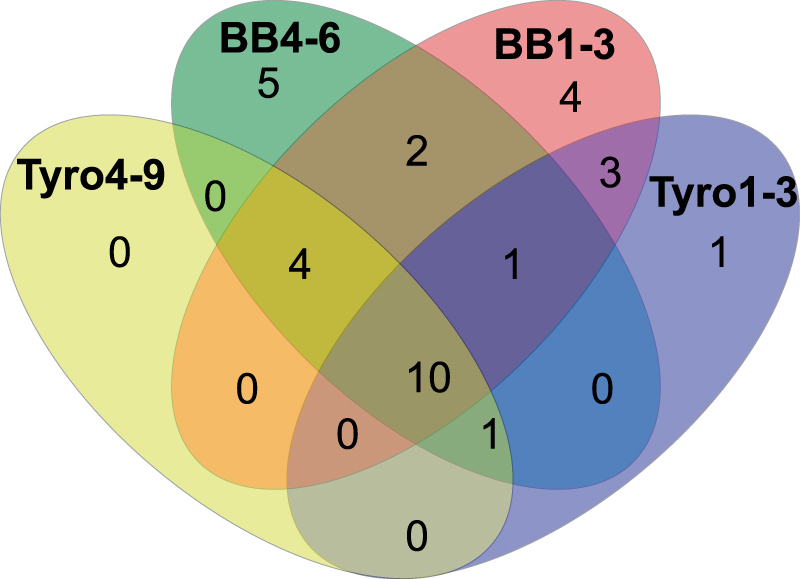
Qualitative comparison of shared and unique bacterial taxa belonging to predatory and prey mite microbiota by Venn diagram. The compared samples include predatory mites (Neoseiulus cucumeris) from the mass-production population (BB1-3) and the population with disease symptoms (BB4-6); the prey mites (Tyrophagus putrescentiae) from pure laboratory cultures without predators (Tyro1-3) and from the mass rearing production population with the presence of predators (Tyro4-9). The diagram was constructed from the core species per samples. The shared OTUs among all samples were the following taxa ordered by decreasing relative abundance: Staphylococcus kloosii (OTU2), Wolbachia (OTU1), Blattabacterium-like (OTU5), Bartonella-like (OTU3), Solitalea-like (OTU6), Staphylococcus saprophyticus (OTU7), Staphylococcus cohnii (OTU19), Bacillus cereus (OTU21), Cardinium (OTU12), Brevibacterium siliguriense (OTU16). Brenneria (OTU9), Staphylococcus lentus (OTU23), Kocuria koreensis (OTU25) and Xenorhabdus innexi (OTU43) were shared by predatory mites (BB1-3 and BB4-6) and prey mites from the mass rearing production population (Tyro4-9). Wolbachia (OTU45) was shared by predatory mites (BB1-3 and BB4-6) and prey mites from pure laboratory cultures without predators (Tyro1-3). Arthrobacter (OTU47) was shared by predatory mite population with disease symptoms (BB4-6) and both populations of the prey mites (Tyro1-3 and Tyro4-9).
From the 75 OTUs analyzed by METASTATS, 10 OTUs had higher relative abundance in predatory mites compared with prey mites (Table 1). OTUs associated with predatory mites were Brevibacterium (OTU18), Staphylococcus (OTU2 and OTU19), Bacillus (OTU21), Kocuria (OTU25 and OTU39), Stenotrophomonas (OTU52), Chryseobacterium (OTU84) and Pantoea (OTU83). Only 2 OTUs were most abundant in prey mites the symbiotic/parasitic bacteria Blattabacterium-like (OTU5) and Solitalea-like (OTU6) (Table 1). The remaining 63 OTUs were not influenced by the prey/predator mites (i.e. factor 1, Table S2).
Table 1.
The list of OTUs in the samples of predatory (Neoseiulus cucumeris) and prey (Tyrophagus putrescentiae) mites and the results of Random Forest and METASTATS analyses describing the means of relative abundance of OTUs (%) for the samples of predatory and prey mites.
| OTU97 | GenBank identification | Random forest | METASTATS | ||||
|---|---|---|---|---|---|---|---|
| aOTU | OTU ID | Taxon | predator/prey | populations | prey | predator | p-value |
| mean ± standard error | |||||||
| OTUs presented in all samples | |||||||
| 456731 | OTU2 | Staphylococcus kloosii (99) | 0.1 | 0.1 | 26.62 ± 10.04 | 66.75 ± 4.01 | 0.001 |
| 180511 | OTU1 | Wolbachia (97) | 0.02 | 14.74 ± 6.77 | 5.97 ± 2.47 | 0.267 | |
| 122903 | OTU5 | Blattabacterium-like | 0.1 | 13.66 ± 3.92 | 1.64 ± 0.48 | 0.003 | |
| 101995 | OTU3 | Bartonella queenslandensis (95) | 0.08 | 0.06 | 14.06 ± 5.90 | 4.72 ± 0.29 | 0.102 |
| 71643 | OTU6 | Solitalea-like | 0.17 | 0.03 | 14.47 ± 3.62 | 3.77 ± 0.59 | 0.004 |
| 64153 | OTU7 | Staphylococcus saprophyticus (99) | 0.13 | 0.13 | 7.97 ± 2.56 | 13.06 ± 1.80 | 0.091 |
| 5593 | OTU19 | Staphylococcus cohnii (99) | 0.16 | 0.01 | 0.32 ± 0.08 | 0.69 ± 0.06 | 0.000 |
| 2402 | OTU21 | Bacillus cereus (99) | 0.07 | 0.04 | 0.21 ± 0.04 | 0.37 ± 0.04 | 0.004 |
| 1730 | OTU12 | Cardinium | 0.06 | 0.06 | 0.12 ± 0.03 | 0.1 ± 0.03 | 0.142 |
| 1569 | OTU16 | Brevibacterium siliguriense (97) | 0.09 | 0.07 | 0.09 ± 0.04 | 0.34 ± 0.16 | 0.148 |
| OTUs presented in the samples of predators and prey from mass rearing | |||||||
| 40567 | OTU9 | Brenneria salicis (91) | 0.13 | 0.15 | 7.28 ± 3.74 | 0.87 ± 0.55 | 0.075 |
| 563 | OTU23 | Staphylococcus lentus (99) | 0.09 | 0.07 | 0.04 ± 0.01 | 0.10 ± 0.02 | 0.007 |
| 366 | OTU25 | Kocuria koreensis (96) | 0.32 | 0.14 | 0.01 ± 0.003 | 0.09 ± 0.04 | 0.015 |
| 350 | OTU43 | Xenorhabdus innexi (92) | 0.07 | 0.02 | 0.03 ± 0.01 | 0.02 ± 0.004 | 0.491 |
| OTUs presented in the samples of predators and prey from laboratory | |||||||
| 133 | OTU45 | Wolbachia (94) | 0.06 | 0.02 | 0.01 ± 0.002 | 0.02 ± 0.01 | 0.283 |
| OTUs presented in the samples of sick predator and both groups of prey | |||||||
| 78 | OTU47 | Arthrobacter russicus (91) | 0.06 | 0.01 | 0.01 ± 0.003 | 0.02 ± 0.01 | 0.492 |
| OTUs presented in the two groups of samples | |||||||
| 389 | OTU26 | Brevibacterium iodinum (98) | 0.06 | 0.02 | 0.03 ± 0.01 | 0.09 ± 0.04 | 0.188 |
| 241 | OTU39 | Kocuria koreensis (99) | 0.07 | 0.04 | 0.02 ± 0.01 | 0.06 ± 0.02 | 0.028 |
| 152 | OTU61 | Pseudomonas monteilii (99) | 0.13 | 0.04 | 0.01 ± 0.003 | 0.08 ± 0.05 | 0.091 |
| 142 | OTU35 | Moraxella osloensis (99) | 0.1 | 0.04 | 0.01 ± 0.003 | 0.05 ± 0.03 | 0.073 |
| 100 | OTU32 | Bartonella coopersplainsensis (95) | 0.07 | 0.02 | 0.01 ± 0.003 | 0.01 ± 0.001 | 0.943 |
| 66 | OTU38 | Bartonella rattaustraliani (98) | 0.07 | 0.03 | 0.004 ± 0.001 | 0.01 ± 0.002 | 0.751 |
| OTUs presented in one group of samples | |||||||
| 953 | OTU11 | Wolbachia (97) | 0.1 | 0.05 | 0.02 ± 0.01 | 0.17 ± 0.11 | 0.189 |
| 565 | OTU18 | Brevibacterium oceani (99) | 0.1 | 0.01 ± 0.01 | 0.16 ± 0.07 | 0.023 | |
| 317 | OTU22 | Corynebacterium variabile (97) | 0.08 | 0.1 | 0.03 ± 0.01 | 0.06 ± 0.03 | 0.271 |
| 166 | OTU31 | Propionibacterium acnes (99) | 0.08 | 0.02 | 0.03 ± 0.02 | 0.02 ± 0.01 | 0.675 |
| 105 | OTU55 | Acinetobacter lwoffii (97) | 0.16 | 0.001 ± 0.001 | 0.05 ± 0.03 | 0.100 | |
| 99 | OTU53 | Lactococcus lactis (99) | 0.08 | 0.01 | 0.01 ± 0.003 | 0.02 ± 0.02 | 0.718 |
| 83 | OTU48 | Leuconostoc gasicomitatum (99) | 0.07 | 0.03 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.779 |
| 81 | OTU52 | Stenotrophomonas maltophilia (99) | 0.07 | 0.01 ± 0.002 | 0.02 ± 0.01 | 0.027 | |
| 61 | OTU96 | Tsukamurella paurometabola (99) | 0.13 | 0.1 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.869 |
| 37 | OTU91 | Streptococcus thermophilus(99) | 0.11 | 0.08 | — | 0.02 ± 0.01 | 0.122 |
| 682 | OTU13 | Wolbachia (99) | 0.08 | 0.001 ± 0.001 | 0.14 ± 0.14 | 0.457 | |
| 149 | OTU50 | Pseudomonas poae (99) | 0.03 | 0.01 ± 0.01 | 0.003 ± 0.002 | 0.622 | |
| 128 | OTU51 | Alcaligenes faecalis (99) | 0.08 | 0.01 | 0.01 ± 0.01 | 0.02 ± 0.02 | 0.868 |
| 115 | OTU34 | Microbacterium indicum (97) | 0.08 | 0.02 | 0.03 ± 0.03 | 0.03 ± 0.02 | 0.980 |
| 92 | OTU60 | Wautersiella falsenii (97) | 0.11 | 0.004 ± 0.003 | 0.02 ± 0.02 | 0.237 | |
| 73 | OTU36 | Paracoccus chinensis (99) | 0.04 | — | 0.03 ± 0.03 | 0.436 | |
| 73 | OTU42 | Prevotella paludivivens (97) | 0.07 | 0.02 | 0.001 ± 0.001 | 0.03 ± 0.03 | 0.461 |
| 72 | OTU62 | Cloacibacterium rupense (96) | 0.04 | 0.01 | 0.01 ± 0.01 | — | 0.410 |
| 70 | OTU49 | Acinetobacter baumannii (99) | 0.17 | 0.04 | — | 0.04 ± 0.02 | 0.077 |
| 69 | OTU40 | Corynebacterium nuruki (99) | 0.07 | 0.02 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.656 |
| 67 | OTU70 | Pseudochrobactrum asaccharolyticum (99) | 0.13 | 0.01 | 0.01 ± 0.01 | 0.001 ± 0.001 | 0.737 |
| 61 | OTU87 | Sphingobacterium multivorum (99) | 0.13 | 0.04 | 0.01 ± 0.01 | 0.001 ± 0.001 | 0.637 |
| 58 | OTU69 | Corynebacterium vitaeruminis (99) | 0.11 | 0.02 | 0.01 ± 0.01 | — | 0.242 |
| 57 | OTU63 | Acinetobacter johnsonii (99) | 0.13 | 0.05 | — | 0.03 ± 0.02 | 0.256 |
| 57 | OTU94 | Staphylococcus aureus (99) | 0.13 | 0.07 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.711 |
| 50 | OTU57 | Paenochrobactrum glaciei (99) | 0.09 | 0.01 | 0.01 ± 0.01 | — | 0.129 |
| 47 | OTU77 | Ralstonia insidiosa (99) | 0.11 | 0.06 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.883 |
| 45 | OTU71 | Leclercia adecarboxylata (99) | 0.1 | 0.02 | 0.001 ± 0.001 | 0.03 | 0.295 |
| 33 | OTU103 | Escherichia coli (99) | 0.08 | 0.01 | 0.001 ± 0.001 | 0.001 ± 0.001 | 1.000 |
| 32 | OTU56 | Lactococcus chungangensis (97) | 0.08 | — | 0.01 ± 0.01 | 0.229 | |
| 31 | OTU79 | Delftia tsuruhatensis (99) | 0.13 | 0.05 | 0.001 ± 0.001 | 0.01 ± 0.01 | 0.323 |
| 30 | OTU82 | Acinetobacter radioresistens (99) | 0.1 | 0.02 | 0.001 ± 0.001 | 0.01 ± 0.01 | 0.370 |
| 27 | OTU95 | Leucobacter denitrificans (97) | 0.13 | 0.05 | 0.01 ± 0.01 | 0.001 ± 0.001 | 0.269 |
| 25 | OTU54 | Pseudomonas caeni (99) | 0.07 | 0.01 | 0.003 ± 0.003 | 0.01 ± 0.01 | 0.562 |
| 25 | OTU64 | Brevundimonas bullata (99) | 0.12 | 0.03 | 0.003 ± 0.003 | 0.01 ± 0.005 | 0.591 |
| 24 | OTU101 | Brevibacterium siliguriense (96) | 0.14 | — | 0.01 ± 0.01 | 0.084 | |
| 22 | OTU68 | Chryseobacterium bernardetii (94) | 0.04 | 0.01 | 0.001 ± 0.001 | — | 0.520 |
| 22 | OTU74 | Sphingobacterium faecium (99) | 0.06 | 0.02 | — | 0.01 ± 0.01 | 0.386 |
| 20 | OTU102 | Alcanivorax dieselolei (94) | 0.01 | 0.003 ± 0.003 | 0.001 ± 0.001 | 0.412 | |
| 18 | OTU104 | Corynebacterium singulare (99) | 0.04 | — | 0.01 ± 0.01 | 0.102 | |
| 18 | OTU72 | Finegoldia magna (99) | 0.09 | 0.05 | — | 0.01 ± 0.01 | 0.211 |
| 14 | OTU88 | Rickettsia bellii (99) | 0.08 | 0.001 ± 0.001 | — | 0.155 | |
| 13 | OTU75 | Acidovorax radicis (99) | 0.05 | 0.03 | — | 0.01 ± 0.01 | 0.115 |
| 13 | OTU92 | Paenibacillus hordei (97) | 0.04 | 0.01 ± 0.01 | — | 0.410 | |
| 12 | OTU84 | Stenotrophomonas rhizophila (99) | 0.1 | 0.03 | — | 0.01 ± 0.01 | 0.292 |
| 12 | OTU89 | Buchnera aphidicola (97) | 0.07 | 0.04 | 0.001 ± 0.001 | 0.01 ± 0.01 | 0.180 |
| 10 | OTU80 | Paracoccus marinus (99) | 0.09 | 0.05 | — | 0.001 ± 0.001 | 0.160 |
| 10 | OTU83 | Pantoea calida (94) | 0.07 | 0.04 | 0.001 ± 0.001 | 0.005 ± 0.005 | 0.041 |
| 10 | OTU93 | Afipia birgiae (99) | 0.07 | 0.07 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.395 |
| 9 | OTU81 | Chryseobacterium balustinum (97) | 0.06 | 0.02 | — | 0.001 ± 0.001 | 0.026 |
| 7 | OTU86 | Lactobacillus paracollinoides (99) | 0.11 | 0.05 | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.309 |
| 6 | OTU97 | Anaerococcus senegalensis (99) | 0.04 | 0.01 | — | 0.001 ± 0.001 | 0.064 |
| 4 | OTU65 | Phyllobacterium myrsinacearum (99) | 0.02 | 0.01 | 0.001 ± 0.001 | 0.001 ± 0.001 | 1.000 |
Supplementary Table S1 provides the extensive identification of OTUs. The random forest analyze was calculated for predatory/prey and populations as the factors (see Table S2), separately. The forest error rates were 0.33 and 0.53 respectively, aOTU - total number of sequnces in dataset, P-values < 0.05 are indicated by bold.
Differences in bacterial microbiota within populations of predatory and prey mites
There were no significant differences in the bacterial composition between lab-reared (showing disease symptoms) or mass-reared populations of N. cucumeris (one way PERMANOVA, F = 2.312, P = 0.198). This was supported by their proximity on the axes of the principle coordinate analysis (PCoA) (Fig. 3). The random forest algorithms (forest error rate = 0.66) indicated the following OTUs as the most important for differences: Stenotrophomonas rhizophila (OTU84) Lactococcus chungangensis (OTU56), Lactococcus lactis (OTU53), Leclercia adecarboxylata (OTU71). These were OTUs present only in healthy predator populations. The Venn diagram showed that 8 OTUs were found only in the populations of mites with disease symptoms: Wolbachia (OTU11), Corynebacterium variabile (OTU22), Brevibacterium oceani (OTU11), Stenotrophomonas maltophilia (OTU52) (Fig. 4, Table 1).
Significant differences were detected between the microbiota composition of T. putrescentiae populations with and without the presence of the predator (one way PERMANOVA; F = 5.337, P = 0.024) according to Bray-Curtis distance matrix. The random forest algorithm (forest error rate = 0.33) revealed that the following OTUs were important for differences and based on METASTATS were abundant in the microbiome of T. putrescentiae from N. cucumeris rearing units compared to T. putrescentiae pure colonies.: Staphylococcus saprophyticus (OTU7), Brenneria (OTU9), Bartonella (OTU3), Kocuria koreensis (OTU25 and OTU39) (Table 1). The Venn diagram (Fig. 4) indicated only 2 OTUs to be unique for T. putrescentiae from mass-rearing system (Leuconostoc gasicomitatum OTU48) and without predator (Lactococcus lactis OTU53). The bacterium similar to Staphylococcus kloosii (OTU2) made up a large percentage of the microbiota from both N. cucumeris populations and the T. putrescentiae population from the mass-rearing where N. cucumeris was present.
Discussion
Bacterial ecology of N. cucumeris and its prey mite T. putrescentiae
To our knowledge this is the first study comparing the entire bacterial microbiome of predatory phytoseiid mites and prey astigmatid mites used together under mass rearing conditions. We found that the predatory mite N. cucumeris and the prey mite T. putrescentiae shared 15% of core bacterial taxa. These taxa differ in relative abundance between predatory and prey mites, and among populations. Similarly, previous studies comparing the bacterial microbiota of M. occidentalis and its spider mite prey Tetranychus urticae (Koch) (Acari: Tetranychidae), provided evidence for a shared microbiota between the predator and the prey22. The dissimilarity in the bacterial community between lab reared and mass-reared populations of T. putrescentiae was higher in prey mites than in predatory mites. The prey mites showed higher diversity in their overall microbiota than predatory mites. The differences in diversity of microbiota among saprophagous compared to predatory groups are known in insects23 and a similar situation is expected in these mites.
The OTU similar to S. kloosii accounted for a large proportion of the total bacterial sequences isolated from both N. cucumeris (72% mass-reared, 61% lab-reared) and T. putrescentiae populations (40%) the latter from the mass rearing where the predator was present. In the T. putrescentiae pure population, where no N. cucumeris was present, this bacterium was still present but yet not so prevalent. A similar pattern was seen for other Staphylococcus OTUs found in this study. This suggests that bacteria might be transmitted from N. cucumeris to its factitious prey, possibly via direct contact or through the feces. Alternatively, the rearing environment of T. putrescentiae is not as optimal for Staphylococcus growth as the habitat in the presence of the predator. Our suggestion is that the predatory mites can alter the rearing environment in such a way that promotes the proliferation of certain bacterial taxa. Taking into account the pattern observed here as well as the previously demonstrated effects of diet and environment on Astigmata mites microbiota17,24,25, it is possible that the microbiota of T. putrescentiae is more influenced by diet and environment than by the microbiota of N. cucumeris.
The bacterial community of T. putrescentiae exhibited higher diversity than that of the predatory mites. Acquisition of bacteria through the prey mites’ diet might explain these results. It is important to highlight that the analyses of differences in the bacterial composition (PCoA) and population level analyses showed that the differences between predatory and prey mites were lower than among the two prey mite populations examined. This is supported by differences between predator and prey in relative numbers of Blattabacterium-like and Solitalea-like symbiotic bacteria, with a higher relative abundance in prey mites. This suggests that the microbiota of different T. putrescentiae populations are more variable than the microbiota of different N. cucumeris populations.
While feeding, it is suggested that T. putrescentiae ingests debris of plant cells, fungi or yeasts which are already colonized by environmental bacteria26–28. The environmental bacteria enter the gut with the food and are passed through the gut or the bacteria adhere to the mite integument in the manner that fungal spores do29. Both modes of transmission resulted in the presence of environmental bacteria in the microbiota of astigmatids17,25. In the present study the mites were surface sterilized with sterilized phosphate saline buffer and tween 20, thus the identified bacteria species should be regarded as colonizing the mites internally. However, we cannot explicitly exclude either the possibility of some levels of surface contamination or random ingestion of environmental bacteria.
Based on previous studies with stored product mites we can identify the bacterial taxa which could be considered as environmental17. For example, Brevibacterium, Staphylococcus, Kocuria and Stenotrophomonas are present in mite laboratory habitats e.g. rearing diets and the feces of mites. However, some bacterial taxa isolated in this study (Table S1) are known to belong to either skin or gut communities in other animals. For example, the genus Corynebacterium includes species that are widely found in the microbiota of vertebrates30,31, Anaerococcus and Finegoldia spp. are commonly found among microbial skin communities in humans and can occasionally become pathogenic32. Bacterial taxa characteristic of mammal gut communities were also found in this study, particularly bacteria of the genera Escherichia, Lactococcus, Leuconostoc, and Propionibacterium. The latter genus contains species often found to have beneficial probiotic and nutritional effects within the gut microbiota of many vertebrate hosts such as humans and ruminants33. It is possible that Propionibacterium has a similar nutritional effect on its mite host, i.e prey mites. Conversely, some Propionibacterium species belong to skin microbiota and may cause disease in some cases34.
Endosymbionts in predatory mite and prey populations
The observed microbiota of the predatory mite N. cucumeris and the prey mite T. putrescentiae consisted of intracellular symbionts as well as gut/environmental bacteria as it is reported for other species of predatory or saprophagous mites35,36. The presence of core bacterial taxa in prey and predatory mites poses the question whether these bacteria are autochthonous or allochthonous, i.e. horizontally transmitted from predator to prey. For example, the intracellular symbiont Wolbachia has been shown to be transmitted across trophic levels. Aedes aegypti (Diptera: Culicidae) larvae infected with the wMelPop strain did not transfer Wolbachia to predators, including copepods (Crustacea: Copepoda) and mosquito species37. In contrast, T. putrescentiae feeding on Wolbachia infested corpses of Drosophila melanogaster Meigen (Diptera: Drosophilidae) resulted in establishment of Wolbachia population in both mites and Drosophila and suggested horizontal transfer38. Generally, the presence of prey DNA is common in predators. For example, aphid nuclear and mitochondrial DNA had a detectability period longer than 23 hours in the harlequin ladybird Harmonia axyridis39. Likewise, the bacteria of prey mites and their DNA can be ingested by the predator mites and may be detected in the predator’s digestive tract13,40,41. Therefore, the sole detection of bacterial DNA in predators does not necessarily mean that these bacteria form part of the predator’s microbiota, e.g. Wolbachia in Phytoseiulus persimilis42 and M. occidentalis40 occurs due to infected prey. Presence of bacteria in the predator’s eggs (and concomitant vertical transfer) would unambiguously confirm the bacterial presence in the microbiota13,25,41. Further studies screening the presence of symbiotic bacteria in the eggs and other life stages are needed in order to solve the question of autochthonous or allochthonous origin of symbiotic or parasitic bacteria in phytoseiid mites.
The co-occurrence of intracellular symbiotic bacteria such as Cardinium and Wolbachia in predatory and prey mites is quite common6,14,22,43,44. Double infections within the same individuals were common in tetranychid mites45. It is likely that similar occurrences are found in N. cucumeris and T. putrescentiae. It is interesting to mention the presence of the symbiotic Blattabacterium-like bacteria found in all populations of mites examined in this study. Blattabacterium-like symbionts were recently identified in some T. putrescentiae populations25. Bayesian analyses of the 16S rRNA gene sequences showed that Blattabacterium-like symbionts clustered as a monophyletic lineage. This cluster is outside Blattabacterium46, Cand. Brownia rhizoecola47, Cand. Uzinora diaspidicola48 and Cand. Sulcia muelleri49. Blattabacterium species are obligate endosymbionts found in all cockroaches. Genome sequencing of these bacteria showed that they have a nutritional role in their host including vitamin synthesis and nitrogen recycling50,51. Given their similarity to Blattabacterium, the bacteria found in N. cucumeris and T. putrescentiae populations in this study may fulfil a similar role in their host mites. These Blattabacterium-like bacteria may be a unique mite specific lineage of Flavobacterium and their presence warrants further study to elicit their role in mites. Given that they were more prevalent in T. putrescentiae populations and have been previously isolated from this mite25, it is likely that this bacterium is found in predatory mites due to ingestion during feeding rather than an obligate interaction.
Solitalea-like bacterium was found in the reproductive tract and parenchymal tissues of A. siro L. and in five populations of T. putrescentiae24,25. The clones formed a new distinct cluster separate from Solitalea and other genera of the Sphingobacteriaceae family. Based on their localization in the gut, fat body, reproductive tissues and eggs of A. siro a symbiotic mode of action has been suggested24. Bartonella-like bacteria were previously identified in Dermatophagoides spp., A. siro and T. putrescentiae16,17,52. However, Bartonella-like bacteria were never amplified from eggs, eliminating the possibility of a vertically transmitted symbiont25. Herein, Bartonella-like bacteria were found in both predatory and prey mites, showing similar proportions to the predator–prey system of Cheyletus eruditus (Schrank) (Acarina: Cheyletidae) – A. siro53. Finally, Spiroplasma a commonly known bacteria from some Phytoseiidae13 was not isolated from either mite species in this study. The original screening never isolated Spiroplasma13 from N. cucumeris.
In this study we found significant differences in themicrobiome of T. putrescentiae from mass rearing units (with predator) and laboratory culture (without predator). The OTUs predominantly responsible for these differences were similar to S. saprophyticus, Brenneria and K. koreensis. These taxa had higher relative abundance in the bacterial microbiota of T. putrescentiae from mass rearing units (predator present) compared to laboratory cultures with no predators present. These taxa are suggested as environmental17 and we can only speculate that the mass rearing conditions with the presence of predators are more favorable for their development, possibly due to the buildup of feces or husks of predated prey mites. Surprisingly we found Bartonella-like bacteria of higher relative abundance in prey mites from the mass rearing units compared to the laboratory cultures. The higher proportion of Bartonella-like bacteria in mass rearing culture with the presence of predators may indicate better conditions for T. putrescentiae.
Potential acaropathogens in mite populations
Not known acaropathogenic bacteria were detected in the natural microbiota of the mites examined in this study. Of special interest is the occurrence of B. cereus in all samples of predatory and prey mites. Some Bacillus taxa such as B. spahericus and B. thuringiensis (which are not distinguishable by 16S rRNA) are known to be acaropathogenic54,55. While B. cereus is suggested to be opportunistic on insects, B. sphaericus, B. papillae and B. thuringiensis are known pathogens56. B. cereus was previously isolated from the feces of a laboratory population of T. putrescentiae reared on dog kernels27; the identification was confirmed by cloning and sequencing of the motB gene57. The addition of B. cereus to this mite diet led to a substantial reduction of population growth. Exo-enzymes of this bacterium have been proposed as aiding mite digestion27 altogether with inhibition of the population growth of mites suggesting a sort of opportunistic mode of pathogenesis56. The presence of B. cereus in all mite samples, coupled with the fact that no differences were detected in the microbiota between healthy and sick populations of predatory mites, suggests that symptoms of illness of the predatory mites in the laboratory colony were not caused by bacteria or that the method we used could not detect the acaropathogenic bacteria. Often, some pathogens make up part of the natural microflora of its host and opportunistically cause disease due to subtle changes in environmental factors, host immune system and even microbiota58. Presence of disease symptoms in the lab-reared N. cucumeris could not be explained by the presence of harmful bacteria. Instead, perhaps it was the result of the lack of beneficial protective microbes as seen in other animals58. Alternatively, the disease symptoms in the laboratory population of the predatory mites might have been caused by viruses, fungi or protozoan pathogens8 or accumulation of toxic metabolites such as guanine10.
Conclusion
The microbiota of the prey mite T. putrescentiae and the predatory mite N. cucumeris consisted of core bacterial taxa present in all prey and predatory mite populations.This core microbiota comprised Wolbachia, Cardinium, Bartonella-like, Blattabacterium-like, Solitalea-like, Brevibacterium, Staphylococcus spp. and B. cereus. Among them Brevibacterium, Staphylococcus and Bacillus were the most abundant in predatory mites, while Blattabacterium-like and Solitalea-like bacteria were the most abundant in prey mites. Significant differences were detected between the bacterial communities of prey mites without predators and prey mites reared with N. cucumeris. S. saprophyticus, Brenneria and K. koreensis were more abundant in the presence of predators. The occurrence of no acaropathogenic bacteria was examined in predatory mite populations with and without disease symptoms. Interestingly, bacterial microbiome of healthy and with disease symptoms predatory mites showed similar diversity, without significant differences between the two groups. This suggests disease symptoms in this case are caused by non-bacterial entities such as other types of microbes, unsuitable environmental conditions or genetic factors. Further study is required to confirm the cause of disease. Ultimately, characterization of predatory mite and prey microbiota may help inform mass rearing practices, but this understanding also required knowledge on the effects of certain microbial taxa on the health of both species of mites. This study can serve to influence the subsequent study of mechanistic studies of the effects of some bacterial taxa on mite hosts of economic importance.
Materials and Methods
Origin of mites
The predatory N. cucumeris and prey T. putrescentiae mites originated from the Biobest rearing facilities in Belgium (Westerlo). T. putrescentiae was reared on a mix of yeast flakes, wheat germ and dried yeasts at a ratio 10:10:1 (w/w)59 in rearing units of 0.2 L kept at 27 °C and 85% relative humidity (r.h.). N. cucumeris was reared on the aforementioned mix plus T. putrescentiae (treatment 4 see below) at a ratio of approximately (1:15; N. cucumeris: T. putrescentiae) in rearing buckets of 5L (mass rearing) or 0.2 L (laboratory population) kept at 25 °C and 70% r.h. The trial included the following treatments: (i) N. cucumeris from a mass-production population (ii) N. cucumeris from a laboratory population with disease symptoms (slower movement and lighter coloration of the mites); (iii) pure T. putrescentiae (with no predators); (iv) T. putrescentiae from rearing units with predatory mites. Three samples were examined for treatments i–iii (see below for sample definition), and six samples for treatment iv (Table S2). Predatory and prey mites from treatment 1 were sampled one week after the addition of the prey mites.
DNA extraction from mites
One sample consisted of about 50 (predator or prey) mites collected in ethanol (90%). Ethanol was removed and samples were washed 3 times in Phosphate Buffered Saline and Tween-20 (PBST). The mites were homogenized in 500 μL of PBST using a Radnoti tissue grinder (Cat No. 440613, Monrovia, CA, USA). Total DNA was extracted using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. The extracted DNA was stored at −20 °C until further analyses.
Amplification, cloning and sequencing
The quality and presence of bacterial DNA in every sample was tested by PCR amplification using eubacterial primers and routinely using protocols60. When amplicons were not obtained, the samples were replaced by new samples positive for amplicons. The DNA samples were sent to MR DNA (http://mrzaqqqsadnalab.com, Shallowater, TX, USA). The sequencing of V1-V3 part of 16S rRNA gene was based by the universal primers 27Fmod and 519Rmod in the Illumina MiSeq platform and the bTEFAP® process61. The length of the read was 300 bp, the reads were forward and reverse. The sequencing reads were preprocessed in MR DNA to contingencies. The sequences in bioproject PRJNA321085 were deposited in GenBank SRP074673, barcodes and biosample codes are given in Table S2.
Data analyses and sequence processing
The contingencies were demultiplexed using MR DNA binning software (http://mrdnalab.com), sequences were renamed and trimmed in MOTHUR v. 1.36.1 software62 according to the MiSeq standard operation procedure MiSeq SOP63. The actual commands used in MOTHUR are available at http://www.mothur.org/wiki/MiSeq_SOP (accession date 2/19/2016). The demultiplexed, renamed and trimmed sequences were processed in UPARSE and USEARCH64, singletons were removed and the sequences were classified using a naive Bayesian classifier with a training set (version 15) made available through the Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu)65. Operational taxonomic units (OTUs) were defined by the clustering of sequences at ≥97% identity. The representative sequences obtained from UPARSE were processed in MOTHUR again, aligned against SILVA reference database66. The aligned sequences were screened for chimeras with UCHIME67. Sequences from chloroplasts, mitochondria, Archaea and Eukaryotes were removed. The representative sequences were analyzed via BLASTin GenBank68,69. Solitalea-like, Blattabacterium-like and Cardinium bacteria were identified by aligning OTUs to previously identified almost full length 16S rRNA sequences of these taxa17,24,25 in Codone Code Aligner (CodonCode Corporation, Centerville, MA, USA). Taxonomic diversity and relative proportions of bacterial taxa were visualized using Krona tools70.
Statistical analyses
The shared file was generated from UPARSE data and was processed in MOTHUR, and PAST 3.0671. The results were visualized by XLSTAT (http://www.xlstat.com/en/, Addinsoft, New York, NY, USA). The subsample data set was constructed in MOTHUR on 17,613 sequences. Alpha-diversity, (OTUs richness) in predatory or prey mites was assessed using the Inverse-Simpson index, the number of OTUs was calculated in MOTHUR from a subsample data set. The Inverse-Simpson index and species level OTUs (sOTU) of the predator and prey samples were compared using a nonparametric Mann-Whitney test. Beta-diversity, (similarity of samples) was assessed using the Bray-Curtis and Jacquard indices and visualized by principal coordinate analyses (PCoA) of the subsample data set. The contribution of OTUs was calculated using Pearson correlation coefficient. We analyzed the effects of two factors on bacterial diversity: (i) mite species (i.e. prey and predator) and (ii) origin of mite population (i.e predator with and without sick symptoms and prey from mass rearing facility and without any predator) (Table S2). The effects of both factors were evaluated with two-way PERMANOVA72 with 100,000 permutations. Calculations were based on Bray-Curtis and Jaccard matrices of the subsample data. The different populations of predatory mite (sick and without disease symptoms) and T. putrescentiae (from mass production and without predatory mites) were also compared separately. Venn diagrams were used to highlight shared bacterial taxa in mites from different treatments, to the analyses we included only those OTUs, which were presented in all three replicates per treatment. The population-level analysis was calculated using METASTATS73 based on 100,000 permutations to compare the effects predator/prey to distribution of bacterial taxa in the subsample data set. The random forest analysis in MOTHUR was applied to compare the differences in bacterial microbiome of sick and healthy predator or T. putrescentiae from mass rearing and laboratory culture.
Electronic supplementary material
Acknowledgements
The authors are obligated to Dr. Jan Kopecky for his kind help and advice in bioinformatics and Nancy Lenaerts for collecting the mite samples and Martin Markovic for technical help. J.H. and M.N. were supported by the projects of the Ministry of Education, Youth and Sports of the Czech Republic No. LD13052 of COST FA1105 and of the Ministry of Agriculture of the Czech Republic No. RO0416.
Author Contributions
J.H. and A.P. wrote the main manuscript and contributed equally. A.P., J.H., and M.N. designed the experiments, M.N. performed the experiments, J.H. performed the data analyses, M.A.P., J.S., A.P. and E.P. contributed writing. J.H. provided grants.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-19514-8.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-00046-6
Change History
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Helle, W. & Sabelis, M. W. Spider mites: their biology, natural enemies and control. World crop pests, vol. 1B. (Elsevier, 1986).
- 2.Nomikou M, Janssen A, Schraag R, Sabelis MW. Phytoseiid predators as potential biological control agents for Bemisia tabaci. Exp. Appl. Acarol. 2001;25:271–291. doi: 10.1023/A:1017976725685. [DOI] [PubMed] [Google Scholar]
- 3.Put K, Bollens T, Wackers F, Pekas A. Non-target effects of commonly used plant protection products in roses on the predatory mite Euseius gallicus Kreiter & Tixier (Acari: Phytoseidae) Pest Manag. Sci. 2016;72:1373–1380. doi: 10.1002/ps.4162. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa MFC, de Moraes GJ. Evaluation of astigmatid mites as factitious food for rearing four predaceous phytoseiid mites (Acari: Astigmatina; Phytoseiidae) Biol. Control. 2015;91:22–26. doi: 10.1016/j.biocontrol.2015.06.010. [DOI] [Google Scholar]
- 5.Gerson, U., Smiley, R. L. & Ochoa, R. Mites (acari) for pest control. 2nd edition. (Blackwell Science, 2003).
- 6.Schutte C, Gols R, Kleespies RG, Poitevin O, Dicke M. Novel bacterial pathogen Acaricomes phytoseiuli causes severe disease symptoms and histopathological changes in the predatory mite Phytoseiulus persimilis (Acari, Phytoseiidae) J. Invertebr. Pathol. 2008;98:127–135. doi: 10.1016/j.jip.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Hoy MA, Jeyaprakash A. Symbionts, including pathogens, of the predatory mite Metaseiulus occidentalis: current and future analysis methods. Exp. Appl. Acarol. 2008;46:329–347. doi: 10.1007/s10493-008-9185-3. [DOI] [PubMed] [Google Scholar]
- 8.Bjornson S. Natural enemies of mass-reared predatory mites (family Phytoseiidae) used for biological pest control. Exp. Appl. Acarol. 2008;46:299–306. doi: 10.1007/s10493-008-9187-1. [DOI] [PubMed] [Google Scholar]
- 9.van der Geest LPS, Elliot SL, Breeuwer JAJ, Beerling EAM. Diseases of mites. Exp. Appl. Acarol. 2000;24:497–560. doi: 10.1023/A:1026518418163. [DOI] [PubMed] [Google Scholar]
- 10.Hubert J, Nesvorna M, Palevsky E, Smrz J. Diseases of prey mites used for mass rearing predatory mites. Acta Hortic. 2014;1041:177–185. doi: 10.17660/ActaHortic.2014.1041.20. [DOI] [Google Scholar]
- 11.Zindel R, Gottlieb Y, Aebi A. Arthropod symbioses: a neglected parameter in pest- and disease-control programmes. J. Appl. Ecol. 2011;48:864–872. doi: 10.1111/j.1365-2664.2011.01984.x. [DOI] [Google Scholar]
- 12.Chiel E, et al. Almost there: transmission routes of bacterial symbionts between trophic levels. PLoS One. 2009;4:e4767. doi: 10.1371/journal.pone.0004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enigl M, Schausberger P. Incidence of the endosymbionts Wolbachia, Cardinium and Spiroplasma in phytoseiid mites and associated prey. Exp. Appl. Acarol. 2007;42:75–85. doi: 10.1007/s10493-007-9080-3. [DOI] [PubMed] [Google Scholar]
- 14.Famah Sourassou N, et al. The endosymbionts Wolbachia and Cardinium and their effects in three populations of the predatory mite Neoseiulus paspalivorus. Exp. Appl. Acarol. 2014;64:207–221. doi: 10.1007/s10493-014-9820-0. [DOI] [PubMed] [Google Scholar]
- 15.Gols R, Schutte C, Stouthamer R, Dicke M. PCR-based identification of the pathogenic bacterium, Acaricomes phytoseiuli, in the biological control agent Phytoseiulus persimilis (Acari: Phytoseiidae) Biol. Control. 2007;42:316–325. doi: 10.1016/j.biocontrol.2007.06.001. [DOI] [Google Scholar]
- 16.Kopecky J, Nesvorna M, Hubert J. Bartonella-like bacteria carried by domestic mite species. Exp. Appl. Acarol. 2014;64:21–32. doi: 10.1007/s10493-014-9811-1. [DOI] [PubMed] [Google Scholar]
- 17.Hubert J, et al. Detection and identification of species-specific bacteria associated with synanthropic mites. Microb. Ecol. 2012;63:919–928. doi: 10.1007/s00248-011-9969-6. [DOI] [PubMed] [Google Scholar]
- 18.Ferragut Perez, F., Perez Moreno, I., Iraola Calvo, V. M. & Escudero Colomar, L. A. Acaros depredadores de la familia Phytoseiidae en las plantas cultivadas. (Ediciones Agrotecnicas, 2010) (in Spanish).
- 19.McMurtry JA, Croft BA. Life-styles of phytoseiid mites and their roles in biological control. Annu. Rev. Entomol. 1997;42:291–321. doi: 10.1146/annurev.ento.42.1.291. [DOI] [PubMed] [Google Scholar]
- 20.van Lenteren JC. The state of commercial augmentative biological control: plenty of natural enemies, but a frustrating lack of uptake. BioControl. 2012;57:1–20. doi: 10.1007/s10526-011-9395-1. [DOI] [Google Scholar]
- 21.Ramakers PMJ. Mass prodution and introduction of Amblyseius mckenziei and A. cucumeris. Bull. SROP. 1983;6:203–206. [Google Scholar]
- 22.Hoy MA, Jeyaprakash A. Microbial diversity in the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae) and its prey, Tetranychus urticae (Acari: Tetranychidae) Biol. Control. 2005;32:427–441. doi: 10.1016/j.biocontrol.2004.12.012. [DOI] [Google Scholar]
- 23.Yun J-H, et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 2014;80:5254–5264. doi: 10.1128/AEM.01226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubert J, Kopecky J, Nesvorna M, Perotti MA, Erban T. Detection and localization of Solitalea-like and Cardinium bacteria in three Acarus siro populations (Astigmata: Acaridae) Exp. Appl. Acarol. 2016;70:309–327. doi: 10.1007/s10493-016-0080-z. [DOI] [PubMed] [Google Scholar]
- 25.Erban T, et al. Populations of stored product mite Tyrophagus putrescentiae differ in their bacterial communities. Front. Microbiol. 2016;7:1046. doi: 10.3389/fmicb.2016.01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erban T, Hubert J. Digestive function of lysozyme in synanthropic acaridid mites enables utilization of bacteria as a food source. Exp. Appl. Acarol. 2008;44:199–212. doi: 10.1007/s10493-008-9138-x. [DOI] [PubMed] [Google Scholar]
- 27.Erban T, Rybanska D, Harant K, Hortova B, Hubert J. Feces derived allergens of Tyrophagus putrescentiae reared on dried dog food and evidence of the strong nutritional interaction between the mite and Bacillus cereus producing protease bacillolysins and exo-chitinases. Front. Physiol. 2016;7:53. doi: 10.3389/fphys.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybanska D, Hubert J, Markovic M, Erban T. Dry dog food integrity and mite strain influence the density-dependent growth of the stored-product mite Tyrophagus putrescentiae (Acari: Acaridida) J. Econ. Entomol. 2016;109:454–460. doi: 10.1093/jee/tov298. [DOI] [PubMed] [Google Scholar]
- 29.Duek L, Kaufman G, Palevsky E, Berdicevsky I. Mites in fungal cultures. Mycoses. 2001;44:390–394. doi: 10.1046/j.1439-0507.2001.00684.x. [DOI] [PubMed] [Google Scholar]
- 30.Collins MD, Hoyles L, Foster G, Falsen E. Corynebacterium caspium sp. nov., from a Caspian seal (Phoca caspica) Int. J. Syst. Evol. Microbiol. 2004;54:925–928. doi: 10.1099/ijs.0.02950-0. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Honda H, Katsumata R. Production of antibacterial compounds analogous to chloramphenicol by a n-paraffin-grown bacterium. Agric. Biol. Chem. 1972;36:2223–2228. doi: 10.1080/00021369.1972.10860545. [DOI] [Google Scholar]
- 32.Murphy EC, Frick I-M. Gram-positive anaerobic cocci – commensals and opportunistic pathogens. FEMS Microbiol. Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 33.Jan G, Leverrier P, Proudy I, Roland N. Survival and beneficial effects of propionibacteria in the human gut: in vivo and in vitro investigations. Lait. 2002;82:131–144. doi: 10.1051/lait:2001012. [DOI] [Google Scholar]
- 34.Foligne B, Breton J, Mater D, Jan G. Tracking the microbiome functionality: focus on Propionibacterium species. Gut. 2013;62:1227–1228. doi: 10.1136/gutjnl-2012-304393. [DOI] [PubMed] [Google Scholar]
- 35.Zindel R, et al. The role of the bacterial community in the nutritional ecology of the bulb mite Rhizoglyphus robini (Acari: Astigmata: Acaridae) FASEB J. 2013;27:1488–1497. doi: 10.1096/fj.12-216242. [DOI] [PubMed] [Google Scholar]
- 36.Hubert J, et al. Assessment of bacterial communities in thirteen species of laboratory-cultured domestic mites (Acari: Acaridida) J. Econ. Entomol. 2016;109:1887–1896. doi: 10.1093/jee/tow089. [DOI] [PubMed] [Google Scholar]
- 37.Hurst TP, et al. Impacts of Wolbachia infection on predator prey relationships: evaluating survival and horizontal transfer between wMelPop infected Aedes aegypti and its predators. J. Med. Entomol. 2012;49:624–630. doi: 10.1603/ME11277. [DOI] [PubMed] [Google Scholar]
- 38.Brown AN, Lloyd VK. Evidence for horizontal transfer of Wolbachia by a Drosophila mite. Exp. Appl. Acarol. 2015;66:301–311. doi: 10.1007/s10493-015-9918-z. [DOI] [PubMed] [Google Scholar]
- 39.Paula DP, et al. Detection and decay rates of prey and prey symbionts in the gut of a predator through metagenomics. Mol. Ecol. Resour. 2015;15:880–892. doi: 10.1111/1755-0998.12364. [DOI] [PubMed] [Google Scholar]
- 40.Wu K, Hoy MA. Cardinium is associated with reproductive incompatibility in the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae) J. Invertebr. Pathol. 2012;110:359–365. doi: 10.1016/j.jip.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 41.Wu K, Hoy MA. Extended starvation reduced and eliminated Wolbachia, but not Cardinium, from Metaseiulus occidentalis females (Acari: Phytoseiidae): a need to reassess Wolbachia’s status in this predatory mite? J. Invertebr. Pathol. 2012;109:20–26. doi: 10.1016/j.jip.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Enigl M, Zchori-Fein E, Schausberger P. Negative evidence of Wolbachia in the predaceous mite Phytoseiulus persimilis. Exp. Appl. Acarol. 2005;36:249–262. doi: 10.1007/s10493-005-6075-9. [DOI] [PubMed] [Google Scholar]
- 43.Zchori-Fein E, Perlman SJ. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 2004;13:2009–2016. doi: 10.1111/j.1365-294X.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y-K, Chen Y-T, Yang K, Hong X-Y. A review of prevalence and phylogeny of the bacterial symbiont Cardinium in mites (subclass: Acari) Syst. Appl. Acarol. 2016;21:978–990. doi: 10.11158/saa.21.7.11. [DOI] [Google Scholar]
- 45.Zhang Y-K, Chen Y-T, Yang K, Qiao G-X, Hong X-Y. Screening of spider mites (Acari: Tetranychidae) for reproductive endosymbionts reveals links between co-infection and evolutionary history. Sci. Rep. 2016;6:27900. doi: 10.1038/srep27900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark JW, Kambhampati S. Phylogenetic analysis of Blattabacterium, endosymbiotic bacteria from the wood roach, Cryptocercus (Blattodea: Cryptocercidae), including a description of three new species. Mol. Phylogenet. Evol. 2003;26:82–88. doi: 10.1016/S1055-7903(02)00330-5. [DOI] [PubMed] [Google Scholar]
- 47.Gruwell ME, Hardy NB, Gullan PJ, Dittmar K. Evolutionary relationships among primary endosymbionts of the mealybug subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae) Appl. Environ. Microbiol. 2010;76:7521–7525. doi: 10.1128/AEM.01354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruwell ME, Morse GE, Normark BB. Phylogenetic congruence of armored scale insects (Hemiptera: Diaspididae) and their primary endosymbionts from the phylum Bacteroidetes. Mol. Phylogenet. Evol. 2007;44:267–280. doi: 10.1016/j.ympev.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 49.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kambhampati S, Alleman A, Park Y. Complete genome sequence of the endosymbiont Blattabacterium from the cockroach Nauphoeta cinerea (Blattodea: Blaberidae) Genomics. 2013;102:479–483. doi: 10.1016/j.ygeno.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Sabree ZL, Kambhampati S, Moran NA. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. USA. 2009;106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valerio CR, Murray P, Arlian LG, Slater JE. Bacterial 16S ribosomal DNA in house dust mite cultures. J. Allergy Clin. Immunol. 2005;116:1296–1300. doi: 10.1016/j.jaci.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 53.Hubert J, et al. Differences in the bacterial community of laboratory and wild populations of the predatory mite Cheyletus eruditus (Acarina: Cheyletidae) and bacteria transmission from its prey Acarus siro (Acari: Acaridae) J. Econ. Entomol. 2016;109:1450–1457. doi: 10.1093/jee/tow032. [DOI] [PubMed] [Google Scholar]
- 54.Saleh SM, Kelada NL, Shaker N. Control of European house dust mite Dermatophagoides pteronyssinus (Trouessart) with Bacillus spp. Acarologia. 1991;32:257–260. [Google Scholar]
- 55.Erban T, Nesvorna M, Erbanova M, Hubert J. Bacillus thuringiensis var. tenebrionis control of synanthropic mites (Acari: Acaridida) under laboratory conditions. Exp. Appl. Acarol. 2009;49:339–346. doi: 10.1007/s10493-009-9265-z. [DOI] [PubMed] [Google Scholar]
- 56.Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat. Rev. Microbiol. 2008;6:302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 57.Oliwa-Stasiak K, Molnar CI, Arshak K, Bartoszcze M, Adley CC. Development of a PCR assay for identification of the Bacillus cereus group species. J. Appl. Microbiol. 2010;108:266–273. doi: 10.1111/j.1365-2672.2009.04419.x. [DOI] [PubMed] [Google Scholar]
- 58.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hubert J, Pekar S, Aulicky R, Nesvorna M, Stejskal V. The effect of stored barley cultivars, temperature and humidity on population increase of Acarus siro, Lepidoglyphus destructor and Tyrophagus putrescentiae. Exp. Appl. Acarol. 2013;60:241–252. doi: 10.1007/s10493-012-9639-5. [DOI] [PubMed] [Google Scholar]
- 60.Lane, D. J. 16S/23S rRNA sequencing In Nucleic acid techniques in bacterial systematics (ed. Stackebrandt, E. & Goodfellow, M.) 115–175 (John Wiley and Sons, 1991).
- 61.Chiodini RJ, et al. Microbial population differentials between mucosal and submucosal intestinal tissues in advanced Crohn’s disease of the ileum. PLoS One. 2015;10:e0134382. doi: 10.1371/journal.pone.0134382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–968. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 65.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 69.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammer, O., Harper, D. A. T. & Ryan, P. D. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 4 (2001). http://palaeo-electronica.org/2001_1/past/issue1_01.htm. Accessed 23 June 2016.
- 72.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 2013;83:557–574. doi: 10.1890/12-2010.1. [DOI] [Google Scholar]
- 73.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.