Abstract
Bagging is an efficient method to improve fruit colour development. This work reported a transcriptome analysis using bagging-treated red Chinese sand pear peels. In total, 8,870 differentially expressed genes were further analysed by a weighted gene co-expression network analysis and early-, middle- and late light-responsive genes were identified. An annotation analysis revealed several pathways involved in the different responsive stages. The presence of LONG HYPOCOTLY 5, CRY-DASH and a CONSTANS-like transcription factors among the early light-responsive genes indicated the pivotal role of light, especially blue light, in the biological changes that occurred after bag removal. Other light-responsive transcription factors were also identified from the three light-responsive stages. In addition, the light-responsive pattern of anthocyanin biosynthetic genes differed among the biosynthetic steps. Although yeast-one hybrid assay showed that most of the structural genes were regulated by PpMYB10, their different temporal expressive pattern suggested that besides PpMYB10, other light-responsive transcriptional factors were also involved in the regulation of anthocyanin biosynthesis. In summary, our transcriptome analysis provides knowledge of the transcriptional regulatory network operating during light responses, which results in anthocyanin accumulation and other significant physiological changes in red Chinese sand pear peels after bag removal.
Introduction
Pear (Pyrus) is an important fruit in China. Traditional pears are usually green or yellow but in recent years, red pears have gained consumer preference because of their attractive appearance and nutritional value. To date, red pear resources (or sports) have been identified in all four cultivation groups in China. Because the colour developmental pattern differs among these groups1, they are good research materials for studying the regulatory mechanism of fruit colouration. The red colouration of red Chinese sand pears (RCSPs, Pyrus pyrifolia) mainly depends on the concentration and composition of anthocyanins in the fruit peel1–3. Anthocyanin is synthesised in the cytosol and transported to the vacuole by a glutathione S-transferase (GST)4. The biosynthesis of anthocyanin involves several well-characterised enzymes, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS) and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT). The spatial and temporal expressions of the genes coding these enzymes are regulated at the transcriptional level by various transcription factors (TFs), especially the well-studied MBW complex, which is composed by MYB, basic helix-loop-helix (bHLH) and WD405, 6.
In many species, R2R3 MYB TFs were characterized as important TFs for the activation of anthocyanin biosynthesis genes. In apple, MdMYB1/A/10 act as positive factors of anthocyanin synthesis in skin and flesh when co-expressed with MdbHLH3 or MdbHLH337–9. In grapes, several R2R3 MYBs were isolated and implicated in anthocyanin biosynthesis10. In strawberry, two R2R3 MYBs are involved in anthocyanin biosynthesis. FaMYB10 acts as an activator, whereas FaMYB1 acts as a repressor. Interestingly, both the activator and the repressor showed high expression levels at the fruit ripening stage, suggesting that they may fine tune gene expression to balance the accumulation of anthocyanin11–13. Pear PpMYB10 (misassigned as PyMYB10 previously) contributes to the red pigmentation of RCSPs14.
In fruit crop cultivation, poor colouration has long been a problem owing to unfavourable environmental conditions and the varietal differences in adaptability. To improve fruit colouration, various management practices, such as leaf removal, fruit bagging, reflective films, evaporative cooling and postharvest artificial irradiation, have been developed15–19. In China, paper bagging during fruit development is used to enhance the red pigmentation of RCSP fruit skins1, 2, as well as to protect fruits from birds and insects, and alleviating fruit russeting. A commercial paper bag is usually composed of two light-blocking layers, while some have an additional translucent inner layer (usually red). The fruits are covered with these bags 1 month after full blossom and the bags are removed 10–15 days before the expected harvest date. This is referred to as the ‘bagging treatment’ in the current report.
The bagging treatment is basically a light re-exposure process, although the air composition, temperature and humidity around the fruit may also change, resulting in various physiological changes in the fruit. Fruit bagging efficiently improves the coloration of RCSPs. Fruit bagging works in the following two ways to improve fruit colouration: (1) The optimum light condition can be controlled by the timing of the bag removal, thereby achieving the desired fruit colour, which often occurs at maturity. (2) Bagging can increase the light sensitivity of the fruit, enhancing the development of an intense and uniform red colour20.
The mechanism of light-induced anthocyanin biosynthesis has been well characterized in model plant Arabidopsis as a part of photomorphogenesis. It requires the light-responsive elements including phytochromes and their downstream factors, such as CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), LONG HYPOCOTYL 5 (HY5) and etc 21. Based on these reports, several attempts have been carried out to explain the biological process of fruit coloration using high-throughput approaches22–24. The involvement of light-responsive genes in fruit coloration was confirmed but the detailed regulatory pathway may be different from that in model plants. For instance, Arabidopsis HY5 directly binds to the promoter of AtMYB75 to activate its expression but there are no evidence showing the activating effect of apple HY5 on the expression of MdMYB1 (unpublished data) although HY5 can bind to its upstream region22. Therefore, the mechanism for the light-induced anthocyanin biosynthesis in fruit peel needs further analysis.
In this study, we analysed the transcriptomes of RCSP fruit peel samples harvested at the initial (0 h)-bag removal stage, along with early (6 h), middle (24 h) and late (144 h) stages after bag removal. Bagged fruit sampled at the same times served as controls. By constructing co-expression networks, we identified the genes involved in early, middle and late light-responsive reactions and further analysed the differential regulation of the structural genes involved in anthocyanin biosynthesis. Our work identified sequentially expressed light-responsive genes, which will help us to further clarify the mechanism of bagging-treatment-induced colouration and enrich our knowledge regarding the regulation of anthocyanin biosynthesis. The data obtained from RNA sequencing (RNA-Seq) will also provide researchers with useful basic information for data mining and will facilitate further experiments on light-induced fruit colouration.
Results
Changes in fruit pigmentation patterns in the peel of RCSP ‘Meirensu’ fruit
Paper bags were removed ten days before predicted harvest day. By visual inspection, the fruits removed the bags were light yellow for 24 h and gradually turned red after 72 h, while the control samples remained light yellow (Fig. 1a and data not shown). The red colouration was accompanied by an elevation in the anthocyanin content. In addition, the carotenoid and chlorophyll contents in the bagged fruit were very low and slightly increased immediately after bag removal, prior to the anthocyanin accumulation (Fig. 1b).
Figure 1.
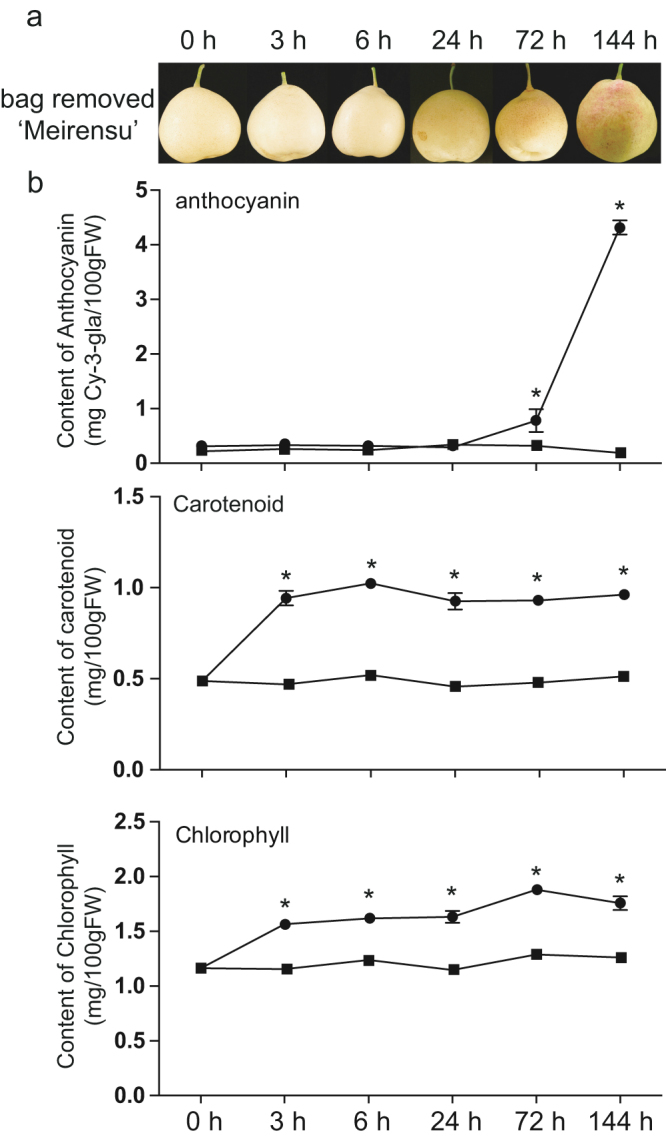
Changes in pigmentation of fruit peel of red Chinese sand pear in bagged and bag-removed samples. (a) Photos for bag-removed pear ‘Meirensu’. (b) Content changes of anthocyanin (up), carotenoid (middle) and chlorophyll (down) in fruit peel after bag removal. The labels of the samples were displayed as “hours after bag-removal” (h). The graphs showed the average values from three independently sampled fruits as biological replicates. Error bars are the standard deviation. Asterisk indicates p < 0.05.
Library construction and sequencing
The fruit peel sampled at 0 h, 6 h, 24 h and 144 h after bag-removal, and their corresponding bagged fruit peel (0H, 6H, 24H, 144H, 6HC, 24HC and 144HC) were subjected to total RNA extraction and an RNA-Seq analysis. High-throughput sequencing generated 20.37–24.11 million (M) 100-bp paired-end reads from each library (Table 1). After a stringent quality filtering process, 94.20 Gb of clean data (96.40% of the raw data) were obtained, with a Q30 percentage ≥80%. The counts of clean reads per library ranged from 19.71 to 23.86 M (Table 1). Reads were mapped to the reference genome sequence of Chinese white pear ‘Dangshansuli’ (Pyrus pyrifolia White pear group, misassigned to Pyrus bretschneideri Rehd.; should be an ecotype of P. pyrifolia)25, 26. The percentages of mapped reads were similar among the 21 libraries (72.75–77.32%), and 35.04–50.29% of the reads were perfectly mapped (Table 1 and Table S1).
Table 1.
Statistics on the quality and output of the RNA-Seq libraries.
| Classification | Maximum | Minimum | Average |
|---|---|---|---|
| No.raw reads | 24,116,813 | 20,373,330 | 22,348,931 |
| No.clean reads | 23,864,720 | 19,711,240 | 21,544,238 |
| No.mapped reads | 18,227,873 | 14,339,927 | 16,141,316 |
| % of mapped reads | 77.32% | 72.75% | 74.87% |
| No.perfect mapped reads | 12,231,683 | 8,349,296 | 9,746,449 |
| % of perfect mapped reads | 50.29% | 35.04% | 37.40% |
| Q30% | 81.86% | 80.06% | 80.68% |
| Total base pair | 91,377,116,282 | ||
| Coverage of the pear transcriptome | 793× | ||
The total clean reads were assembled into transcripts and compared with the reference gene model (including 42,812 genes). In total, 31,581 known genes (73.57% of the total model genes) and 4,616 new transcripts were identified. After filtering out new transcripts shorter than 150 bp and those with only one exon, 1,198 transcripts were predicted as new genes. To obtain comprehensive information on the detected transcripts, both known and newly predicted genes were annotated based on five public protein databases. Differentially expressed genes (DEGs) were identified in each pair of samples using the criteria listed in the Material and Methods section. The genes with reads per kilobase of transcript per million mapped reads (RPKM) value in more than 12 samples lower than 0.3 were filtered out. Finally, 8,870 DEGs remained for further analysis (Fig. 2, Table S2).
Figure 2.
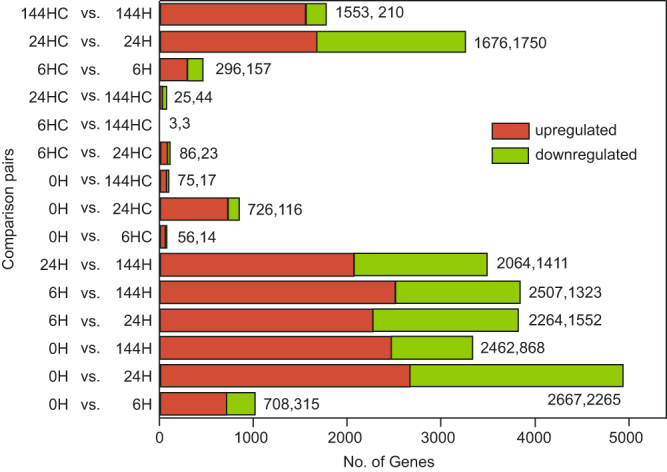
Comparison of the differential expressed genes of each pairs. The labels of the samples are displayed as “hours after bag-removal” (H) or “hours without bag-removal” (HC) Note that bag removal significantly changes the gene expression pattern in the fruit peel, while genes of the control had an expression pattern similar to the 0 h sample.
Validation of DEGs identified using RNA-Seq
Eight DEGs were randomly selected to have their transcript levels measured by qPCR. Although the exact fold change of the DEGs at several data points varied between RNA-Seq and qPCR, the differential expression trends detected by the two approaches were largely consistent (Fig. S1), indicating the reliability of the RNA-Seq results.
Co-expression network analysis identified anthocyanin-related DEGs
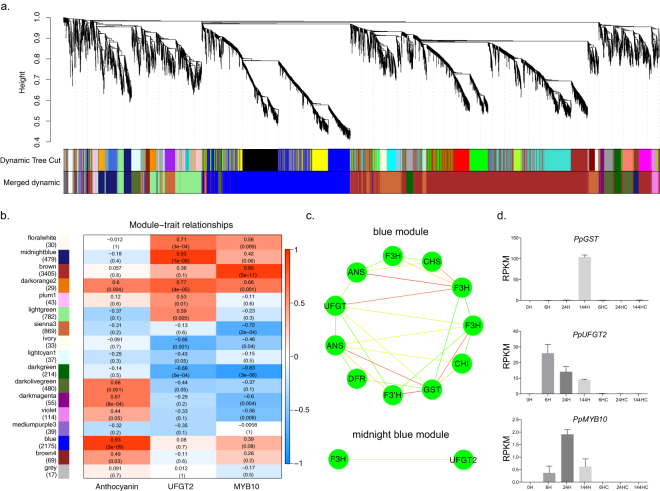
Anthocyanin started to accumulate 72 h after bag removal (Fig. 1). To identify genes related to anthocyanin biosynthesis, we performed a weighted gene co-expression network analysis (WGCNA) using non-redundant DEGs. The analysis identified 13 WGCNA modules (Fig. 3a,b). An analysis of the module-trait relationships revealed that the “blue” module was highly correlated with the total anthocyanin content (r = 0.93, p = 2 × 10−9) in the 21 samples. In this module, we found 11 structural genes involved in anthocyanin biosynthesis and transportation. The expression patterns of these genes were highly connected even when we used a relatively stringent edge weight cut-off of 0.4 (Fig. 3c). Specifically, the Glutathione S-transferase(GST) gene had the greatest number of edges connecting to other genes, followed by a UFGT and an F3H. In addition, two structural genes, an F3H and an UFGT2, fell into the module of “midnight blue”. Interestingly, in our dataset, the main regulatory TF PpMYB10 did not belong to these modules but to the “brown” module. A further module-trait relationship analysis, using the RPKMs of Pp UFGT2 and Pp MYB10 as the trait data, revealed that their expression patterns were indeed highly related to the modules “midnight blue” and “brown”, respectively. We further revealed the expression patterns represented in these three modules (Fig. 3d). Genes belonging to module “blue” had an expression peak 144 h after bag removal, similar to the anthocyanin accumulation pattern, while genes in module “midnight blue” were highly expressed from 6 h after bag removal and then gradually declined. The expression pattern of the “brown” module, containing PpMYB10, had an expression peak at 24 h after bag removal (Fig. 4). We focused on these three modules in our further analysis.
Figure 3.
Weighted gene co-expression network analysis (WGCNA) of DEGs identified from ‘Meirensu’ pear peel after bag removal. (a) Hierarchical cluster tree showing 17 modules of co-expressed genes. Each of the 8,870 DEGs is represented by a tree leaf and each of the modules by a major tree branch. The lower panel shows modules in designated colours. (b) Module–trait correlations and corresponding p-values (in parentheses). The left panel shows the 17 modules and the number of member genes. The colour scale on the right shows module–trait correlations from −1 (blue) to 1 (red). The left panel “Anthocyanin” represents anthocyanin biosynthesis as a trait. The middle panel “UFGT2” represents the expression changes of PpUFGT2, which encodes the enzyme that catalyses the last step in anthocyanin biosynthesis, as a trait. The right panel “MYB10” represents the expression changes of PpMYB10, which is the key transcriptional factor activating anthocyanin biosynthesis, as a trait. (c) Cytoscape representation of co-expressed genes with edge weights ≥0.40 in module ‘blue’ and ‘midnight blue’. (d) RNA-Seq expression patterns of PpGST, Pp UFGT2 and PpMYB10.
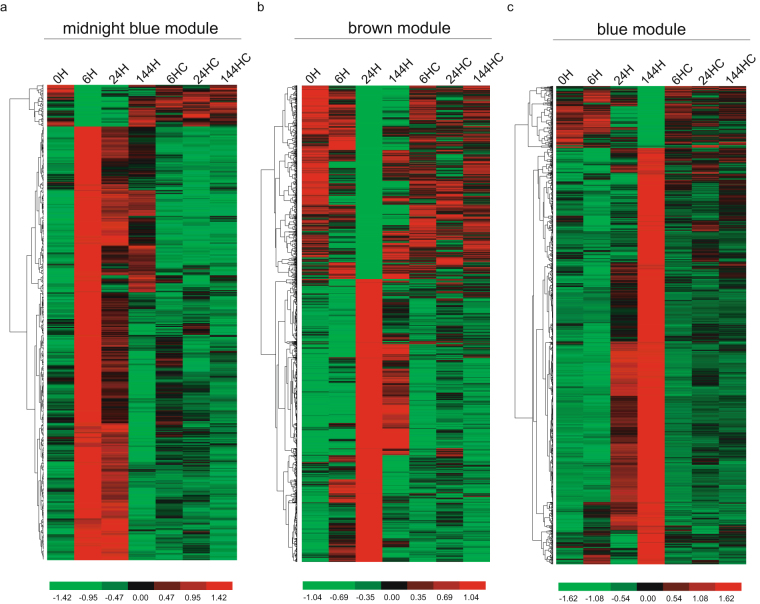
Figure 4.
Heat maps showing the expression patterns of modules from the light-responsive genes.
Compared with the control samples, including the 0 h sample, the expression levels of the genes belonging to the “midnight blue” module significantly changed by 6 h after bag removal, and were considered early light-responsive genes. The genes in the “brown” module were considered middle light-responsive genes, while the genes in the “blue” module were late light-responsive genes.
Functional annotations of light-responsive genes
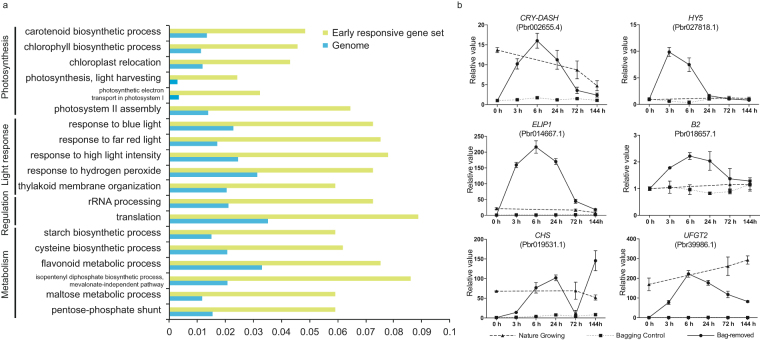
A Gene Ontology (GO) enrichment analysis of the early light-responsive genes identified 19 significantly enriched GO terms. Most of them were related to photosynthesis and light response, which are the typical light-induced pathways (Fig. 5, Table S3). In addition, some metabolism terms were also enriched (Fig. 5, Table S3). To obtain more detailed information, a pathway analysis was carried out using MapMan.
Figure 5.
Analysis of the early-responsive genes. (a) Gene ontology enrichment analysis of early-responsive genes. (b) qPCR confirmation of the expression patterns of selected genes.
Based on the MapMan analysis, some primary metabolism-related pathway genes, including those involved in photosynthesis, light reaction, lipid biosynthesis and cell wall degradation, were in this gene set. However, genes belonging to carotenoid, flavonoid and lignin biosynthesis pathways were also observed in this gene set.
Because light was the most important environmental change after bag removal, a gene annotated as CRY-DASH and seven light-responsible genes, three EARLY LIGHT-INDUCIBLE PROTEIN genes, three SUPPRESSOR PHYA-105 genes, and one light-inducible phototropism gene, were identified in the fruit’s early light-responsive gene set. However, other light photoreceptors were not found. In addition, some TFs that are involved in light signal transduction, including two HY5 homologs and one CONSTANS-like (COL) B-box gene, were identified in this gene set (Fig. 5b).
The GO enrichment analyses of the middle and late light-responsive genes revealed distinct enrichment patterns, although both groups contained over 2,000 genes. In the middle light-responsive gene set, most of the enriched terms belonged to “cellular biosynthetic process”, including “pyrimidine ribonucleotide biosynthetic process” and “RNA methylation”. The enrichment of terms related to photosynthesis and light response pathways was not found at this stage (Table S3).
The functional annotation of the late light-responsive genes revealed that the most highly enriched GO terms were “defense response”, “response to chitin” and “immune response”. Most of these terms were involved in the responsive pathways. In the “metabolic process” category, “flavonoid biosynthesis”, “ethylene biosynthesis process”, “jasmonic acid metabolic process” and “cGMP biosynthetic process” were overrepresented. In addition, in the “biological regulation” category, “negative regulation of programmed cell death”, “regulation of hydrogen peroxide metabolic process” and “negative regulation of defense response” were highly enriched (Table S3).
Genes involved in the biosynthesis and signal transduction of plant hormones
To determine the functions of plant hormones in the bagging-induced colouration, the expression patterns of genes involved in their biosynthesis and signal transduction were analysed. Significant expression changes were mostly observed in the middle- or late-light responsive stages, but a few genes, such as a 1-amino-cyclopropane-1-carboxylate synthase (PpACS), responded to the light condition change at the early stage. In addition, several genes related to the signal transduction of phytohormones were identified, and these were involved in auxin, ethylene, jasmonic acid (JA), abscisic acid (ABA), gibberellic acid (GA), brassinosteroid (BR) and cytokinin signalling pathways (Table 2).
Table 2.
Selected differentially expressed genes related to the phytohormone biosynthesis and signalling pathways during light-responsive reactions.
| Gene ID | Pathway | Module | Peak orientation | Annotation |
|---|---|---|---|---|
| Pbr041497.1 | ABA | midnight blue | Up | protein phosphatase 2C 77-like isoformX1 |
| Pbr026127.1 | ABA | midnight blue | Up | magnesium-chelatase subunit ChlH, chloroplastic-like |
| Pbr019415.1 | ABA | brown | Up | abscisic acid receptor PYL4-like |
| Pbr010794.1 | ABA | brown | Up | abscisic acid receptor PYL8-like |
| Pbr003860.1 | ABA | brown | Up | abscisic acid 8′-hydroxylase 2 |
| Pbr007589.1 | ABA | brown | Down | protein ABSCISICACID-INSENSITIVE 5-like |
| Pbr006776.1 | ABA | blue | Up | abscisic acid 8′-hydroxylase1-like |
| Pbr030688.1 | Auxin | midnight blue | Up | probable indole-3-acetic acid-amido synthetase GH3.5 |
| Pbr008163.2 | Auxin | brown | Down | auxin transporter-like protein 3 |
| Pbr013531.1 | Auxin | brown | Up | auxin-induced protein 15A-like |
| Pbr004491.1 | Auxin | brown | Up | indole-3-acetic acid-induced protein ARG7-like |
| Pbr022122.1 | Auxin | brown | Down | auxin-induced protein15A |
| Pbr021158.1 | Auxin | brown | Down | indole-3-acetic acid-amido synthetase GH3.6-like |
| Pbr000415.1 | Auxin | brown | Down | auxin response factor 3- like |
| Pbr021934.1 | BR | brown | Down | brassinosteroid LRR receptor kinase-like |
| Pbr021939.1 | BR | brown | Down | brassinosteroid LRR receptor kinase-like |
| Pbr001823.1 | BR | brown | Down | cytochrome P450 734A1 |
| Pbr004288.1 | BR | blue | Up | BRASSINOSTEROID INSENSITIVE1-associated receptor kinase 1-like |
| Pbr012231.1 | BR | blue | Up | delta(24)-sterol reductase |
| Pbr023118.1 | BR | blue | Up | receptor-like protein kinase BRI1-like3 |
| Pbr011993.1 | Cytokinin | brown | Down | two-component response regulator ARR1-like isoform X2 |
| Pbr005849.1 | Cytokinin | brown | Up | cytokinin riboside 5′-monophosphate phosphoribohydrolase LOG3-like |
| Pbr009698.1 | Cytokinin | brown | Up | cytokinin dehydrogenase 6-like |
| Pbr015617.1 | Cytokinin | brown | Up | cytokinin dehydrogenase 3-like |
| Pbr015575.1 | Ethylene | midnight blue | Up | 1-aminocyclopropane-1-carboxylate synthase 7 |
| Pbr023044.1 | Ethylene | brown | Up | ethylene-responsive transcription factor 1B-like |
| Pbr008360.1 | Ethylene | brown | Down | ethylene-insensitive protein 2 |
| Pbr005179.1 | Ethylene | brown | Up | 1-aminocyclopropane-1-carboxylate oxidase |
| Pbr011802.1 | Ethylene | brown | Down | 5′-3′ exoribonuclease 3-like isoform X1 |
| Pbr004323.1 | Ethylene | blue | Down | protein EIN4-like |
| Pbr004403.1 | Ethylene | blue | Up | 5′-3′ exoribonuclease 4 isoform X1 |
| Pbr015589.1 | Ethylene | blue | Up | 1-aminocyclopropane-1-carboxylate oxidase 1 |
| Pbr015895.1 | Ethylene | blue | Down | probable amino transferase ACS10 |
| Pbr036605.1 | GA | brown | Down | transcription factor TGA2-like |
| Pbr017104.1 | GA | brown | Down | gibberellin receptor GID1C-like |
| Pbr013018.1 | GA | brown | Down | gibberellin 2-beta-dioxygenase 8-like |
| Pbr014064.1 | GA | brown | Down | DELLA protein GAI-like |
| Pbr039229.1 | JA | brown | Up | protein TIFY10B-like |
| Pbr003675.1 | JA | blue | Up | protein TIFY9-like |
| Pbr027730.1 | JA | blue | Up | protein TIFY10A |
| Pbr011711.1 | JA | blue | Up | protein TIFY3B-like |
| Pbr037418.1 | JA | blue | Up | protein TIFY10A-like |
Transcription factors of light-responsive genes
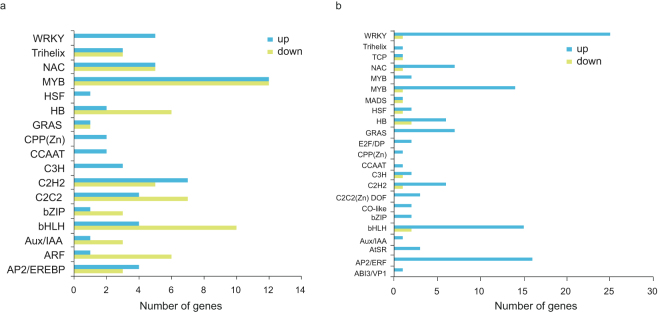
In the early light-responsive gene set, only 10 TFs were identified. In addition to the two HY5s and one COL mentioned above, one bHLH, two C2H2, two CCAAT box-binding factors, one MYB gene and one WRKY gene were identified. Interestingly, unlike other TFs that were upregulated at 6 h, the expression levels of MYB and WRKY were down-regulated, suggesting that they had distinct regulatory functions during the light-response process.
In the middle stage, 134 genes were assigned to the MapMan “transcription factors” bin and more than half were down-regulated. Members of the WRKY, CCAAT, C3H, HSF and CPP families were upregulated, while other TF families were regulated in both directions. Of these TFs, MYB was the largest family with 24 members, including PpMYB10, which is the key regulator of anthocyanin biosynthesis (Fig. 6a).
Figure 6.
Transcription factor analysis of middle- (a) and late- (b) responsive genes. The numbers of up-regulated genes and down-regulated genes comparing to 0 h were showed with blue (up) and green (down), respectively.
Although there were similar numbers of TFs in the late stage as in the middle stage, most of them were upregulated. In this gene set, WRKY was the largest family with 26 members, followed by the bHLH, AP2/ERF and MYB families, with 17, 16 and 15 members, respectively (Fig. 6b).
Various expression patterns of anthocyanin biosynthesis structural genes
Because the biosynthesis of anthocyanin is catalysed by multiple enzymes that are encoded by structural genes in the phenylpropanoid pathway (Fig. 7), we mapped the light-responsive genes to this pathway to identify the expression patterns of the anthocyanin biosynthesis-related genes. The genes in the early biosynthesis stage, such as PpCHSs and PpCHIs, were upregulated relatively early, while other genes, such as PpF3Hs, PpF3′Hs, PpDFRs and PpANSs, were upregulated later. Interestingly, PpUFGT2, the last enzyme in anthocyanin biosynthesis, was upregulated 6 h after bag removal. The key regulatory gene PpMYB10, together with some PpCHSs, PpF3Hs and PpF3′Hs, peaked at 24 h after bag-removal, which was prior to the anthocyanin accumulation. In addition, the members of the bHLH family had different expression patterns, as shown in Fig. 6.
Figure 7.
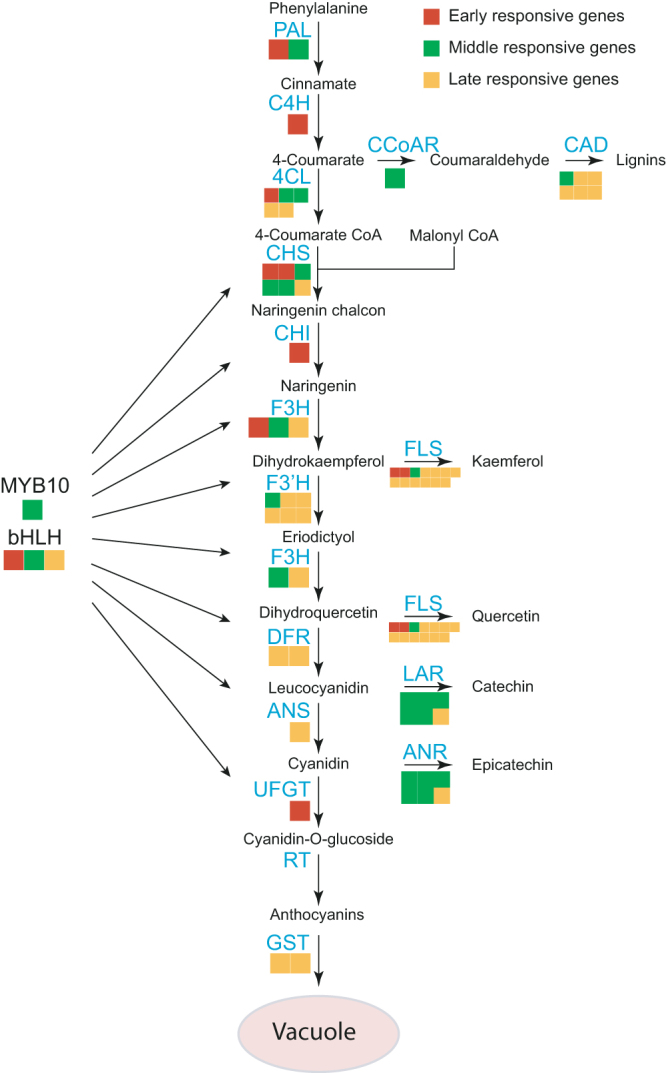
Categorisation of genes belonging to the phenylalanine pathway. Structural genes within the phenylalanine pathway were assigned to different light-responsive gene sets.
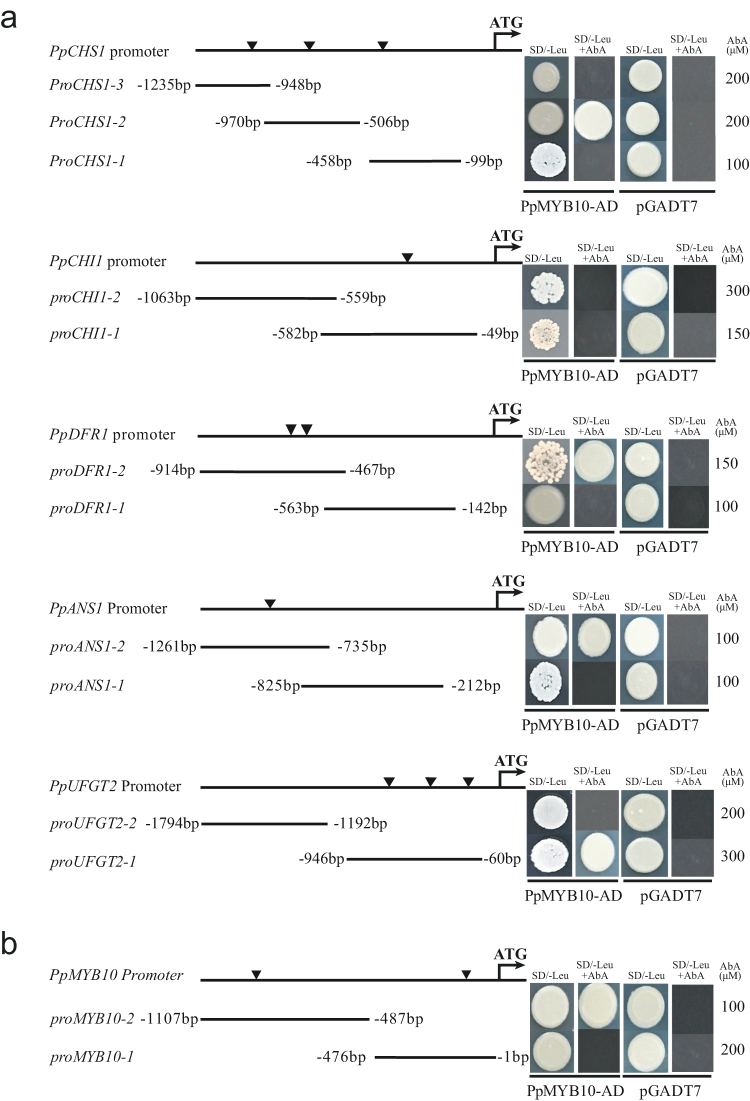
Previous reports showed that most anthocyanin biosynthetic genes were regulated by MYB family, potentially PpMYB10 in pear. As PpMYB10 is the middle light-responsive gene while some anthocyanin biosynthetic genes were late-responsive genes, their direct interaction were further tested in yeast-one hybrid system. PpMYB10 directly bound the promoters of PpCHS1 (Pbr019531.1), PpDFR1 (Pbr038148.1), PpANS1 (Pbr001543.2) and PpUFGT2 (Pbr039986.1) in yeast-one hybrid system (Fig. 8a) while the interaction between PpMYB10 and PpCHI promoter were not detected. In all these binding events, the presence of conserved MYB-binding motif were identified. In addition, PpMYB10 also bound its own promoter (Fig. 8b), suggesting a self-enhanced regulatory pathway during the pear fruit colouration.
Figure 8.
Direct binding of PpMYB10 on the promoter of anthocyanin biosynthetic structural genes (a) and PpMYB10 itself (b) of ‘Meirensu’ in yeast-one hybrid system. The tri-angle marked the conserved MYB binding domain predicted using PlantCare.
Discussion
The responses to bagging treatment and postharvest irradiation in the RCSP are distinct from those of European pears. During post-harvest irradiation after bag removal, RCSPs gradually accumulate anthocyanin in the fruit skin, while European pears show almost no change1, 2, 27. In addition, the accumulation of anthocyanin in the RCSPs accompanies the maturation process, whereas European pears lose some of the red colour as the harvest time approaches, indicating that different triggers mediate the red colouration in these two species. In apple, bagging-induced colouration is mainly mediated by light and enhanced by a relatively low ambient temperature7, 28. This probably involves light-responsive genes, such as HY5 and COP1, and the flavonoid biosynthesis pathway22, 23. In our results, the red colour of ‘Meirensu’ only developed in the fruits that were removed from bags, confirming that the red colouration of RCSP is a light-dependent process, similar to the colouration of apple.
The bagging, followed by bag removal, is a transition from long-term darkness to light, which triggers a response to oxidative stress in the fruit peel that includes the accumulation of lignin and flavonoids, in addition to the scavenging enzymes of reactive oxygen species29. Based on the gene expression patterns during this process, we identified three responsive stages to light in pear fruit peels. In the early-responsive stage, GO terms related to light response and photosynthesis were highly enriched (Fig. 5). In the middle responsive stage, most of these GO terms were not enriched, while the “cellular biosynthetic process” was highly enriched. In the late responsive stage, “flavonoid biosynthesis”, “hormone metabolism” and “defense response” GO terms were overrepresented (Table S3). These results indicated a sequential light-responsive reaction in fruit peel. However, whether these pathways contribute to anthocyanin biosynthesis needs further investigation.
Upstream trans-regulator of PpMYB10
MYB10 is the critical light-inducible TF for anthocyanin biosynthesis7–9, 30. However, how MYB10 is transcriptionally regulated is still not clear, except that it involves HY5 and the B-box gene MdCOL11 22, 28. In our dataset, PpMYB10 was categorised in the middle light-responsive gene set that had peaked expression levels 24 h after bag removal. As high expression levels of HY5 and COL5, which are both light-related genes, were observed in the early-responsive stage, it is reasonable to speculate that both of these genes directly, or indirectly, induced the expression of PpMYB10, which further triggered the expression of downstream structural genes.
In Arabidopsis, HY5 is a pivotal TF for photomorphogenesis that is regulated by multiple photoreceptors and the COP1 protein degradation machinery31–33, and directly binds to the G-box in the promoter of AtMYB75, the Arabidopsis homolog of PpMYB10, to induce its expression34. In apple, MdHY5 also bound on the 5′ upstream region of MdMYBA, at least in a yeast system22, although the direct activation effects were still not observed. The high expression of PpHY5 immediately after bag removal is in accordance with our expectations. However, more studies are needed to characterize whether there is direct interaction between PpHY5 and PpMYB10 to unveil the regulatory pathway and further explain the light-induced anthocyanin biosynthesis.
The CO-like family, also known as the BBX family, contains an N-terminal zinc-binding B-box motif and functions as a transcriptional regulator in response to light, circadian cues and brassinosteroid-light crosstalk signalling. Some members of the COL family have been characterized as positive regulators of anthocyanin biosynthesis. The overexpression of apple MdCOL11 in Arabidopsis increased anthocyanin biosynthesis and promoted earlier flowering under long-day conditions28. PpCOL5 is also a member of the COL family. In Arabidopsis, AtCOL5, the homolog of PpCOL5, functions in controlling the flowering time under short-day conditions in a CO-independent pathway35. In our dataset, PpCOL5 was significantly upregulated immediately after bag removal (Fig. 5), suggesting it has a potential role in the light-responsive pathway. However, owing to limited reports on the COL5 gene in model plants, we are unable to conclude that PpCOL5 is involved in the regulation of anthocyanin biosynthesis. However, because MdCOL11 can simultaneously induce biosynthesis and flowering, PpCOL5 may also participate in light-induced anthocyanin biosynthesis. In addition, because HY5 lacks the trans-activation domain and is, therefore, unable to activate transcription, at least in yeast36 and in plant cells37, its trans-activity must require a cofactor31, 37. Although BBX21 and BBX22 have been proposed as cofactors for HY5, no solid evidence has been provided38, 39. The similar expression patterns of PpHY5 and PpCOL5 suggest that COL5 may be another candidate cofactor of HY5.
Other TFs related to anthocyanin biosynthesis
The differential expression patterns of many other TFs, especially some MADS-box, NAC and MYB genes, were recorded in our datasets, suggesting that they had potential roles in anthocyanin biosynthesis. MADS-box genes were also involved in the anthocyanin biosynthesis in European pear in a MYB-independent manner, especially during early development40, but their functions in the light-responsive process still needs further studies. NAC also participates in the modulation of anthocyanin biosynthesis by directly regulating MYB10 genes in peach41. Here, although we identified many light-induced or -suppressed TFs, their functions in anthocyanin biosynthesis need further investigation.
Phytohormones are involved in bagging-induced fruit colouration
Applying exogenous hormones affects the light-dependent accumulation of anthocyanin and the signalling pathways triggered by plant hormones interact with those triggered by light42. We also identified genes, several enriched GO terms, and DEGs related to phytohormones mostly during the late-responsive stage (Table 2). Although we cannot conclude that the differentially expressed hormone-related genes contributed to anthocyanin biosynthesis, it is reasonable to speculate that some phytohormones indeed affected the bagging-induced anthocyanin biosynthesis.
Exogenous methyl jasmonate (MeJA) can induce the fruit colouration in apple and pear3, 43, 44. Coronatine insensitive1 (COI), an F-box protein that forms a complex with ASK1/ASK2, Cullin1 and Rbx1, is the key player in diverse JA responses45. In its Arabidopsis mutant, coi-1, anthocyanin fails to accumulate in response to MeJA owing to the reduced expression of three anthocyanin regulatory factors, MYB75, MYB90 and GLABRA 3 46–48. In apple, the mechanism by which MeJA induces the anthocyanin accumulation is also well characterised44. The apple jasmonate zim domain (JAZ) protein interacts with bHLH3 to attenuate the formation of an MdMYB9-involved MBW complex that promotes the transcription of structural genes. Upon the perception of a JA signal, the JAZ protein is degraded through the COI-mediated 26S proteosome pathway, releasing bHLH for the formation of MBW, which activates the downstream structural genes44. In our results, we found six differentially expressed JAZ genes (Table 2), most of which were upregulated in the middle- or late-responsive stages, opposite to our expectations. Two possible explanations for this result are as follows: 1) Because JAZs are also subjected to post-translational regulation, the increased transcription level did not equal the high level of translated protein48, or 2) The JA-induced and light-induced anthocyanin biosyntheses activated different MYB proteins44, and the increase of JAZ did not affect the expression and function of PpMYB10, which is the essential light-inducible anthocyanin-related MYB protein.
The effects of ethylene on the anthocyanin accumulation vary among species. As ethylene biosynthesis and anthocyanin accumulation are coherently regulated during fruit ripening, it is thought that ethylene is a positive regulator of anthocyanin biosynthesis. The treatment of grape berries with 2-chloroethylphosphonic acid activated the transcription of structural genes encoding the key enzymes of anthocyanin biosynthesis and increased anthocyanin accumulation49. A similar phenomenon was also observed in apple50. However, ethylene plays a negative regulatory role in anthocyanin biosynthesis in some other species, such as red cabbage, tobacco and corn51–54. Specifically, sugar-induced anthocyanin accumulation is suppressed by ethylene in the model plant Arabidopsis. Ethylene suppresses anthocyanin accumulation by binding to redundant receptors, such as ETHYLENE RESPONSE 1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR 1 (ERS1) and ERS2, with ETR1 possibly playing a dominant regulatory role55. Ethylene tends to enhance the nuclear localisation of COP1, thus inhibiting the activity of HY5 and promoting hypocotyl growth56 under light conditions or seed germination under salt stress conditions57. The antagonistic regulation between ethylene and light on COP1-HY5 pathway is essential for anthocyanin biosynthesis, suggesting that ethylene may suppress anthocyanin biosynthesis through the COP1-HY5 pathway. However, we observed that the anthocyanin accumulation, along with the increased transcription of PpHY5 and PpACSs (Fig. 4, Table 2), suggested that the anthocyanin accumulation in the pear peel may be subjected to complex regulation and that ethylene may have different functions in the regulation of anthocyanin biosynthesis through various pathways.
GA suppresses anthocyanin biosynthesis through the DELLA proteins58–60. DELLA proteins are negative regulators of GA signalling and positively regulate anthocyanin biosynthesis. In DELLA protein quadruple mutants (gai-t6/rga-t2/rgl1-1/rgl2-1), the expression of anthocyanin biosynthesis genes, such as ANS and F3′H, decreased, while the expression of UFGT and PAP1 is independent of the DELLA proteins58. The DELLA-promoted anthocyanin biosynthesis is partially mediated by tMYBL2, an inhibitor of transcription, and JAZ regulatory proteins60. In this sense, GA-mediated anthocyanin inhibition is at least partially independent of the light-induced anthocyanin biosynthesis pathway in which PAP1 was involved. However, GA is also involved in the HY5-mediated skotomorphogenesis by reducing the abundance of the HY5 protein through the modulation of COP1 activity61, potentially indirectly regulating anthocyanin biosynthesis. In the present study, we observed the downregulation of two DELLA proteins, suggesting that GA was also involved in the light-induced anthocyanin biosynthesis in pear peel. However, how GA is involved in the fine-tuned regulation of light-induced anthocyanin biosynthesis needs to be studied further.
Differential regulation of structural genes for anthocyanin biosynthesis
Several reports proposed that both the early and late anthocyanin biosynthetic genes could be activated by MYB107, 8. However, from the temporal expression patterns of these genes, it appears that these genes are differentially regulated, which were further confirmed by yeast-one hybrid assay. Some lines of evidence show that the expression of UFGT is coupled with that of MYB10 in other plants, while the expression of other genes are not30, 62. In our work, the early-stage biosynthetic genes, such as PpCHS1 and PpCHI1, are upregulated immediately after bag removal (Fig. 7), which is much earlier than the anthocyanin accumulation and the high expression of PpMYB10, but PpMYB10 is capable to bind the promoters of PpCHS1 but not PpCHI1 (Fig. 8). These results suggested that PpMYB10 directly regulate PpCHS1 but other regulatory were also involved in. Indeed, HY5 directly activates AtCHS by binding to the G-box motif in the Arabidopsis promoter36, 63, suggesting a function for PpHY5 in the light-induced flavonoid biosynthesis in RCSP. On the other hand, some of the late-stage biosynthetic genes, such as PpDFR and PpANS, were induced in the late-responsive stage, along with the accumulation of anthocyanin, are mainly under the regulation of PpMYB10 (Fig. 8). In our dataset, the expression of PpUFGT2 peaked 6 h after bag removal, much earlier than PpMYB10, which was further confirmed by qRT-PCR, indicating a novel regulation of PpUFGT2 in RCSP ‘Meirensu’, although it is still under the regulation of PpMYB10. Because the expression pattern of PpUFGT2 is similar to that of HY5, and a G-box motif is found in the 5′ upstream region of PpUFGT2, HY5 may directly activate the expression of PpUFGT2 by binding the G-box. Interestingly, we observed the direct binding of PpMYB10 on the promoter of itself, which may explain the later appearance of the peak than PpHY5.
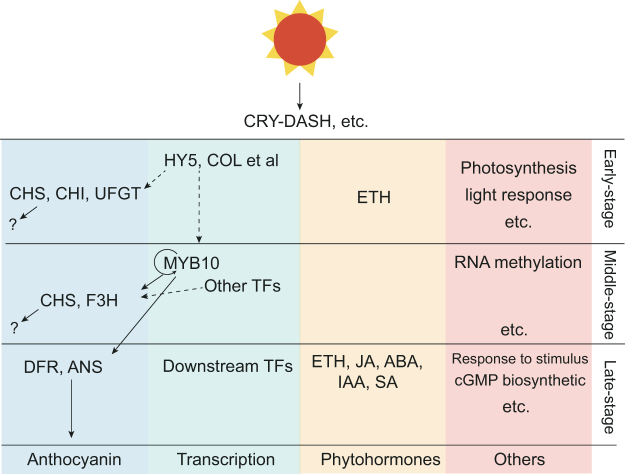
In summary, a transcriptional regulatory model (Fig. 9) showing the light-responsive reactions in pear fruit peel was inferred from the transcriptome analysis. After bag removal, photoreceptors, such as CRY-dash, received the light, triggering the light-responsive pathway, activating the transcription of HY5 and COL5, and inducing the transcription of downstream TFs, such as MYB10, and structural genes, CHS, F3H and UFGT2, which resulted in the accumulation of anthocyanin in the fruit peel. In addition, plant hormone- and other biological process-related pathways were activated by the light-responsive pathway, resulting in significant physiological changes in fruit after bag removal, including anthocyanin accumulation in the fruit peel.
Figure 9.
Putative model for bag removal-induced anthocyanin biosynthesis in ‘Meirensu’ pear peels. The phytohormone and other pathways based on the gene ontology enrichment analysis.
Anthocyanin biosynthesis is a complex process involving various regulatory pathways. Our work provided a transcriptomic view on the light-induced anthocyanin biosynthesis that occurs after bagging treatment. However, owing to technical limitations, our work did not provide any post-transcriptional or (post)-translational evidence for anthocyanin biosynthesis, which are also importation regulatory pathways for red pigmentation64, 65. Whether these pathways are also involved in the bagging-improved colouration needs further analysis.
Materials and Methods
Plant materials and experimental treatments
The fruit of RCSP ‘Meirensu’ were obtained from a commercial orchard in Kunming, Yunnan Province, China. In total, 500 fruitlets were covered with double layers of yellow-black paper bags [Kobayashi (Qingdao) Co., Ltd., Qingdao China] 40 days after full blossom (DAFB). Half of the fruits had their bags removed 10 days before harvest (155 DAFB), and the others remained bagged as controls. 30 fruits were randomly sampled at 0, 3, 6, 24, 72 and 144 h after bag removal and divided to three biological replicates. Control fruit were sampled at the same times and named as 3 h-C, 6 h-C, 24 h-C, 72 h-C and 144 h-C (C, control; the 0 H sample did not require a control). Fruit peel was scraped and immediately frozen in liquid nitrogen, and stored at −80 °C until use.
Measurements of total anthocyanin, chlorophyll and carotenoid concentrations
The anthocyanin concentration was measured according to a methanol-HCL method and was presented as mg cyanidin-3-galactoside per 100 g fresh tissue. One gram of fruit peel was mixed with methanol containing 0.1% HCl, followed by centrifugation at 4 °C and 18,000 × g for 20 min. The absorbance of each 100-μL extract was assessed using a spectrophotometer (DU800; Beckman, IN, USA) at 510 and 700 nm, in buffers of pH 1.0 and 4.5, respectively. The anthocyanin concentration was calculated using the equation: A = [(A510–A700)pH1.0 − (A510–A700)pH4.5] with a molar extinction coefficient of 3.02 × 104 cyanidin-3-galactoside66. One gram of ground fruit peel was homogenized in 6 mL of 80% cold acetone and centrifuged at 4 °C and 12,000 rpm for 20 min. The extract’s absorbance was measured using a spectrophotometer (DU800; Beckman) at 440, 645 and 663 nm. The chlorophyll concentration was calculated using the equation Ct = 20.2 A645 + 8.02 A663, and the carotenoid concentration was calculated based on the equation Ck = 4.7 A440 − 0.27 Ct67.
RNA extraction, library preparation and RNA-Seq
Total RNA from each sample was isolated three times, as biological replications, using a modified cetyltrimethylammonium bromide method68. The RNA quality was assessed using an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Only the samples with RNA integrity numbers >7.5 were used for deep sequencing. mRNA was enriched and purified, and then cleaved into small pieces. Short fragments were purified with a QiaQuick PCR extraction kit (Qiagen, Venlo, Netherlands) and dissolved with the EB buffer supplied in the kit for end preparation and poly(A) addition. Then, the short fragments were connected with sequencing adapters. After agarose gel electrophoresis, suitable fragments were selected as templates for PCR amplification, and the library was sequenced using an Illumina HiSeq™2000 (San Diego, CA, USA) by Biomarker (Beijing, China).
Sequence data processing and mapping reads to the pear genome
To obtain high-quality clean read data for sequence analysis, adaptor sequences, empty reads, low-quality sequences with greater than 5% N (the percentage of nucleotides in the reads that could not be sequenced) and those containing more than 10% ambiguous bases were removed. Clean reads were mapped to the pear genome sequence (http://peargenome.njau.edu.cn/) using TopHat69 with default parameters. The reads were then assembled into transcripts and compared with reference gene models using Cufflinks69. Transcripts that did not exist in the CDS database of the pear genome were extracted to predict new genes using the EMBOSS package (http://emboss.open-bio.org/).
Functional annotations of known and new genes
All of the known and newly predicted genes were annotated with protein databases, including the NCBI nonredundant, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes and Cluster of Orthologous Groups databases, using the BLASTX algorithm with a cutoff e-value of 10−5. Blast2GO (http://www.blast2go.com) was used to obtain GO annotation of the transcripts. The GO enrichment analysis of DEGs was implemented by the Cytoscape plugin BinGO70. GO terms with corrected p-values < 0.05 were considered significantly enriched.
Identification of significant DEGs
A statistical analysis of the frequency of each gene in each library was performed to compare gene expression levels. All of the clean reads were aligned to genes using SOAP aligner71 and then normalized into RPKM values72. Transcript abundance differences between each pair of samples were then calculated based on the ratios of the RPKM values, and the false discovery rate control method was used to identify the p-value thresholds in multiple tests to compute the significance of any differences in transcript abundance73. Genes with an absolute log2ratio ≥ 2 and a false discovery rate significance score < 0.001 were considered significantly differentially expressed.
Construction and visualization of co-expression network
The DEGs were then used to perform the co-expression network analysis using the R package WGCNA74. The co-expression network was visualized using the free software Cytoscape75. The expression data were clustered with Cluster 3.076 and displayed using Java Treeview77.
Real-time quantitative RT-PCR (qPCR) analysis
Total RNAs were reverse transcribed to cDNAs using an RTase (Takara Bio, Kusatsu, Japan), following the manufacturer’s instructions, and subjected to qPCR as described in Zhang, et al.27. Primers used for qRT-PCR was listed in Table S4.
Yeast One-hybrid assay
The yeast one-hybrid assays were performed using the Matchmaker Gold Yeast One-Hybrid System Kit (Clontech) according to the manufacturer’s instruction. The primers used for construction were listed in Table S4.
Statistical analysis
t-test (p = 0.05) were performed using the Data Processing System (DPS, version 3.01; Zhejiang University, Hangzhou, China).
Data accessibility
The sequencing data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/), with sample accession numbers SAMN03857509, SAMN03857510, SAMN03857511, SAMN03857512, SAMN03857513, SAMN03857514 and SAMN03857515.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos 31471852, 31272141 and 31301753) and the earmarked fund for Modern Agro-industry Technology Research Systems (CARS-29).
Author Contributions
D.Z. and Y.T. conceived and planned the study. Y.S., M.Q., L.L. and Q.S. collected the samples and extracted total RNAs for next-generation sequencing. S.B. and Y.S. analysed the sequencing data, F.Y., J.N. and R.T. performed the qRT-PCR. S.B., Y.S., D.Z. and Y.T. wrote the manuscript. All of the authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Songling Bai and Yongwang Sun contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00069-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong Zhang, Email: afant@nwsuaf.edu.cn.
Yuanwen Teng, Email: ywteng@zju.edu.cn.
References
- 1.Qian M, et al. Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci Hortic. 2013;151:75–82. doi: 10.1016/j.scienta.2012.12.020. [DOI] [Google Scholar]
- 2.Huang C, et al. Effects of fruit bagging on coloring and related physiology, and qualities of red Chinese sand pears during fruit maturation. Sci Hortic. 2009;121:149–158. doi: 10.1016/j.scienta.2009.01.031. [DOI] [Google Scholar]
- 3.Qian M, et al. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia Nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/Vis conditions. Plant Mol Biol Report. 2013;32:428–437. doi: 10.1007/s11105-013-0652-6. [DOI] [Google Scholar]
- 4.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 5.Hichri I, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 6.Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005;8:272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Takos AM, et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ban Y, et al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- 9.Espley RV, et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982–982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 11.Medina-Puche L, et al. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria ananassa fruits. J Exp Bot. 2014;65:401–417. doi: 10.1093/jxb/ert377. [DOI] [PubMed] [Google Scholar]
- 12.Aharoni A, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 13.Kadomura-Ishikawa Y, Miyawaki K, Takahashi A, Noji S. RNAi-mediated silencing and overexpression of the FaMYB1 gene and its effect on anthocyanin accumulation in strawberry fruit. Biologia Plantarum. 2015;59:677–685. doi: 10.1007/s10535-015-0548-4. [DOI] [Google Scholar]
- 14.Feng S, Wang Y, Yang S, Xu Y, Chen X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 2010;232:245–255. doi: 10.1007/s00425-010-1170-5. [DOI] [PubMed] [Google Scholar]
- 15.Bai, S. et al. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 244, 573–586 (2016). [DOI] [PubMed]
- 16.Layne DR, Jiang ZW, Rushing JW. Tree fruit reflective film improves red skin coloration and advances maturity in peach. HortTechnology. 2001;11:234–242. [Google Scholar]
- 17.Wand, S. J. E. et al. Use of evaporative cooling to improve ‘Rosemarie’ and ‘Forelle’ pear fruit blush colour and quality. Acta Hortic. 103–111 (2005).
- 18.Sun YW, et al. Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biology and Technology. 2014;91:64–71. doi: 10.1016/j.postharvbio.2013.12.015. [DOI] [Google Scholar]
- 19.Ju Z, Duan Y, Ju Z. Effects of covering the orchard floor with reflecting films on pigment accumulation and fruit coloration in ‘Fuji’ apples. Sci Hortic. 1999;82:47–56. doi: 10.1016/S0304-4238(99)00038-2. [DOI] [Google Scholar]
- 20.Steyn WJ, Holcroft DM, Wand SJE, Jacobs G. Regulation of pear color development in relation to activity of flavonoid enzymes. J Am Soc Hortic Sci. 2004;129:6–12. [Google Scholar]
- 21.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Peng T, et al. Screening of UV-B-induced genes from apple peels by SSH: possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiol Plant. 2013;148:432–444. doi: 10.1111/j.1399-3054.2012.12002.x. [DOI] [PubMed] [Google Scholar]
- 23.Vimolmangkang S, et al. Transcriptome analysis of the exocarp of apple fruit identifies light-induced genes involved in red color pigmentation. Gene. 2014;534:78–87. doi: 10.1016/j.gene.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Feng S, et al. Differential expression of proteins in red pear following fruit bagging treatment. The Protein Journal. 2011;30:194–200. doi: 10.1007/s10930-011-9320-6. [DOI] [PubMed] [Google Scholar]
- 25.Bao L, Chen K, Zhang D, Li X, Teng Y. An assessment of genetic variability and relationships within Asian pears based on AFLP (amplified fragment length polymorphism) markers. Sci Hortic. 2008;116:374–380. doi: 10.1016/j.scienta.2008.02.008. [DOI] [Google Scholar]
- 26.Wu J, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, et al. Effects of high temperatures on UV-B/visible irradiation induced postharvest anthocyanin accumulation in ‘Yunhongli No. 1’ (Pyrus pyrifolia Nakai) pears. Sci Hortic. 2012;134:53–59. doi: 10.1016/j.scienta.2011.10.025. [DOI] [Google Scholar]
- 28.Bai S, et al. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta. 2014;240:1051–1062. doi: 10.1007/s00425-014-2129-8. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M, et al. Identification of Arabidopsis genes regulated by high light stress using cDNA microarray. Plant Cell Physiol. 2002;43:S159–S159. doi: 10.1093/pcp/pcf019. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. The methylation of the PcMYB10 promoter is associated with Green-skinned sport in max red bartlett pear. Plant Physiol. 2013;162:885–896. doi: 10.1104/pp.113.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm M, Ma L, Qu L, Deng X. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin D, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Hassidim M, Harir Y, Yakir E, Kron I, Green RM. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta. 2009;230:481–491. doi: 10.1007/s00425-009-0958-7. [DOI] [PubMed] [Google Scholar]
- 36.Ang L, et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/S1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 37.Li J, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of Phytochrome A signaling. Plant Cell. 2010;22:3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-Box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–3255. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta S, et al. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, et al. Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.) Tree Genetics & Genomes. 2013;9:75–83. doi: 10.1007/s11295-012-0534-3. [DOI] [Google Scholar]
- 41.Zhou H, et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant Journal. 2015;82:105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
- 42.Das PK, Shin DH, Choi S-B, Park Y-I. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells. 2012;34:501–507. doi: 10.1007/s10059-012-0151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudell DR, Mattheis JP, Fan X, Fellman JK. Methyl jasmonate enhances anthocyanin accumulation and modifies production of phenolics and pigments in ‘Fuji’ apples. J Am Soc Hortic Sci. 2002;127:435–441. [Google Scholar]
- 44.An XH, et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–662. doi: 10.1093/pcp/pcu205. [DOI] [PubMed] [Google Scholar]
- 45.Xu L, et al. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. The Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devoto A, et al. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol. 2005;58:497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- 47.Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/S1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 48.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 49.El-Kereamy A, et al. Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol Plant. 2003;119:175–182. doi: 10.1034/j.1399-3054.2003.00165.x. [DOI] [Google Scholar]
- 50.Whale SK, Singh Z. Endogenous ethylene and color development in the skin of ‘pink lady’ apple. J Am Soc Hortic Sci. 2007;132:20–28. [Google Scholar]
- 51.Craker LE, Wetherbee PJ. Ethylene, light, and anthocyanin synthesis. Plant Physiol. 1973;51:436–438. doi: 10.1104/pp.51.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang BG, Burg SP. Role of ethylene in phytochrome-induced anthocyanin synthesis. Planta. 1973;110:227–235. doi: 10.1007/BF00387635. [DOI] [PubMed] [Google Scholar]
- 53.Takada K, Ishimaru K, Minamisawa K, Kamada H, Ezura H. Expression of a mutated melon ethylene receptor gene Cm-ETR1/H69A affects stamen development in Nicotiana tabacum. Plant Sci. 2005;169:935–942. doi: 10.1016/j.plantsci.2005.06.012. [DOI] [Google Scholar]
- 54.Rengel Z, Kordan HA. Effects of growth regulators on light-dependent anthocyanin production in Zea mays seedlings. Physiol Plant. 1987;69:511–516. doi: 10.1111/j.1399-3054.1987.tb09233.x. [DOI] [Google Scholar]
- 55.Jeong SW, et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 2010;154:1514–1531. doi: 10.1104/pp.110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, Y. W. et al. Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet9 (2013). [DOI] [PMC free article] [PubMed]
- 57.Yu YW, et al. Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 2016;170:2340–2350. doi: 10.1104/pp.15.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang C, Gao X, Liao L, Harberd NP, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Van den Ende W, Rolland F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Molecular Plant. 2014;7:570–572. doi: 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]
- 60.Xie Y, Tan HJ, Ma ZX, Huang JR. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/BHLH/WD40 complex in Arabidopsis thaliana. Molecular Plant. 2016;9:711–721. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Alabadi D, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 62.Tuan PA, et al. The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 2015;15:280. doi: 10.1186/s12870-015-0664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, et al. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012;160:1011–1022. doi: 10.1104/pp.112.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia, R., Zhu, H., An, Y., Beers, E. & Liu, Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol13 (2012). [DOI] [PMC free article] [PubMed]
- 66.Wrolstad, R. E. Color and pigment analyses in fruit products. Corvallis Or. Agricultural Experiment Station.oregon State University (1993).
- 67.Huang CH, et al. Effects of fruit bagging on coloring and related physiology, and qualities of red Chinese sand pears during fruit maturation. Sci Hortic. 2009;121:149–158. doi: 10.1016/j.scienta.2009.01.031. [DOI] [Google Scholar]
- 68.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 69.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 71.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 72.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 73.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of statistics. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 74.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics9 (2008). [DOI] [PMC free article] [PubMed]
- 75.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 77.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/), with sample accession numbers SAMN03857509, SAMN03857510, SAMN03857511, SAMN03857512, SAMN03857513, SAMN03857514 and SAMN03857515.