Abstract
Bacterial species constituting the intestinal microbiota are implicated in maintenance of health but also pathogenesis of inflammatory disease. The compositional complexity of the microbiota and metabolic interdependencies of microbial species challenge our ability to attribute host responses to specific bacterial strains. Studies using gnotobiotic mice, however, are providing important insights.
The hundreds of bacterial species and strains residing in the mammalian intestinal tract compete and cooperate, in the process forming a complex ecosystem that interfaces with the gut mucosa and enhances host health by optimizing nutrient absorption, producing essential vitamins, providing colonization resistance against pathogens and promoting regulatory immune responses that limit inflammation. The intestinal microbiota, however, is also implicated in the pathogenesis of a wide range of inflammatory diseases. Bacterial species residing in the intestine are influenced by factors such as nutrient availability, mucin production, antimicrobial peptide secretion and intestinal peristalsis. Interactions between bacterial species in the intestine remain incompletely defined but examples of cooperation have been discovered, such as reciprocity between Bacteroides species during carbohydrate metabolism (Rakoff-Nahoum et al., 2016). The host immune system responds to microbial colonization of the intestine and, over the past decade, we have learned about the impact of various bacterial species on T lymphocyte differentiation, mucin production and expression of antimicrobial factors.
In part because of their therapeutic potential, identifying and characterizing bacterial species and the immune mechanisms they trigger have become areas of intense and exciting research. However, despite the widely recognized positive and negative microbial contributions to human health, our ability to determine the specific impact of different bacterial species remains limited. The interdependencies of different bacterial taxa constituting the microbiota, and the overall complexity of microbial populations in the intestine make it difficult to attribute specific immune effects to individual bacterial species.
Although transfer of human microbiota into germ free mice has been revealing (Smith et al., 2013), incomplete interspecies transfer of symbiotic bacterial populations and differences between human and murine symbionts can result in incomplete or altered host immune activation (Chung et al., 2012). For now, however, gnotobiotic mouse models provide the most tractable in vivo system to characterize the impact of specific human commensal bacterial species on mammalian physiology. A seminal study measured gene expression in intestinal epithelial cells and bacteria of mice that were mono-colonized with Bacteroides thetaiotaomicron or Bifidobacterium longum (Sonnenburg et al., 2006). On the transcriptional level, host and colonizing bacteria responded to each other, and the epithelial response to the two bacterial species was distinct (Sonnenburg et al., 2006). Segmented filamentous bacterium was discovered to drive the differentiation of Th17 T cells in the mouse gut (Gaboriau-Routhiau et al., 2009, Ivanov et al., 2009), and furthermore, a Bacteroides fragilis derived polysaccharide was shown to induce T regulatory cells (Round and Mazmanian, 2010). Characterization of bacterial species derived from a human fecal sample revealed that regulatory T cells can be induced in the colon of mice by a consortium of 17 commensal Clostridia strains (Atarashi et al., 2013) while another study focusing on non-clostridial human commensal species demonstrated that multiple species of Bacteroides also induce regulatory T cells (Faith et al., 2014). These studies reveal that vastly different commensal bacterial species can drive T cell differentiation along similar or distinct pathways and that some bacterial species can function as lone rangers while others function only as members of a posse composed of multiple species. These varied findings raise important questions about the molecular mechanisms that are triggered by different intestinal microbes.
A recent and important contribution to this field catalogues activation of the murine immune system by 53 distinct commensal bacterial species (Geva-Zatorsky et al., 2017). The findings are interesting in terms of the breadth of bacterial species that were used to mono-colonize mice, the depth of the immunologic analyses and the overall consistency of their results with previously published work. Analyses of these different bacterial species demonstrated that similar immune responses can be initiated by diverse microbial species while closely related bacterial species often differ in terms of immune activation. Surprisingly, the majority of bacterial species were found to disseminate from the intestinal lumen to mesenteric lymph nodes and systemic lymphoid organs. The finding that it is not possible to predict the impact of a bacterial strain on the immune system on the basis of responses to closely related strains is an important message, emphasizing the need to go beyond simply classifying symbionts on the basis of 16S rRNA gene similarity.
As the Geva-Zatorsky paper in Cell becomes a much-cited resource, it will be important for readers to remember two qualifications, emphasized by the authors. First, they use an “unabashedly reductionist experimental strategy” to identify microbial interactions with the mammalian immune system. Second, mono-colonization with individual bacterial strains results in unnaturally high colonization densities. Thus, mono-colonization may induce host immune responses that do not occur in the presence of companion microbes, and markedly increased densities of specific organisms may lead to microbial dissemination and inflammatory responses that are not detected with physiologic microbial densities. Reducing complexity by colonizing mice with a single bacterial strain, while simplifying data interpretation, comes at the cost of lost inter-microbial interactions that almost certainly create a “sauce” with distinct qualities that impact the immune system. Indeed, assessing and appreciating the complex flavor of a gourmet meal would be very challenging if one were limited to only tasting individual ingredients in isolation.
Understanding the interactions of different bacterial species with each other and with the mammalian host will facilitate development of commensal bacteria as therapeutic agents. Microbiota stimulation of the immune system establishes the host’s immune tone, which determines the threshold for immune activation. In the absence of the microbiota, many of the host’s immune defenses are in a state of dormancy, which can delay responses to an invading pathogen. While increasing the host’s innate immune tone enhances resistance against infection, the price of greater resistance against pathogens may be an increased risk of autoimmune and inflammatory diseases, and perhaps even cancer. How tightly resistance to infection and vulnerability to inflammatory states are interdependent is unclear. Nevertheless, the recent discoveries that specific bacterial species can sway immune development and alter the risk of inflammation and reduce the risk of certain infections suggest that microbiota manipulation has great potential as a medical intervention for a wide range of diseases that includes cancer, inflammatory bowel disease, arthritis and microbial infection. Understanding the impact of individual microbes on the mammalian immune system is an important and essential step on the path forward.
Figure 1. Reducing Microbial Complexity to Decode Complex Signals.
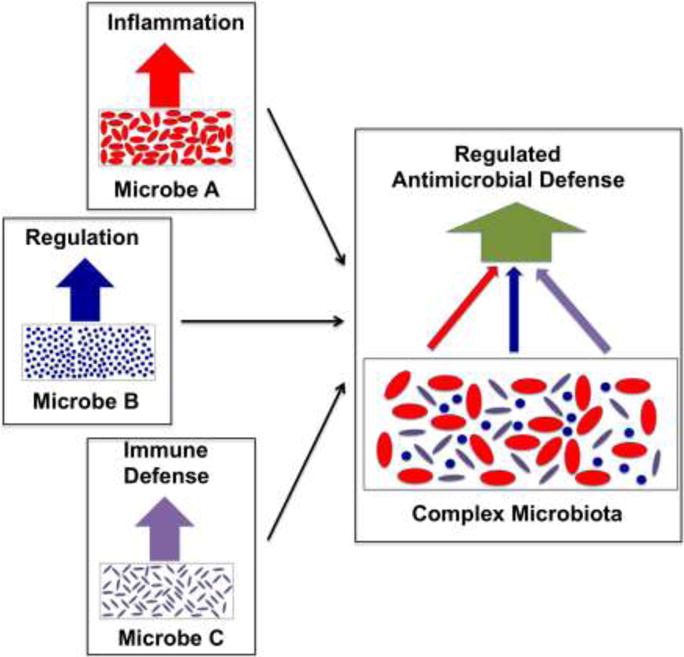
Mono-colonization of mice with different symbiotic bacterial strains (shown in the left three panels) leads to high colonization densities that trigger distinct mucosal and systemic immune responses (indicated by the red, blue and purple arrows). In mice colonized with a complex microbiota (right panel), interactions between different bacterial species will lead to a composite signal (green arrow) that will provide a broader and more complex host response.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 2.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Science translational medicine. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Kasper DL. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928–943 e911. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]


