Abstract
Microvesicle biogenesis is a highly regulated process. Aberrant release of microvesicles from cancer cells have been associated with their invasiveness and prognosis. However, the mechanism of aberrant release remains poorly understood. Herein, we found that hepatocellular carcinoma cells shed more microvesicles than normal hepatocytes and miR-200a were shown to inhibit the release of microvesicles in hepatocellular carcinoma cells. Then, we confirmed that miR-200a might target Gelsolin and change cytoskeleton to regulate microvesicles secretion. Further miR-200a may inhibit the proliferation of adjacent cells by inhibiting the release of microvesicles. Collectively, our findings indicate that miR-200a regulated the microvesicle biogenesis involved in the hepatocellular carcinoma progression.
Keywords: microRNA, microvesicle biogenesis, cytoskeleton, hepatocellular carcinoma
Introduction
Microvesicles (MVs) are small vesicular structures (30–1,000 nm in diameter) that are formed and shed directly from the surfaces of cells under physiological and pathological conditions (1). They play vital roles in intercellular communication. Many studies suggest that de-regulated biogenesis of MVs is responsible for several diseases, especially cancers such as hepatocellular carcinoma (HCC) (2).
The formation of MVs involves tightly regulated processes. There are generally two types of MVs depending on the membrane vesicles from which they are derived, microparticles (100–1000 nm) and exosomes (30–100 nm). Microparticles bud from micro-domains within the plasma membrane (known as lipid rafts), then are loaded with the specific cargo and are finally released into the extracellular environment (3). In contrast, the biogenesis of exosomes uses the multivesicular body (MVB) sorting pathway, in which proteins and lipids are initially destined for degradation in lysosomes. The biogenesis of MVs results in dramatic morphological changes in the cells. RhoA signaling, which is responsible for actin-cytoskeletal rearrangements and cell morphology, is able to stimulate the formation of MVs in different cancer cell lines (4). An increase of the intracellular Ca2+ has also been shown to promote the formation of MVs (5,6). Gelsolin (GSN) is a potent actin-severing protein that removes the capping proteins and regulates the assembly and disassembly of the actin filaments (F-actin) (7). The breakdown of actin filaments by GSN depends on the cytosolic Ca2+ concentration (8). Ca2+ serves as a rapid activator for GSN and assists GSN in participating in the regulation of cell cytoskeleton and vesicle secretion. However, thus far, the molecular aspects associated with the regulation of gene expression of GSN, especially at post-transcriptional level, have not been clearly elucidated.
MVs can transport several types of cargo including proteins, lipid, and nucleic acids (mRNA, microRNA, and DNA) to recipient cells to regulate the functional or behavioral aspects of the recipient cells. The biological function of MVs appears to be dependent on the content transported by them. Our previous study suggested that overexpression of miR-483 in HL7702 cells might lead to an intercellular transfer of miR-483 between HL7702 and LX-2 cells (9). Chen et al also confirmed that miR-214 could be transferred by exosomes from hepatocytes to hepatic stellate cells and be involved in fibrogenesis (10). However, it is unclear whether microRNA regulates the secretion of MVs.
Increased MV levels have been reported in patients with HCC and might serve as a potential biomarker for the diagnosis and prognosis of HCC (2,11). Kogure et al have shown that HCC-derived MVs can carry a specific subset of miRNAs that can be internalized by other cells to regulate transforming growth factor-β (TGF-β)-activated kinase-1 (TAK1) expression and enhance transformed cell growth in recipient cells (12). Furthermore, the expression of miRNAs in serum exosomes could be a potential diagnostic marker for HCC. Serum exosomal miR-21 expression in patients with HCC has been reported to be higher than healthy volunteers (13). In the past decade, extensive and in-depth studies on microRNAs in liver diseases, especially in HCC, have been conducted. miR-21, miR-122, miR-200, and miR-221 have been reported to be de-regulated in HCC progression and implicated in cancer cell proliferation and metastasis (14–17). Nevertheless, whether de-regulated microRNA could regulate the secretion of MVs in HCC is unclear.
In this study, we proposed that miR-200a could inhibit the secretion of MVs derived from liver cancer cell lines and regulate the proliferation of the adjacent cells. Herein, we first identified the amount of MVs released from normal liver cell lines and HCC cells. Second, we explored the effect of miR-200a on the secretion of MVs. Subsequently, GSN was predicted and identified as the functional target of miR-200a. Finally, the upregulation of miR-200a was able to regulate the proliferation of the adjacent cells. To the best of our knowledge, this is the first study to propose that microRNA regulates the secretion of MVs. The results of this study can help in understanding the functions of microRNA and the secretion of MVs in liver diseases.
Materials and methods
Cell culture
The human HCC cell line HepG2 and Huh7 and human normal hepatocyte cell line HL7702 were conserved in our laboratory. HCC cell lines and normal hepatocyte cell line were cultured in Dulbecco's modified Eagle's medium and RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA), respectively, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a humid atmosphere containing 5% CO2 at 37°C.
Transfection and luciferase assay
Cells were seeded in 6-well plates and transfected with miR-200a or miR-483 mimics, or non-targeting control (NC) (GenePharma Co., Ltd., Shanghai, China) using Lipofectamine-2000 (Invitrogen). The NC was synthetic scrambled double oligonucleotide, non-targeting against any mRNA. Protein and mRNA expression levels were analyzed 48-h post-transfection. For the MV isolation, we used 10-cm plates to do the transfection. The luciferase assay was performed in HEK-293T cells. Briefly, we generated two Luc-M 3′-UTR constructs, including the potential biding sequence (GSN-WT-UTR) and mutated the potential biding sequence (GSN-MUT-UTR). Cells were seeded in 24-well plates and co-transfected with 100 ng Luc-GSN 3′-UTR reporter vector, 40 ng TK and 30 nM miR-200a mimics and incubated overnight. Luciferase activity was measured using the Dual-Glo Luciferase assay system (Promega, Madison, WI, USA).
Isolation of MVs and electron microscopy
MVs were purified from cell culture medium as previously described (18). Cell culture medium was centrifuged at 300 × g for 10 min, at 1200 × g for 10 min, and then at 10,000 × g for 30 min. The supernatant was ultracentrifuged at 110,000 × g for 2 h at 4°C. Pelleted MVs were resuspended in culture medium and sent to the Center for Electron Microscopy, Harbin Medical University for transmission electron microscopy (TEM) analysis.
MVs labeling and flow cytometry
MVs were labeled with PE anti-human CD9 (#312105, BioLegend, London, UK) for 30 min at room temperature in the dark according to the manufacturer's instructions. CD9 is a specific molecular marker for exosomes (19). After incubation with CD9, the samples were immediately analyzed using flow cytometry. The procedure was carried out as previously described (20). To establish a MV gate, we used size-calibrated fluorescent beads with a size of 1 µm (#L2778, Sigma, St. Louis, MO, USA). The upper limit of the MV gate was established using the 1 µm beads, while the lower limit was set by the medium (Fig. 1B). We defined MVs as particles within the MV gate, which had positive staining for CD9. The formula, number of events of defined MVs/number of cells used for the MVs isolation, was used for MV quantification.
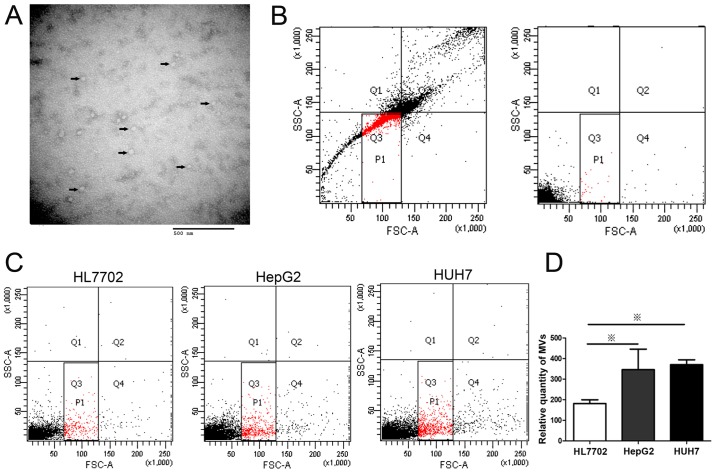
Figure 1.
HepG2 and Huh7 cells release more MVs than HL7702 cells. MVs were isolated from the supernatant of cultured cells through ultracentrifugation. (A) Representative image of isolated MVs observed by transmission electron microscopy. Small 40- to 90-nm vesicles are indicated with arrows. (B) Construction of MV gates of flow cytometry using size-calibrated fluorescent beads (left) and medium (right). The solid square (P1) is the total MV gate. (C) MVs released from liver cell lines (HepG2, Huh7, and HL7702 cells) measured by flow cytometry. (D) Statistical analysis of MV release. HepG2 and Huh7 cells secreted more MVs than HL7702. ※P<0.05.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). Reverse-transcribed complementary DNA was synthesized with random primers or microRNA specific stem-loop primers (Applied Biosystems, Foster City, CA, USA). Subsequently, the cDNA was subjected to real-time PCR on a 7500 real-time PCR system (Applied Biosystems). The 2−∆∆Ct method was used to calculate the relative expression levels of the genes of interest. GAPDH and U6 were used as internal controls. The primer sequences used were as follows: GSN sense: 5′-CAGACAGCCCCTGCCAGCACCC-3′, antisense: 5′-GAGTTCAGTGCACCAGCCTTAGGC-3′; GAPDH sense: 5′-AGCCTCCCGCTTCGCTCTCT-3′, antisense: 5′-GCGCCCAATACGACCAAATCCGT-3′; miR-200a sense: 5′-GGCGTAACACTGTCTGGTAA-3′, antisense: 5′-CGTATCCAGTGCGTGTCGTG-3′; U6 sense: 5′-GCTTCGGCAGCACATATACTAAAAT-3′, antisense: 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
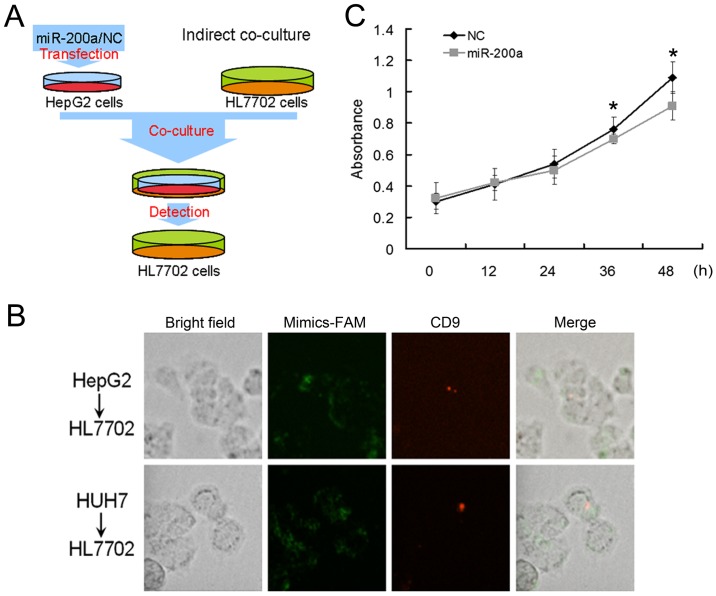
Cell indirect co-culture and MTT assay
For the indirect co-culture assay, polycarbonate membrane inserts in multidishes (Nunc, Beijing, China) were used. The HepG2 or Huh7 cells transfected with carboxyfluorescent labeled miR-200a mimics or NC were seeded in the upper transwell inserts, and the HL7702 cells were seeded in the lower 3.5-cm plates. HepG2 or Huh7 cells expressing green fluorescence was visualized directly by fluorescence microscopy and at least twenty fields were recorded per experiment. The formula - % transfection efficiency = (number of cells stained with fluorescent positive-green/total number of cells per field) was used to calculate the transfection efficiency. The HepG2 or Huh7 cells with transfection efficiency exceeding 70% was used in the indirect co-culture experiment. After transfection, we changed the culture medium with medium containing CD9 in the upper transwell inserts. The CD9+ HL7702 cells were observed by fluorescence microscopy and we calculated the percentage of CD9+ HL7702 cells by choosing ten fields for each experiment. Cell viability was assessed using the MTT assay. To each well of cells 100 µl of 5 mg/ml solution of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) was added and incubated at 37°C for 4 h and then lysed in 700 µl of dimethyl sulfoxide (DMSO) at room temperature for 15 min. The absorbance of each well at a wavelength of 492 nm was read on a spectrophotometer. Three independent experiments were performed in quadruplicate.
Western blotting
Total proteins from cells were extracted using RIPA buffer. Cellular protein extracts were separated in a 10% SDS-polyacrylamide gel and electrophoretically transferred onto a PDVF membrane (Millipore, Bedford, MA, USA). Membranes were blocked with 5% non-fat dried milk and incubated with antibodies against GSN (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and GAPDH (Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. After washing with PBST, the membranes were incubated with horseradish peroxidase-linked secondary antibody. The proteins were visualized using ECL chemiluminescence and detected by X-ray film. Bands were quantified with ImageJ (National Institutes of Health, Bethesda, MD, USA).
F-actin staining
The F-actins in fixed cells were stained by CytoPainter F-actin Staining kit (Abcam) according to the manufacturer instructions. The cells were rinsed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature followed by permeabilizing with 0.1% Triton X-100 in PBS for 5 min. The cells were rinsed in PBS and 1X red fluorescent phalloidin conjugate working solution was added at room temperature for 45 min.
Statistical analysis
All experiments were repeated at least three times before statistical analysis. All experimental data are shown as the mean ± SEM. Differences between samples were analyzed using the two-tailed Student's t-test. Statistical significance was accepted at P<0.05.
Results
Hepatocellular carcinoma cells release more microvesicles than normal hepatocytes
MVs are one of the key vectors of intercellular communication and transport regulatory molecules from donor cells to recipient cells. Our previous study suggested that hepatocytes might secrete MVs containing miR-483 and transferred the MVs to hepatic stellate cells (9). However, the amount of MVs released by normal hepatocytes and hepatocellular carcinoma cells remains unclear. The presence of exosomes was confirmed by several criteria including i) appearance as 50–90 nm bi-membrane vesicles assessed by electron microscopy ii) complementary biochemical (immunoblotting), mass spectrometry, and imaging techniques to identify exosomal proteins including tetraspanins (CD9, CD63), Alix, TSG101 and ESCRT (12,13).
In our study, we chose CD9 for exosomal identification because CD9 is a well-characterized marker of exosomes and is highly expressed on the surface of exsomes. MVs extracted from the media of HCC cells, HepG2, were visualized by transmission electron microscopy (TEM) (Fig. 1A). The results showed that most MVs from supernatants were membrane particles with a diameter of approximately 50 nm. The size and ultrastructural morphology suggested that the extracted MVs were predominantly exosomes (21). To quantify the MVs released from HCC cells, the number of MVs from different liver cell lines was determined by flow cytometry after labeling with PE-conjugated CD9 antibody (Fig. 1C). The MV levels of HepG2 and Huh7 cells were higher than that of normal liver cell line, HL7702 (Fig. 1D). Considering that MVs derived from cancer cells are capable of transferring oncogenic cargo to recipient cancer cells (3), this result suggested that high level of cellular MV secretion might be associated with the abnormal cell behavior growing out of control in liver cells. Further studies were conducted to determine the mechanism of the de-regulation of MV levels.
Overexpression of miR-200a suppresses the secretion of HCC cell-derived microvesicles
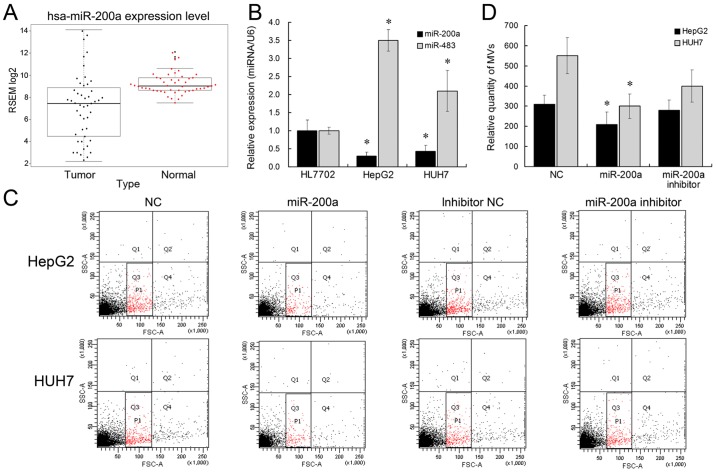
We have previously showed that miR-483 might be transported by MVs to influence adjacent cells (9). Other microRNAs with significantly differential expression in HCC, such as miR-200a, are downregulated in HCC (22). Herein, we aimed to determine whether miR-483 or miR-200a could be involved in the MV biogenesis. We initially analyzed miR-200a expression pattern in TCGA data and found that miR-200a expression was significantly low (Fig. 2A). We subsequently identified the expression levels of miR-483 and miR-200a by qRT-PCR. miR-483 expression was >3-fold higher, and miR-200a expression was 2-fold lower in HepG2 and Huh7 cells, compared with HL7702 (Fig. 2B), which is also in accordance with previous studies (22,23). Further, we tested the effect of ectopic miR-483 or miR-200a expression on liver cell MV secretion. The results showed that miR-200a overexpression was able to inhibit the release of MVs from liver cancer cell lines (Fig. 2C and D). miR-200a inhibitor treatment did not have a significant effect on MV release, which might be caused by the low expression level of endogenous miR-200a in HCC cells. Nevertheless, the number of MVs derived from HL7702 showed no differences between the miR-200 overexpression group and negative control group. This might be due to the de-regulated expression of miR-200a in liver cancer cells. However, miR-483 did not affect the MV release in liver cells. Collectively, these results suggested that miR-200a overexpression could suppress the release of MVs in liver cancer cells.
Figure 2.
miR-200a inhibits the release of MVs from HepG2 and Huh7 cells. (A) hsa-miR-200a expression level of 48 HCC tissues from TCGA database was significantly lower compared with corresponding non-tumor normal tissues. P<0.0001. (B) miR-200a expression level was downregulated in HepG2 and Huh7 cells compared with HL7702 cells, while miR-483 was upregulated. (C) miR-200a suppressed the release of MVs derived from HepG2 and Huh7 cells by flow cytometry. (D) Statistical analysis of MV release. miR-200a inhibited the MV release from HepG2 and Huh7 cells. *P<0.05.
miR-200a inhibits the release of MVs of HCC cells by functionally regulating GSN
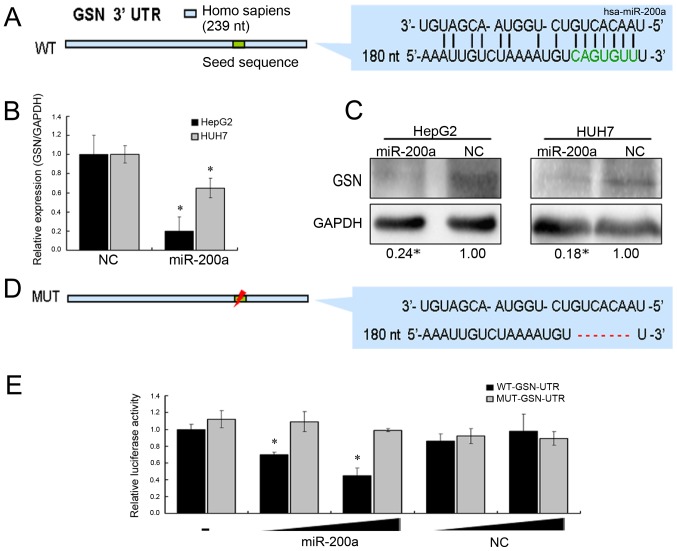
Many studies have shown that miR-200a could regulate the proliferation, migration, and epithelial-mesenchymal transition (EMT) of cancer cells (16,24–26). These reports revealed the functional targets of miR-200a, such as ZEB1/ZEB2, DEK, SPAG9, and EPHA2. To identify the target of miR-200a in the secretion of MVs, we used bioinformatics tools (miRanda and PicTar) to predict the potential target of miR-200a, which might be involved in the stability of cell membrane (Fig. 3A). Among these potential targets, GSN has been reported to play an important role in regulating the assembly and disassembly of actin filament (F-actin). Actin-cytoskeletal rearrangements have been implicated in the release of MVs (4,27,28). Additionally, we found that GSN expression was upregulated in HCC by analyzing TCGA data (Fig. 4A). mRNA and protein levels of GSN were investigated after transfection with miR-200a mimics or negative control sequences by qRT-PCR and western blotting. The results suggested that miR-200a could inhibit the expression of GSN gene (Fig. 3B and C). Next, we proved that miR-200a directly targeted the 3′-UTR of GSN using the dual luciferase reporter assay (Fig. 3D). Further, the lack of GSN induced by miR-200a overexpression resulted in marked stabilization of the polymerized actin networks, as shown by F-actin staining in HCC cells (Fig. 4B). In summary, we assumed that a miR-200a-GSN-F-actin pathway might promote the release of liver cancer cell MVs.
Figure 3.
GSN is the direct target of miR-200a involved in cytoskeleton regulation. (A) The base-pairing interaction of miR-200a seed sequences and GSN, as predicted by bioinformatics analysis. (B) GSN mRNA level measured by quantitative real-time PCR (normalized to GAPDH) and (C) GSN protein level measured by western blotting (normalized to GAPDH) at 48 h after transfection with miR-200a mimic or NC in HepG2/Huh7 cells. (D) Luciferase assay. HEK293T cells were co-transfected with a luciferase construct fused with the wild-type (WT) or mutanted 3′-UTR of GSN and miR-200a mimic or NC. (E) The relative luciferase activity was detected. *P<0.05.
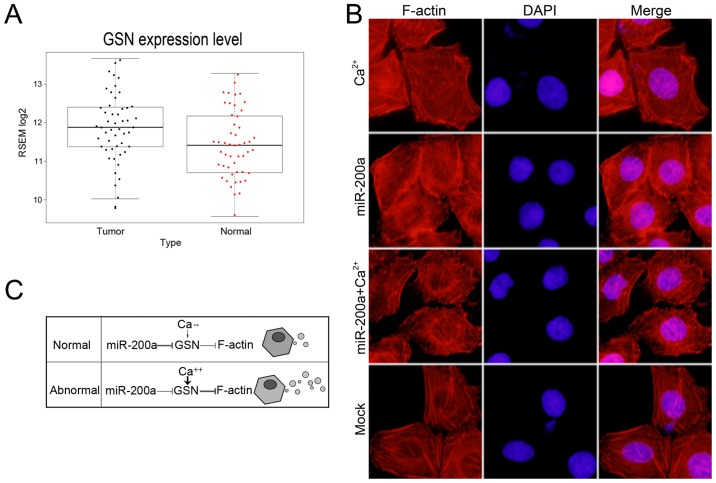
Figure 4.
Function of GSN is regulated by miR-200a and calcium. (A) GSN expression level of 50 HCC tissues from TCGA database was higher compared with corresponding non-tumor normal tissues. P<0.05. (B) Representative images of HepG2 cell F-actin staining. Overexpression of miR-200a resulted in much more polymerized actin filaments. In the presence of Ca2+, the final average length of actin filaments was shorter. Treating cells with miR-200a and Ca2+, more polymerized actin filaments were observed. Magnification, ×400. (C) Roles of miR-200a and Ca2+ in regulating GSN function, in normal cells miR-200a may be the predominant regulator of GSN function and governs the secretion of MVs, while in abnormal cells the expression of miR-200a is inhibited and high level cellular Ca2+ activates GSN and leads to the release of more MVs.
Function of GSN is dually controlled by miR-200a and calcium
In the presence of micromolar Ca2+ concentrations, GSN is activated to bind and sever assembled actin filaments (29). To determine whether GSN could be regulated by both miR-200a and Ca2+, we treated the cells with 1 mM CaCl2 for 30 min to increase the intracellular Ca2+ concentration. We observed more short actin filaments in the presence of Ca2+ (Fig. 4B). However, overexpression of miR-200a resulted in considerably more polymerized and elongated actin filaments. To further confirm whether high level of cellular Ca2+ concentrations could recover the function of GSN in miR-200a mimic-transfected cells. We added 1 mM CaCl2 for 30 min after transfection of miR-200a mimics for 48 h and observed the cytoskeletal change. We observed the stabilization of the polymerized actin networks and more elongated actin filaments, following the ectopic expression of miR-200a and CaCl2 treatment. Collectively, we assumed that in normal cells miR-200a is the predominant regulator of GSN function and governs the secretion of MVs, while in abnormal cells the expression of miR-200a is inhibited and high level cellular Ca2+ activates GSN and leads to the release of more MVs (Fig. 4C). Take together, the results suggested that the function of GSN was controlled by miR-200a and calcium.
Overexpression of miR-200a inhibits the proliferation of adjacent cells
It has been reported that MVs derived from cancer cells can promote the proliferation of other cancer cells and enable normal fibroblasts and epithelial cells to have the transformed characteristics of cancer cells (30). To further explore the role of hypothetic miR-200a-GSN-F-actin pathway in normal liver cells, we performed indirect co-culture experiments in HL7702 cells (Fig. 5A). The HepG2 or Huh7 cells transfected with carboxyfluorescent labeled miR-200a mimics or NC were seeded in the upper transwell inserts, and the HL7702 cells were seeded in the lower 3.5-cm plates. After transfection, we changed the culture medium with medium containing CD-9 in the upper transwell inserts. We observed that CD-9 labeled MVs could be found in the HL7702 cells, which suggested that MVs could be transported from HepG2 or Huh7 cells to HL7702 cells in the lower 3.5-cm plates of indirect co-culture experiments.
Figure 5.
Overexpression of miR-200a inhibits the proliferation of adjacent HL7702 cells. (A) Illustration of indirect co-culture. (B) After the HepG2 and Huh7 cells were transfected with carboxyfluorescent (FAM) labeled miR-200a mimics, the media of HepG2 and Huh7 cells were labeled with CD9. The CD9 (red) and miR-200a mimics (green) were observed in HL7702 cells. (C) The proliferation of HL7702 cells in the lower 3.5-cm plates were measured by MTT. The proliferation of HL7702 cells were inhibited at 36 h (*P=0.048) and 48 h (*P=0.033).
We also observed the existence of fluorescently labelled mir-200a in the HL7702 cells, which suggested that miR-200a may serve as the cargo of MVs and be transported into the HL7702 cells (Fig. 5B). Further, we calculated the percentages of CD9+ HL7702 cells in the lower plates and found that approximately 9% of HL7702 cells were CD9-positive. Later, the cell viability of HL7702 cells in the lower 3.5-cm plates were measured by MTT assay and the proliferation of HL7702 cells cocultured with miR-200a mimic-transfected Huh7 cells was inhibited (Fig. 5C). We assumed that miR-200a might inhibit the release of HCC cell MVs and effect the proliferation of adjacent cells. In summary, MVs could be an important vector for intercellular communication among cancer cells.
Discussion
It is widely recognized that almost all human cancer cells and normal cells are able to generate MVs. However, the amounts of MVs generated by the cells are different. The higher grade and more aggressive cancer cells generate higher number of MVs compared with the lower grade and less aggressive cancer cells (31). However, the cellular mechanism of the increased MV release in cancer cells is not clearly understood.
In this study, we demonstrated that HCCs secreted more MVs than normal hepatocytes by using flow cytometry. We further confirmed that miR-200a was downregulated in HCC cells, which were reported to be correlated with tumor progression, metastasis, and poor prognosis. Furthermore, we found that miR-200a overexpression in HepG2 and Huh7 cells could inhibit the MV secretion possibly by targeting GSN.
Previous studies have demonstrated that peripheral blood MV levels are elevated in HCC patients and tumor-derived exosomes promote tumor progression and tumor cell migration (2,32). Wei et al reported that exosomes derived from HCC cells promoted growth, migration, and invasion of parental cells (33). Our results showed that liver cancer cells could release more MVs and regulate the proliferation of adjacent cells. Considering the fundamental role of MVs in cancer, more researchers have begun to focus on the biogenesis of MVs. Vps4A and Annexin A2 were reported to function in affecting migration and invasion of HCC cells by regulating the secretion of cancer-derived MVs (33,34).
Given the importance of microRNA in HCC progression, we assumed that microRNA might be associated with increased MVs release in cancer cells. We found that miR-200a was able to inhibit the release of MVs in liver cancer cells. To the best of our knowledge, this is the first study to demonstrate that the microRNA miR-200a, can regulate the production of liver cancer-derived MVs. miR-200a overexpression could lead to actin remodeling by suppressing GSN expression and cause dramatic morphological changes, which might be a potential pathway to affect the MV secretion. GSN was first discovered as a Ca2+-dependent actin filament severing and capping protein (8). GSN also plays crucial roles in many different physiological and pathological processes, such as apoptosis, familial amyloidosis of the Finnish type, inflammation, Alzheimer's disease, and even cancer-high expression level of GSN was observed in a subset of non-small cell lung cancers, and a gradual increase of GSN occurred with an increase in the tumor grade (35). It is of great significance to explore the mediators of GSN expression. Besides the activity and promoter regulation of GSN (36), the regulation of GSN expression at post-transcriptional level has not been reported. We first revealed that 3′-UTR of GSN can be targeted by microRNA, miR-200a, and resulted in the decreased expression of GSN and the changes to cell cytoskeleton.
With the formation of MVs, several cargoes are loaded into the MVs, which are released when they arrive at the recipient cells. The function of MVs relies on the cargoes they carry. However, in our study, we did not address whether miR-200a could affect the loaded cargo of cancer cell-derived MVs, which is vital to the understanding of MV function and needs to be further investigated. In addition, we suggested that miR-200a could serve as cargo of MVs and be transferred to recipient cells. Considering the selective packaging of cargo into MVs, whether the packaging of miR-200a into MVs and the release of miR-200a targeting to recipient cells are of specificity needs further investigation. Moreover, it would be of significance to know whether miR-200a-GSN pathway serves as a general mechanism for MV release. Gelsolin overexpression and silencing would provide direct link between gelsolin and MV secretion. In future, further studies will be conducted to determine the mechanism of MV biogenesis, selective cargo loading and MV traffic.
In summary, our results suggested that regulating the secretion of MVs by microRNA-target pathway is feasible for further tumor treatment. Second, we improved the understanding of how GSN expression is regulated. Increased cytosolic Ca2+ could rapidly activate the activity of GSN, while miR-200a can act as a post-transcriptional regulator for GSN. Third, GSN serving as a novel target of miR-200a may provide an additional mechanism explaining the role of miR-200a in EMT. These findings may contribute to a better understanding of MV biogenesis and the identification of new therapeutic targets for cancer.
Acknowledgements
This study was supported by Yu Weihan Academician Fund for distinguished young Scholars of Harbin Medical University, the Postdoctoral Fund of Heilongjiang Province (LBH-Q15108), the National Natural Science Foundation of China (81270511, 81570534, 81611130072) and the Application Technology Research and Development Plan Major Project of Heilongjiang (GA16C105).
References
- 1.Antonyak MA, Wilson KF, Cerione RA. R(h)oads to microvesicles. Small GTPases. 2012;3:219–224. doi: 10.4161/sgtp.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Li H, Zhou Y, Jie S. Peripheral blood microvesicles are potential biomarkers for hepatocellular carcinoma. Cancer Biomark. 2013;13:351–357. doi: 10.3233/CBM-130370. [DOI] [PubMed] [Google Scholar]
- 3.Antonyak MA, Cerione RA. Microvesicles as mediators of intercellular communication in cancer. Methods Mol Biol. 2014;1165:147–173. doi: 10.1007/978-1-4939-0856-1_11. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Antonyak MA, Zhang J, Cerione RA. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31:4740–4749. doi: 10.1038/onc.2011.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner-Britz L, Wang J, Kaestner L, Bernhardt I. Protein kinase Cα and P-type Ca channel CaV2.1 in red blood cell calcium signalling. Cell Physiol Biochem. 2013;31:883–891. doi: 10.1159/000350106. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen DB, Ly TB, Wesseling MC, Hittinger M, Torge A, Devitt A, Perrie Y, Bernhardt I. Characterization of microvesicles released from human red blood cells. Cell Physiol Biochem. 2016;38:1085–1099. doi: 10.1159/000443059. [DOI] [PubMed] [Google Scholar]
- 7.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 8.Yin HL, Stossel TP. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Ma N, Zhao R, Wu G, Zhang Y, Qiao Y, Han D, Xu Y, Xiang Y, Yan B, et al. Overexpression of miR-483-5p/3p cooperate to inhibit mouse liver fibrosis by suppressing the TGF-β stimulated HSCs in transgenic mice. J Cell Mol Med. 2014;18:966–974. doi: 10.1111/jcmm.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Charrier A, Zhou Y, Chen R, Yu B, Agarwal K, Tsukamoto H, Lee LJ, Paulaitis ME, Brigstock DR. Epigenetic regulation of connective tissue growth factor by microRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2014;59:1118–1129. doi: 10.1002/hep.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu Z, Jiang C, Wu J, Ding Y. Exosomes as potent regulators of HCC malignancy and potential bio-tools in clinical application. Int J Clin Exp Med. 2015;8:17088–17095. [PMC free article] [PubMed] [Google Scholar]
- 12.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res Int. 2014;2014:864894. doi: 10.1155/2014/864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 16.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm Vienna. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen TB, Nielsen MH, Handberg A. In vitro incubation of platelets with oxLDL does not induce microvesicle release when measured by sensitive flow cytometry. Front Cardiovasc Med. 2015;2:37. doi: 10.3389/fcvm.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 22.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 23.Ma N, Li F, Li D, Hui Y, Wang X, Qiao Y, Zhang Y, Xiang Y, Zhou J, Zhou L, et al. Igf2-derived intronic miR-483 promotes mouse hepatocellular carcinoma cell proliferation. Mol Cell Biochem. 2012;361:337–343. doi: 10.1007/s11010-011-1121-x. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Wu G, Wu Z, Yao X, Li G. MiR-200a suppresses the proliferation and metastasis in pancreatic ductal adenocarcinoma through downregulation of DEK Gene. Transl Oncol. 2016;9:25–31. doi: 10.1016/j.tranon.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Jiang F, Song H, Li X, Xian J, Gu X. MicroRNA-200a-3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochem Biophys Res Commun. 2016;470:620–626. doi: 10.1016/j.bbrc.2016.01.095. [DOI] [PubMed] [Google Scholar]
- 26.Tsouko E, Wang J, Frigo DE, Aydoğdu E, Williams C. miR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis. 2015;36:1051–1060. doi: 10.1093/carcin/bgv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kholia S, Jorfi S, Thompson PR, Causey CP, Nicholas AP, Inal JM, Lange S. A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. J Extracell Vesicles. 2015;4:26192. doi: 10.3402/jev.v4.26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccin A, Murphy WG, Smith OP. Circulating microparticles: Pathophysiology and clinical implications. Blood Rev. 2007;21:157–171. doi: 10.1016/j.blre.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Yin HL, Hartwig JH, Maruyama K, Stossel TP. Ca2+ control of actin filament length. Effects of macrophage gelsolin on actin polymerization. J Biol Chem. 1981;256:9693–9697. [PubMed] [Google Scholar]
- 30.Du T, Ju G, Wu S, Cheng Z, Cheng J, Zou X, Zhang G, Miao S, Liu G, Zhu Y. Microvesicles derived from human Wharton's jelly mesenchymal stem cells promote human renal cancer cell growth and aggressiveness through induction of hepatocyte growth factor. PLoS One. 2014;9:e96836. doi: 10.1371/journal.pone.0096836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 32.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 33.Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Zhao P, Xu XL, Cai L, Song ZS, Cao DY, Tao KS, Zhou WP, Chen ZN, Dou KF. Annexin A2 promotes the migration and invasion of human hepatocellular carcinoma cells in vitro by regulating the shedding of CD147-harboring microvesicles from tumor cells. PLoS One. 2013;8:e67268. doi: 10.1371/journal.pone.0067268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li GH, Arora PD, Chen Y, McCulloch CA, Liu P. Multifunctional roles of gelsolin in health and diseases. Med Res Rev. 2012;32:999–1025. doi: 10.1002/med.20231. [DOI] [PubMed] [Google Scholar]
- 36.Eun DW, Ahn SH, You JS, Park JW, Lee EK, Lee HN, Kang GM, Lee JC, Choi WS, Seo DW, et al. PKCepsilon is essential for gelsolin expression by histone deacetylase inhibitor apicidin in human cervix cancer cells. Biochem Biophys Res Commun. 2007;354:769–775. doi: 10.1016/j.bbrc.2007.01.046. [DOI] [PubMed] [Google Scholar]