Abstract
Cytochrome P450, a family of monooxygenase enzymes, is organized as a catalytic metabolon, which requires enzymatic partners as well as environmental factors that tune its complex dynamic. P450 and its reducing counterparts—cytochrome P450-reductase and cytochrome b 5—are membrane-bound proteins located in the cytosolic side of the endoplasmic reticulum. They are believed to dynamically associate to form functional complexes. Increasing experimental evidence signifies the role(s) played by both protein-protein and protein-lipid interactions in P450 catalytic function and efficiency. However, the biophysical challenges posed by their membrane-bound nature have severely limited high-resolution understanding of the molecular interfaces of these interactions. In this article, we provide an overview of the current knowledge on cytochrome P450, highlighting the environmental factors that are entwined with its metabolic function. Recent advances in structural biophysics are also discussed, setting up the bases for a new paradigm in the study of this important class of membrane-bound enzymes.
Keywords: cytochrome P450, NADPH-P450 reductase, cytochrome b5, membrane, structure, sold-state NMR
Introduction
Cytochrome P450 (CYP) is a superfamily of heme-containing enzymes responsible for insertion of molecular oxygen into inactivated C-H and C-C bonds. CYPs are ubiquitous in human tissues, being responsible for the metabolism of drug and xenobiotic compounds 1; several CYPs are also involved in the biosynthesis of steroids 2, 3. In humans, 57 genes that code for various CYP enzymes were identified; about 13 isoforms are responsible for the metabolism of more than 80% of the clinically used drugs 1. Human CYPs that typically mediate metabolic clearance of drugs are targeted to the endoplasmic reticulum (ER) by an N-terminal domain that spans the membrane bilayer with the catalytic domain residing in the cytosol and partially embedded in the membrane 1, 4.
For its catalytic function, CYP needs two electrons, provided by its redox partners, cytochrome P450-reductase (CPR) and cytochrome b 5 ( b 5) ( Figure 1). CPR and b 5 are also found in the cytoplasmic side of the ER of eukaryotic cells and also possess a hydrophobic transmembrane binding domain (TM) 5, 6. CPR is a large flavoprotein (approximately 77 kDa), containing FAD and FMN domains, linked by a “hinge” region which provides conformational plasticity. On the other hand, b 5 is a small hemoprotein (15 kDa), which promotes catalysis not only via electron transfer, but also through allosteric regulation 7. A schematic of P450, CPR, and b 5 incorporated in a lipid bilayer is depicted in Figure 2.
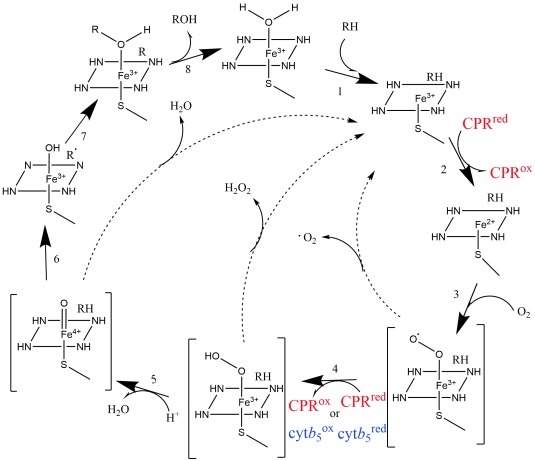
Figure 1. The catalytic cycle of cytochrome P450.
The binding of substrate R-H (1) causes a decrease of the redox potential of about 100 mV, which allows the first electron transfer from cytochrome P450-reductase (CPR) (2). The reduction of Fe 3+ to Fe 2+ makes suitable O 2 binding (3), which now can accept a second electron from either CPR or cytochrome b 5 (cyt b 5) (4), to form a hydroperoxyl intermediate known as compound 0. The O 2 −2 complex reacts with surrounding protons to form the highly reactive oxyferryl intermediate, also known as compound I (5). The Fe-ligated O atom (6) is transferred to the substrate forming a hydroxylated form of the substrate (7). The product is finally released (8), replaced by a molecule of water. Three uncoupling reactions are shown as dashed lines, with the respective products: the autoxidation shunt O 2 −2, the peroxide shunt (H 2O 2), and the oxidase shunt (H 2O).
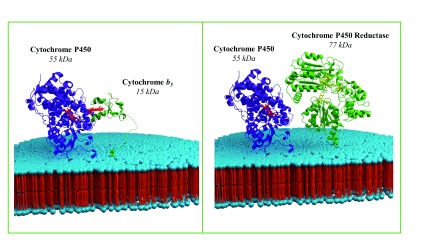
Figure 2. Model structures of cytochrome P450 with cytochrome b 5 (left) and cytochrome P450 with cytochrome P450 NADPH-reductase (right) in a lipid bilayer.
The models were constructed from the crystallographic or solution-state nuclear magnetic resonance (NMR) structures of the soluble domains of the proteins. The structure, dynamics, and topology of the invisible transmembrane domains of the full-length cytochrome proteins have been determined for the first time by static solid-state NMR experiments on magnetically aligned bicelles. These studies reported that the helical structures of the transmembrane domains are significantly tilted away from the lipid bilayer normal and undergo motion in the millisecond (or slower) timescale that is far slower than that of the residues in the soluble domain 17, 18, 45.
The interactions between CYPs and the redox counterparts as well as their catalytic activities—including substrate binding and metabolite release—occur in the membrane landscape. The biphasic amphiphilic environment sets up the molecular requirements for cytochrome P450 metabolon, as observed since the dawn of P450 research 8, 9. Since then, efforts have been made to reconstitute CYP’s catalytic activity in membrane mimic systems, from simple binary 9 or ternary 10 lipid mixtures to the more sophisticated nanodiscs 11, 12, which consist of a lipid patch surrounded by a polypeptide or polymeric chain. More recently, P450 activity has been reconstituted in biomimetic of the ER to study protein-lipid interaction at the single-molecule level 13.
Paradoxically, if much is known about the effects of proteins and lipids turning on CYP function from an ensemble perspective, the molecular details of these protein-protein and protein-lipid interactions are mostly unexplored 14. The intrinsic properties of the cell membrane and the innumerous interactions among molecules—amphipathic lipids, polysaccharides, cholesterol, proteins, and water—provide paramount challenges for biophysics 15. Traditionally, several structures of truncated versions of CYPs have been elucidated at high resolution by x-ray crystallography; CPR and b 5 have also been resolved 5, 16. In all of these structures, the membrane binding domain has been cleaved to allow complete solubility and subsequent crystallization; the lipids were not present in these structures. As a result, the published structures are devoid of any information pertaining to the molecular organization of these enzymes in the membrane. Nuclear magnetic resonance (NMR) spectroscopy has played a pivotal role in the structure determination of biomacromolecules, providing scientists with detailed structural and dynamical information that is inaccessible through other biophysical means 15. In this respect, NMR spectroscopy is both an alternative and a complement to x-ray crystallography. In the last decade, both solution- and solid-state NMR spectroscopy have offered new and detailed insights into the structure, dynamics, and topology of cytochrome b 5 in lipid bilayers and its interface with P450 ( Figure 3) 6, 17– 19.
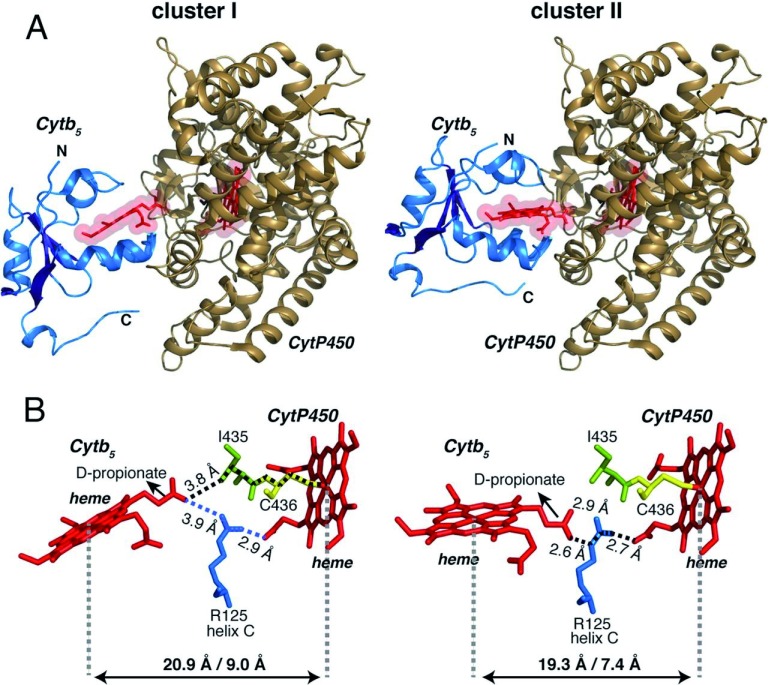
Figure 3. First high-resolution structure of the membrane-bound cytochromes-b5-P450 complex revealing electron transfer pathway.
( A) Three-dimensional structure of the membrane-bound rabbit cytochrome P4502B4–cytochrome b 5 complex reproduced from Ahuja et al. 19. It was obtained by using the high-resolution (solution- and solid-state) nuclear magnetic resonance (NMR) structure of membrane-bound rabbit cytochrome b 5 (PDB code is 2M33), crystal structure of the soluble domain of cytochrome P4502B4 (PDB code is 1SUO), and NMR and mutational constraints to obtain the structure of the membrane-bound complex. ( B) Electron transfer pathway revealed by HARLEM 63 in the complex structure. PDB, Protein Data Bank.
This short review will discuss several catalytic aspects of cytochrome P450 machinery that are intrinsically linked to their membrane-bound nature. We will underline novel mechanistic insights that structural NMR and other biophysical tools have provided, stressing the need for a new paradigm in the structural studies of this important class of membrane-bound enzymes.
Membrane and membrane-anchor domains modulate P450 metabolism and its catalytic efficiency
CYP-mediated oxidation of drugs and xenobiotic in the ER is made possible by the sequential donation of two electrons by CPR and b 5 20. The electron transfer is believed to occur through the dynamic interactions between these proteins. For the electron to be shuttled, it needs to overcome a redox potential gradient. As depicted in Figure 1, b 5 is unable to donate the first electron since it cannot overcome the redox potential barrier. The composition of phospholipids can modulate the midpoint potentials for both CYPs and CPR, as demonstrated by Das and Sligar using nanodiscs 21. When CPR was incorporated in nanodiscs, the redox potentials of both the FAD and FMN domains were shifted to more positive values, compared with CPR lacking the TM domain. Also, the presence of anionic phospholipids made the redox potential suitable for electron transfer from CPR to P450 21, a result that confirmed kinetics observations made by other groups 22. The catalytic activity and membrane insertion of CYP3A4, which metabolize more than 50% of commercial drugs 23, increased as a function of anionic phospholipid concentration 22. It is recognized that membrane composition and polarity can modify both V max and K M 22, 24, which are respectively a measure of catalytic rate and binding affinity under the steady-state assumption of the Michaelis-Menten equation. Phospholipid composition has also been related to protein folding and stability, as reported by Jang et al. for full-length and TM-truncated CYP1B1 24, as well as CYP3A4 25. In regard to the putative role of b 5 as electron donor and allosteric effector for CYP-mediated drug clearance, it is well known that it is isoform-dependent. Allosteric modulations have been observed for CYP3A4 hydroxylation of testosterone and nifedipine 26 but not for CYP1A1 and CYP2D6 1′-hydroxylation of bufuralol 27. Deletion of amino acids of the TM α-helix of rabbit b 5 decreased the binding affinity for CYP2B4 by several folds, as well as the catalytic turnover 28.
The efficiency of cytochrome P450 catalysis is measured in terms of “coupling”, meaning the amount of transferred electron that is committed to the monooxygenase activity versus the extent of the “uncoupled” (or unproductive) pathways. Three unproductive pathways exist ( Figure 1): (a) release of superoxide from the ferrous dioxygen, (b) release of hydrogen peroxide, and (c) the 4e − reduction of heme to produce water 20. Recent studies have postulated that both TM domains and the nature of the membrane may have a role in the coupling efficiency. McDougle et al. 29 found a correlation between the coupling efficiencies of CYP2J2 with the length of the TM domain of CPR: the more extensive the deletion of the TM helix, the lesser the coupling efficiencies. Similar results have been reported for CYP2C19 coupling with several substrates: full-length versus truncated CPR, as well as the presence or absence (or both) of lipids, was related to the overall catalytic efficiency 30. Grinkova et al. 31 observed an increase of coupling when CYP3A4 was incorporated in nanodiscs containing a higher amount of anionic phospholipids, which the authors attributed to the changes in the redox potential of CYP3A4 and reductase. On the P450 side, it is believed that the TM domain of CYPs serves as an “anchor” to the membrane, playing a pivotal role in protein orientation. In CYP1A2 and CYP2D6, the hydrophobicity of the TM domain has been shown to regulate the efficiency of interaction of CYPs with CPR or phospholipids or both and therefore their catalytic activities 32– 35. Similar results have been reported for CYP1B1, and decreased catalytic activity was observed when the TM domain was partially cleaved 24.
Specific interactions between the P450 metabolon and the membrane are believed to play a major role in this catalytic modulation. Traditional x-ray crystallographic studies on P450-CPR and P450- b 5 complexes provided snapshots of their quaternary organization and have been accompanied by specific amino acid mutations able to disrupt/enhance functionality 4, 36– 38. Recently, solution NMR experiments have complemented those initial efforts, providing realistic insights of the complex interface as well as of ligand-induced modifications 39. Other studies have explored the potential of this technique for studying protein complexes between the steroidogenic CYP17A1 and both b 5 and the FMN domain of CPR in the absence of membrane 3, 14. However, complete solubilization and subsequent crystallization of protein-protein complexes have been obtained only by removing the 60-residue segment containing the hydrophobic transmembrane domain of the proteins. Since membrane was not present in these structure studies of protein-protein complexes, the reported results do not contain any information about the structure and topology of the transmembrane domains of these proteins.
Our lab is the first in the field to have used both solution- and solid-state NMR (ssNMR) techniques in studying P450 and related proteins 6 in a functional amphiphilic bilayer, relying on the capacity of both bicelles 15, 40 and nanodiscs 41 to incorporate and preserve the functionality of these protein. The high-resolution structure of the membrane-bound cytochrome-P450- b 5 complex is shown in Figure 3 19. Solid-state NMR experiments on magnetically aligned bicelles revealed the presence of transmembrane helix and its topology in CYP2B4, CPR, and b 5 and further revealed the significant difference in the timescales of motion of residues in the soluble linker and TM domains of b 5 that is crucial in the formation of the productive protein-protein complex 17, 42. Studying isotopically labeled b 5 in bicelles by ssNMR has shown how the mobility of the TM domain of b 5 is significantly reduced by the presence of CYP2B4 without altering its geometry and helical structure 17. In addition, ssNMR experiments on aligned bicelles revealed the direct interactions between the TM domains of CYP2B4 and b 5, in which the “leucine zipper” in the TM domain of b 5 plays a very important role. More importantly, NMR confirmed the electrostatic nature of the b 5-CYP2B4 interactions and that the presence of substrate promotes specific interactions between the two proteins 43. CYP2B4- b 5 complex was more recently studied in nanodiscs, in a detergent-free environment that better simulates the native membrane 41, obtaining high-resolution structural interactions between the proteins. In addition, interactions between membrane-bound CYP2B4, b 5, and CPR have been measured with and without the presence of ligands by using high-resolution NMR experiments 44. The FMN-binding domain (FBD) of CPR has also been studied in bicelles in solid-state NMR experiments 45. Our group was also able to characterize for the first time the interplay among proteins in a tertiary FBD-P450- b 5 complex by NMR spectroscopy in lipid bilayers 44. Notably, we showed that substrate binding to P450 can disrupt P450-CPR complex, facilitating the association of b 5, and that the extent of this equilibrium shift is highly dependent on the substrate. This result can potentially explain the observed difference in the catalytic modulation provided by b 5.
In addition, the structural interaction of FBD with cytochrome c—a small hemoprotein essential for the electron transport chain in mitochondria—has been reported by using cytochrome c as an efficient model system to test the electron transfer process 46. Solution NMR titration experiments showed the formation of a dynamic complex between the two proteins, on a fast exchange timescale. NMR restraints were implemented in molecular docking to generate structural models and map the binding interface on the FBD. The proposed structural model of the FBD–cytochrome c complex suggested potential electron pathways that provide strong electronic coupling between the redox centers 46. More high-resolution structure-based functional and dynamical studies on the binary and ternary P450-redox complexes and their interactions with drugs are in progress in our lab.
Access path channels, substrate availability, and membrane partitioning
As from the above discussion, the membrane and the membrane domains can play a substantial role in P450 catalysis, modulating both electron transfer between the redox couple(s), as well as the overall efficiency of the catalysis. The membrane is also a thick interface between the surroundings (that is, cytosol) and CYPs. Indeed, it has been shown that the membrane can slow down the access of water as well as substrate to the active site 47. Also, lipophilic compounds that are poorly soluble are predominantly partitioning in the membrane, allowing P450 recruiting of hydrophobic substrates directly from the lipid phase 21, 48, 49. For example, Murtazina et al. 50 demonstrated that CYP27A1 affinities for 5α-cholestane-3α,7α,12α-triol and cholesterol were decreased in the presence of the negatively charged phosphatidylglycerol, which can potentially reflect either a different partitioning of these steroidal molecules or an easier access to substrate due to protein/membrane interactions. Differences in the binding affinities and spin equilibrium in soluble versus membrane-anchored P450 have already been reported 25, 49, 51– 53, and their significance is relevant given how crucial the affinity parameters are for pharmacokinetic/pharmacodynamic models 54. In this regard, Denisov et al. 49 have demonstrated the presence of an allosteric site at the membrane interface of CYP3A4, emphasizing how crucial the presence of membrane is for the evaluation of drug-drug interactions in pharmacological studies. In addition, recent studies from Atkins’s group have provided experimental insights into substrate modulation of CYP3A4 when in nanodiscs 25, 53. The embedded portion of the protein can dynamically interact with the membrane, allowing the opening of substrate channels and a water “aqueduct” 25. These properties—along with substrate equilibrium binding—seem also to be modulated by membrane fluidity 53.
It is not completely understood how lipids can perturb P450 spin equilibrium 48, but the observed differences prove the intrinsic kinetic peculiarities of each system used to test chemicals. Molecular dynamic (MD) simulations and H/D exchange studies on CYP3A4 have shown that the interaction with the membrane occurs through specific lipid-protein interactions 25, 55 and is able to affect the opening/closing of the access tunnel 48, confirming experimental evidence obtained two decades before 56. Computational MD studies have also elucidated how phospholipids can induce an opening of membrane-facing tunnels in CYP1A2 57. The speculated mechanism relies on the ability of the upper part of the TM helix to interact with a proline-rich segment of the catalytic domain that along with the FG loop is immersed in the membrane 57. This gave rise to several access channels from both solvent and membrane-facing tunnels. Similar studies conducted on CYP2C9 revealed the opening of additional access tunnels in the presence of membrane 58, 59. MD simulation performed by Fishelovitch et al. 60 has shown that the FMN binding domain of CPR regulates the water channel of CYP3A4 because of an overlapping of specific residues. When the FMN domain of CPR binds to CYP3A4, the water channel fully opened up, thereby allowing a flow of water molecules into the active site. However, the absence of membrane in this computational study raises the question of whether this conformational flexibility also occurs in the presence of membrane.
Though experimental demonstrations are essential, the computational studies have provided useful information, as well as quantitative thermodynamic parameters, that may potentially explain several observations in P450 kinetics. Nevertheless, given the importance of substrate availability and disposition for drug-metabolizing enzymes, there is a lack of compelling experimental evidence that can fulfill the requirement of detailed molecular descriptions. If structural x-ray crystallography has laid the groundwork for determining enzyme channels and quantifying active site volumes 4, it is still unable to look beyond the polypeptide architecture. On the other hand, recent progress in the use of solution- and solid-state NMR techniques is opening the way to a broader inspection of membrane-associated phenomena. Particularly, the high-resolution dynamics of membrane-bound P450 in the presence and absence of ligands to be measured from NMR will be highly valuable in fully understanding the function of P450.
Current challenges and future developments
Membrane protein structure determination is in general an extremely challenging task because of the lack of stability of the protein outside its native environment. In cytochrome P450, the presence of interactions with the reducing counterparts, as well as the pivotal role of membrane in catalysis, adds fascinating complexities to the ongoing efforts. Given the limitations of the x-ray structural characterization discussed above, NMR—both solution- and solid-state approaches—is an excellent alternative for the structural and dynamic studies on cytochrome P450. As demonstrated for b 5 and its interactions with CYP2B4, NMR offers well-tested and sophisticated tools to probe MDs over a wide range of timescales. Through NMR, motions from nanosecond to millisecond timescales can be probed. Nevertheless, the overall size of the membrane-protein complex is still the main drawback in terms of spectral resolution; the reduced molecular mobility of proteins embedded in a membrane environment also contributes to poor sensitivity and resolution for traditional solution NMR experiments 6. For cytochrome P450, a further complication dwells in the paramagnetic nature of the iron in the heme prosthetic group, which quenches NMR signals in the proximity of the active site. In the coming years, NMR will be able to overcome several of its intrinsic limitations by the introduction of labeling strategies, paramagnetic relaxation enhancement effects, ultrafast magic-angle spinning, and sophisticated pulse sequences. Also, reconstitution of P450 and its redox partners now can be performed in more reliable and versatile model systems, such as nanodiscs that are used for solution NMR studies and macrodiscs for solid-state NMR studies.
Many scientific questions regarding the P450 metabolon require answers because of the innumerable biological and pharmacological implications. The ongoing structural studies would offer valid tools and provide intimate insights into biological interfaces. Nevertheless, the authors consider that the combined use of complementary biophysical techniques and biochemical approaches would create unique avenues to successfully overcome the challenges in fully understanding the enzymatic function of a variety of P450s. The acquired knowledge derived from these studies will set an experimental platform to investigate other electron-transfer protein complexes, such as the mitochondrial electron transport chain 61 and the nitric oxide synthase complex 62, which share several biochemical similarities and complexities with the P450 metabolon.
Acknowledgments
We thank Dr. Thirupathi Ravula from our research group for preparing the membrane protein-protein complex structure presented in Figure 2. The authors thank all the past and present members of the Cytochrome subgroup of the Ramamoorthy lab for their contributions.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Gianluigi Veglia, Department of Chemistry, University of Minnesota, Minneapolis, MN, USA
Yechiel Shai, Weizmann Institute for Science, Rehovot, Israel
Giuseppe Melacini, Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, ON, Canada; Department of Chemistry and Chemical Biology, McMaster University, Hamilton, ON, Canada
Stephen Sligar, Department of Biochemistry, School of Molecular and Cellular Biology, University of Illinois, Urbana, Illinois, USA
Ilia Denisov, Department of Biochemistry, School of Molecular and Cellular Biology, University of Illinois, Urbana, Illinois, USA
Funding Statement
This study was supported by funds from the National Institutes of Health (GM084018 and GM095640 to AR). Ayyalusamy Ramamoorthy would also like to acknowledge NIH funding support for the purchase of a 600 MHz solid-state NMR spectrometer and to upgrade a 400 MHz solid-state NMR spectrometer.
[version 1; referees: 4 approved]
References
- 1. De Montellano PR: Cytochrome P450: structure, mechanism, and biochemistry.Springer,2005. 10.1007/b139087 [DOI] [Google Scholar]
- 2. Bernhardt R: Mammalian and Bacterial Cytochromes P450 Involved in Steroid Hydroxylation: Regulation of Catalysis and Selectivity, and Potential Applications. Fifty Years of Cytochrome P450 Research. 2014;135–151. 10.1007/978-4-431-54992-5_8 [DOI] [Google Scholar]
- 3. Estrada DF, Laurence JS, Scott EE: Cytochrome P450 17A1 Interactions with the FMN Domain of Its Reductase as Characterized by NMR. J Biol Chem. 2016;291(8):3990–4003. 10.1074/jbc.M115.677294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson EF, Stout CD: Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem. 2013;288(24):17082–90. 10.1074/jbc.R113.452805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M, Roberts DL, Paschke R, et al. : Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94(16):8411–6. 10.1073/pnas.94.16.8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Durr UH, Waskell L, Ramamoorthy A: The cytochromes P450 and b 5 and their reductases--promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim Biophys Acta. 2007;1768(12):3235–59. 10.1016/j.bbamem.2007.08.007 [DOI] [PubMed] [Google Scholar]
- 7. Porter TD: The roles of cytochrome b 5 in cytochrome P450 reactions. J Biochem Mol Toxicol. 2002;16(6):311–6. 10.1002/jbt.10052 [DOI] [PubMed] [Google Scholar]
- 8. Coon MJ, Autor AP, Strobel HW: Role of phospholipid in electron transfer in a reconstituted liver microsomal enzyme system containing cytochrome P-450. Chem Biol Interact. 1971;3(4):248–50. 10.1016/0009-2797(71)90046-9 [DOI] [PubMed] [Google Scholar]
- 9. Ingelman-Sundberg M, Glaumann H: Reconstitution of the liver microsomal hydroxylase system into liposomes. FEBS Lett. 1977;78(1):72–6. 10.1016/0014-5793(77)80275-5 [DOI] [PubMed] [Google Scholar]
- 10. Shaw PM, Hosea NA, Thompson DV, et al. : Reconstitution premixes for assays using purified recombinant human cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b 5. Arch Biochem Biophys. 1997;348(1):107–15. 10.1006/abbi.1997.0378 [DOI] [PubMed] [Google Scholar]
- 11. Denisov IG, Sligar SG: Cytochromes P450 in nanodiscs. Biochim Biophys Acta. 2011;1814(1):223–9. 10.1016/j.bbapap.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denisov IG, Sligar SG: Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev. 2017;117(6):4669–713. 10.1021/acs.chemrev.6b00690 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Barnaba C, Martinez MJ, Taylor E, et al. : Single-Protein Tracking Reveals that NADPH Mediates the Insertion of Cytochrome P450 Reductase into a Biomimetic of the Endoplasmic Reticulum. J Am Chem Soc. 2017;139(15):5420–5430. 10.1021/jacs.7b00663 [DOI] [PubMed] [Google Scholar]
- 14. Scott EE, Wolf CR, Otyepka M, et al. : The Role of Protein-Protein and Protein-Membrane Interactions on P450 Function. Drug Metab Dispos. 2016;44(4):576–90. 10.1124/dmd.115.068569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durr UH, Gildenberg M, Ramamoorthy A: The magic of bicelles lights up membrane protein structure. Chem Rev. 2012;112(11):6054–74. 10.1021/cr300061w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Q, Modi S, Smith G, et al. : Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution. Protein Sci. 1999;8(2):298–306. 10.1110/ps.8.2.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamamoto K, Dürr UH, Xu J, et al. : Dynamic interaction between membrane-bound full-length cytochrome P450 and cytochrome b 5 observed by solid-state NMR spectroscopy. Sci Rep. 2013;3:2538. 10.1038/srep02538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamamoto K, Gildenberg M, Ahuja S, et al. : Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci Rep. 2013;3:2556. 10.1038/srep02556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahuja S, Jahr N, Im SC, et al. : A model of the membrane-bound cytochrome b 5-cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem. 2013;288(30):22080–95. 10.1074/jbc.M112.448225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Denisov IG, Makris TM, Sligar SG, et al. : Structure and chemistry of cytochrome P450. Chem Rev. 2005;105(6):2253–77. 10.1021/cr0307143 [DOI] [PubMed] [Google Scholar]
- 21. Das A, Sligar SG: Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer. Biochemistry. 2009;48(51):12104–12. 10.1021/bi9011435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim K, Ahn T, Yun C: Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome P450 3A4. Biochemistry. 2003;42(51):15377–87. 10.1021/bi035280k [DOI] [PubMed] [Google Scholar]
- 23. Guengerich FP: Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. 10.1146/annurev.pharmtox.39.1.1 [DOI] [PubMed] [Google Scholar]
- 24. Jang HH, Kim DH, Ahn T, et al. : Functional and conformational modulation of human cytochrome P450 1B1 by anionic phospholipids. Arch Biochem Biophys. 2010;493(2):143–50. 10.1016/j.abb.2009.10.012 [DOI] [PubMed] [Google Scholar]
- 25. Treuheit NA, Redhair M, Kwon H, et al. : Membrane Interactions, Ligand-Dependent Dynamics, and Stability of Cytochrome P4503A4 in Lipid Nanodiscs. Biochemistry. 2016;55(7):1058–69. 10.1021/acs.biochem.5b01313 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Yamazaki H, Johnson WW, Ueng YF, et al. : Lack of electron transfer from cytochrome b 5 in stimulation of catalytic activities of cytochrome P450 3A4. Characterization of a reconstituted cytochrome P450 3A4/NADPH-cytochrome P450 reductase system and studies with apo-cytochrome b 5. J Biol Chem. 1996;271(44):27438–44. 10.1074/jbc.271.44.27438 [DOI] [PubMed] [Google Scholar]
- 27. Yamazaki H, Nakamura M, Komatsu T, et al. : Roles of NADPH-P450 reductase and apo- and holo-cytochrome b 5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr Purif. 2002;24(3):329–37. 10.1006/prep.2001.1578 [DOI] [PubMed] [Google Scholar]
- 28. Clarke TA, Im S, Bidwai A, et al. : The role of the length and sequence of the linker domain of cytochrome b 5 in stimulating cytochrome P450 2B4 catalysis. J Biol Chem. 2004;279(35):36809–18. 10.1074/jbc.M406055200 [DOI] [PubMed] [Google Scholar]
- 29. McDougle DR, Palaria A, Magnetta E, et al. : Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Sci. 2013;22(7):964–79. 10.1002/pro.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Miyamoto M, Yamashita T, Yasuhara Y, et al. : Membrane anchor of cytochrome P450 reductase suppresses the uncoupling of cytochrome P450. Chem Pharm Bull (Tokyo). 2015;63(4):286–94. 10.1248/cpb.c15-00034 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Grinkova YV, Denisov IG, McLean MA, et al. : Oxidase uncoupling in heme monooxygenases: human cytochrome P450 CYP3A4 in Nanodiscs. Biochem Biophys Res Commun. 2013;430(4):1223–7. 10.1016/j.bbrc.2012.12.072 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Kim HJ, Lee SB, Guengerich FP, et al. : Effects of N-terminal modification of recombinant human cytochrome P450 1A2 on catalytic activity. Xenobiotica. 2007;37(4):356–65. 10.1080/00498250601178189 [DOI] [PubMed] [Google Scholar]
- 33. Larson JR, Coon MJ, Porter TD: Purification and properties of a shortened form of cytochrome P-450 2E1: deletion of the NH2-terminal membrane-insertion signal peptide does not alter the catalytic activities. Proc Natl Acad Sci U S A. 1991;88(20):9141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong MS, Bell LC, Guo Z, et al. : Identification of retained N-formylmethionine in bacterial recombinant mammalian cytochrome P450 proteins with the N-terminal sequence MALLLAVFL…: roles of residues 3–5 in retention and membrane topology. Biochemistry. 1996;35(31):10031–40. 10.1021/bi960873z [DOI] [PubMed] [Google Scholar]
- 35. Hanna IH, Kim MS, Guengerich FP: Heterologous expression of cytochrome P450 2D6 mutants, electron transfer, and catalysis of bufuralol hydroxylation: the role of aspartate 301 in structural integrity. Arch Biochem Biophys. 2001;393(2):255–61. 10.1006/abbi.2001.2510 [DOI] [PubMed] [Google Scholar]
- 36. Bridges A, Gruenke L, Chang YT, et al. : Identification of the binding site on cytochrome P450 2B4 for cytochrome b 5 and cytochrome P450 reductase. J Biol Chem. 1998;273(27):17036–49. 10.1074/jbc.273.27.17036 [DOI] [PubMed] [Google Scholar]
- 37. Sevrioukova IF, Li H, Zhang H, et al. : Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci U S A. 1999;96(5):1863–8. 10.1073/pnas.96.5.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Im S, Waskell L: The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b 5. Arch Biochem Biophys. 2011;507(1):144–53. 10.1016/j.abb.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colthart AM, Tietz DR, Ni Y, et al. : Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Sci Rep. 2016;6:22035. 10.1038/srep22035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Xu J, Dürr UH, Im S, et al. : Bicelle-enabled structural studies on a membrane-associated cytochrome B 5 by solid-state MAS NMR spectroscopy. Angew Chem Int Ed Engl. 2008;47(41):7864–7. 10.1002/anie.200801338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang M, Huang R, Ackermann R, et al. : Reconstitution of the Cyt b 5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angew Chem Int Ed Engl. 2016;55(14):4497–9. 10.1002/anie.201600073 [DOI] [PubMed] [Google Scholar]
- 42. Soong R, Smith PE, Xu J, et al. : Proton-evolved local-field solid-state NMR studies of cytochrome b 5 embedded in bicelles, revealing both structural and dynamical information. J Am Chem Soc. 2010;132(16):5779–88. 10.1021/ja910807e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang M, Le Clair SV, Huang R, et al. : Insights into the role of substrates on the interaction between cytochrome b 5 and cytochrome P450 2B4 by NMR. Sci Rep. 2015;5:8392. 10.1038/srep08392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang M, et al. : Incorporation of the cyt b 5-cyt P450 complex in nanodiscs characterized by solution NMR Ph.D. thesis, University of Michigan,2016. [Google Scholar]
- 45. Huang R, Yamamoto K, Zhang M, et al. : Probing the transmembrane structure and dynamics of microsomal NADPH-cytochrome P450 oxidoreductase by solid-state NMR. Biophys J. 2014;106(10):2126–33. 10.1016/j.bpj.2014.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang R: Structural Study of Interaction between the FMN Binding Domain of Cytochrome P450 Reductase and Its Redox Partners-Cytochrome P450/Cytochrome c by NMR and NMR Characterization of Monomeric and Oligomeric Conformations of Human Calcitonin and Its Interaction with EGCG Ph.D. thesis, University of Michigan,2014. Reference Source [Google Scholar]
- 47. Cojocaru V, Winn PJ, Wade RC: The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770(3):390–401. 10.1016/j.bbagen.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 48. Denisov IG, Shih AY, Sligar SG: Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem. 2012;108:150–8. 10.1016/j.jinorgbio.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Denisov IG, Grinkova YV, Baylon JL, et al. : Mechanism of drug-drug interactions mediated by human cytochrome P450 CYP3A4 monomer. Biochemistry. 2015;54(13):2227–39. 10.1021/acs.biochem.5b00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murtazina DA, Andersson U, Hahn I, et al. : Phospholipids modify substrate binding and enzyme activity of human cytochrome P450 27A1. J Lipid Res. 2004;45(12):2345–53. 10.1194/jlr.M400300-JLR200 [DOI] [PubMed] [Google Scholar]
- 51. Kiselev PA, Garda G, Finch SA, et al. : [Regulation of the catalytic activity of the monooxygenase enzyme system depending of the substrate structure and phospholipid composition of the model membrane]. Biokhimiia. 1990;55(11):2058–71. [PubMed] [Google Scholar]
- 52. Barnaba C, Humphreys SC, Barden AO, et al. : Substrate Dependent Native Luminescence from Cytochromes P450 3A4, 2C9, and P450cam. J Phys Chem B. 2016;120(12):3038–47. 10.1021/acs.jpcb.5b11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McClary WD, Sumida JP, Scian M, et al. : Membrane Fluidity Modulates Thermal Stability and Ligand Binding of Cytochrome P4503A4 in Lipid Nanodiscs. Biochemistry. 2016;55(45):6258–68. 10.1021/acs.biochem.6b00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nagar S, Korzekwa K: Commentary: nonspecific protein binding versus membrane partitioning: it is not just semantics. Drug Metab Dispos. 2012;40(9):1649–52. 10.1124/dmd.112.046599 [DOI] [PubMed] [Google Scholar]
- 55. Baylon JL, Lenov IL, Sligar SG, et al. : Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J Am Chem Soc. 2013;135(23):8542–51. 10.1021/ja4003525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ingelman-Sundberg M, Hagbjörk AL, Ueng YF, et al. : High rates of substrate hydroxylation by human cytochrome P450 3A4 in reconstituted membranous vesicles: influence of membrane charge. Biochem Biophys Res Commun. 1996;221(2):318–22. 10.1006/bbrc.1996.0593 [DOI] [PubMed] [Google Scholar]
- 57. Jeřábek P, Florián J, Martínek V: Lipid molecules can induce an opening of membrane-facing tunnels in cytochrome P450 1A2. Phys Chem Chem Phys. 2016;18(44):30344–56. 10.1039/c6cp03692a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cojocaru V, Balali-Mood K, Sansom MS, et al. : Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput Biol. 2011;7(8):e1002152. 10.1371/journal.pcbi.1002152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berka K, Hendrychová T, Anzenbacher P, et al. : Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A. 2011;115(41):11248–55. 10.1021/jp204488j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fishelovitch D, Shaik S, Wolfson HJ, et al. : How does the reductase help to regulate the catalytic cycle of cytochrome P450 3A4 using the conserved water channel? J Phys Chem B. 2010;114(17):5964–70. 10.1021/jp101894k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lapuente-Brun E, Moreno-Loshuertos R, Acín-Pérez R, et al. : Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–70. 10.1126/science.1230381 [DOI] [PubMed] [Google Scholar]
- 62. Kone BC, Kuncewicz T, Zhang W, et al. : Protein interactions with nitric oxide synthases: controlling the right time, the right place, and the right amount of nitric oxide. Am J Physiol Renal Physiol. 2003;285(2):F178–90. 10.1152/ajprenal.00048.2003 [DOI] [PubMed] [Google Scholar]
- 63. Kurnikov, I: HARLEM molecular modeling package.Pittsburgh, PA: Department of Chemistry, University of Pittsburgh,2000. [Google Scholar]