Abstract
Mushrooms are the most conspicuous fungal structures. Transcription factors (TFs) Bri1 and Hom1 of the model fungus Schizophyllum commune are involved in late stages of mushroom development, while Wc-2, Hom2, and Fst4 function early in development. Here, it is shown that Bri1 and Hom1 also stimulate vegetative growth, while biomass formation is repressed by Wc-2, Hom2, and Fst4. The Δbri1Δbri1 and the Δhom1Δhom1 strains formed up to 0.6 fold less biomass when compared to wild-type, while Δwc-2Δwc-2, Δhom2Δhom2, and Δfst4Δfst4 strains formed up to 2.8 fold more biomass. Inactivation of TF gene tea1, which was downregulated in the Δwc-2Δwc-2, Δhom2Δhom2, and Δfst4Δfst4 strains, resulted in a strain that was severely affected in mushroom development and that produced 1.3 fold more biomass than the wild-type. In contrast, introducing a constitutive active version of hom2 that had 4 predicted phosphorylation motifs eliminated resulted in radial growth inhibition and prompt fructification in both Δhom2 and wild-type strains, even in sterile monokaryons. Together, it is concluded that TFs involved in mushroom formation also modulate vegetative growth. Among these TFs is the homeodomain protein Hom2, being the first time that this class of regulatory proteins is implicated in repression of vegetative growth in a eukaryote.
Introduction
Mushroom forming fungi degrade plant waste, establish mutual beneficial interactions with plants (mycorrhiza), and can be pathogens. Moreover, they are an important food source and produce bioactive components. Mushrooms are the most conspicuous fungal structures. Their formation involves a complex developmental program that has been studied in the model systems Coprinopsis cinerea and Schizophyllum commune 1, 2. Germination of a basidiospore of S. commune results in a monokaryotic mycelium that can fuse with another monokaryon with compatible mating types. Blue light is required to initiate fruiting in the resulting dikaryotic mycelium3, whereas high CO2 levels repress this developmental program4, 5. Initiation of mushroom formation starts with asymmetrical colony growth, followed by aggregation of aerial hyphae, and subsequent formation of primordia2. Primordia develop into fruiting bodies that form basidia within the hymenium. Karyogamy, meiosis, and one round of mitosis in the basidia results in haploid, binucleate basidiospores.
The blue light receptor complex of S. commune consists of Wc-1 that has a blue light sensing domain and the transcription factor Wc-2. Inactivation of wc-1 and/or wc-2 results in a blind phenotype6. Dikaryotic colonies of the homozygous deletion strains grow symmetrically in blue light (similar to dark-grown wild-type dikaryons) and do not produce aggregates, primordia, and fruiting bodies. Deletion of the homeodomain gene hom2 and the DNA binding Bright domain protein gene bri1 shows a similar phenotype7. Inactivation of the zinc finger transcription factor gene fst4 results in dikaryons that still grow irregular in the light under low CO2 conditions but aggregates, primordia, and fruiting bodies are not produced8. Strains in which the Cys2His2 zinc finger protein gene c2h2 has been inactivated are arrested at the aggregate stage7, while deletion strains of fst3, gat1and hom1 form smaller fruiting bodies but in higher numbers7, 8. The zinc finger protein Fst3 was proposed to play a role in repression of outgrowth of fruiting bodies from primordia, while the GATA type zinc finger protein Gat1 and the homeodomain protein Hom1 have been proposed to play a role in expansion of the fruiting body. Homologues of the S. commune transcription factors have been identified in Agaricus bisporus, Laccaria bicolor and C. cinerea. Expression studies suggest the existence of a core regulatory program for fruiting body development in these and other basidiomycetes8–10, which is supported by the finding that over-expression of the c2h2 homologue of A. bisporus results in faster fruiting body development11.
In this study, it is shown that transcription factors that are involved in mushroom formation in S. commune also modulate vegetative growth. Bri1 and Hom1 were shown to stimulate biomass formation, while Wc-1, Wc-2, Hom2, Fst4, as well as the newly identified transcription factor Tea1 repress vegetative growth. The interplay between vegetative growth and mushroom formation is illustrated by a constitutive version of Hom2 that strongly reduces colony formation and immediately induces mushroom formation under ambient CO2 conditions, even in a sterile strain.
Results
Genes involved in mushroom development also control vegetative growth
Dikaryotic colonies of wild-type H4-8, and the transcription deletion strains Δwc-2Δwc-2, Δhom2Δhom2, Δbri1Δbri1Δfst4Δfst4, Δc2h2Δc2h2, Δfst3Δfst3, Δhom1Δhom1, and Δgat1Δgat1 were grown for 6 days on glucose MM in the dark. The Δhom2Δhom2 strain formed 1.4-fold more biomass when compared to H4-8 (Fig. 1), which was accompanied by increased radial growth (not shown). In contrast, Δbri1Δbri1 and Δhom1Δhom1 formed 0.4 and 0.6-fold less biomass, respectively, while biomass formation of the other strains was not significantly different from H4-8. Strain Δhom2Δhom2Δfst4Δfst4 also formed more biomass than the wild-type. There was a trend that it also formed more biomass when compared to Δhom2Δhom2 (Supplementary Fig. 1), which was confirmed in liquid shaken cultures where it formed 1.7 fold more biomass when compared to the hom2 deletion strain (Supplementary Fig. 1). These data show that bri1 and hom1 stimulate vegetative growth, while hom2 and fst4 repress biomass formation of dikaryotic strains when glucose is used as a carbon source. Microscopical analysis revealed that wild-type formed two types of hyphae at the colony periphery. Vacuole-rich hyphae with a diameter of 2.68 µm ± 0.14 were 5,7-fold less abundant than hyphae with few vacuoles and a diameter of 1.72 µm ± 0.12. Strain Δhom1Δhom1 formed 5-fold less thick hyphae when compared to the wild-type, while Δbri1Δbri1, Δfst4Δfst4Δhom2Δhom2 formed 1.3-, 2.0-, and 3.3-fold more vacuole-rich, wide hyphae (Supplementary Fig. 2).
Figure 1.
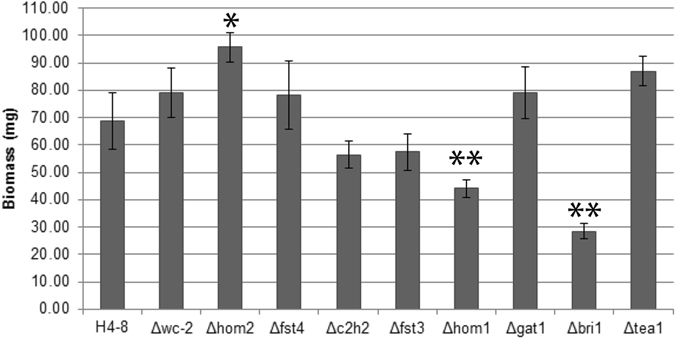
Biomass of biological triplicates of 6-day-old dark-grown agar cultures of the wild-type dikaryon and transcription factor deletion strains using glucose as carbon source. An independent sample t-test (p-value ≤ 0.05) was used to identify strains forming more (*) or less (**) biomass when compared to wild-type H4-8 (lines 506-507).
Biomass formation of Δwc-2Δwc-2, Δhom2Δhom2and Δfst4Δfst4 was assessed on xylose, pectin, and sucrose (Supplementary Fig. 3). The Δfst4Δfst4 strain formed 2.8-fold more biomass on xylose when compared to H4-8, while Δwc-2Δwc-2, Δhom2Δhom2and Δfst4Δfst4 formed more biomass on sucrose (1.2, 1.4, and 1.6-fold, respectively) and pectin (1.3, 1.4, and 1.6-fold, respectively) (Supplementary Fig. 3). Together, these data show that Wc-2, Hom2, and Fst4 repress vegetative growth on different carbon sources.
Reduced vegetative growth of the Δbri1Δbri1 and Δhom1Δhom1 strains may slow down fruiting body development. Therefore, fruiting was monitored after 7 days and 15 days of culturing. The Δhom1Δhom1 strain had formed more but smaller mushrooms both after 7 and 15 days (Fig. 2). In contrast, Δbri1Δbri1 had not formed fruiting bodies after 7 days7 but did so after 15 days showing that fruiting in this strain is delayed and not abolished as reported previously7.
Figure 2.
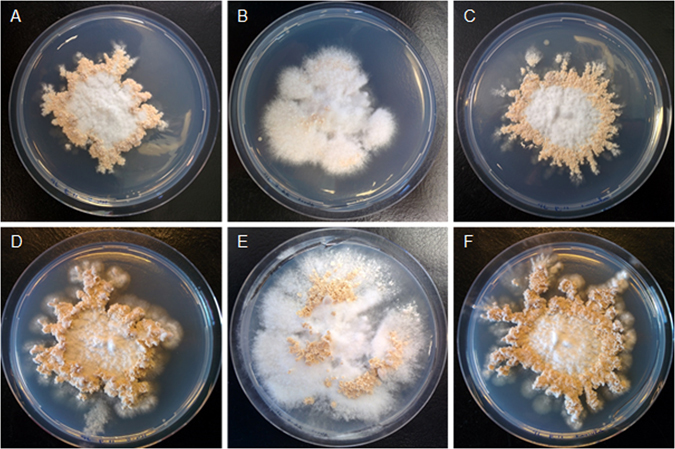
Fruiting body development of the wild-type dikaryon (A,D), Δbri1Δbri1 (B,E), and Δhom1Δhom1 (C,F) after 7 (A–C) and 15 (D–F) days of growth.
Genome-wide expression analysis
RNA composition of the wild-type H4-8 dikaryon was determined during vegetative growth in the dark, after transfer to the light, and during aggregate, primordium, and fruiting body formation. Expression of wc-1, wc-2, hom2, fst4, fst3, gat1, and bri1 in whole cultures increased or decreased ≤2-fold during development when compared to the vegetative mycelium grown in the dark (Table 1). In contrast, c2h2 and hom1 expression increased gradually with a maximum fold change of 4.7 and 2, respectively, during the fruiting body stage.
Table 1.
Temporal expression of the blue light sensor gene wc-1 and transcription factor genes involved in fruiting body development in the wild-type dikaryon based on biological duplicates. Values are expressed in FKPM. Bold font indicates a significant ≥2-fold up-regulation when compared to dark grown vegetative mycelium.
| Vegetative Mycelium Dark | Vegetative mycelium Light (induced) | Aggregate | Primordia | Fruiting body | |
|---|---|---|---|---|---|
| wc-1 | 37.30 | 40.06 | 54.83 | 51.97 | 62.23 |
| wc-2 | 38.29 | 46.19 | 39.88 | 61.66 | 47.47 |
| hom2 | 107.64 | 135.84 | 115.99 | 104.15 | 58.63 |
| tea1 | 9.39 | 14.73 | 18.88 | 62.40 | 10.48 |
| fst4 | 112.88 | 159.43 | 140.70 | 178.65 | 119.08 |
| c2h2 | 22.06 | 29.61 | 47.48 | 87.84 | 105.36 |
| fst3 | 103.23 | 97.82 | 102.70 | 119.53 | 115.64 |
| gat1 | 75.01 | 65.00 | 72.79 | 61.87 | 126.09 |
| hom1 | 114.38 | 108.69 | 149.72 | 200.48 | 234.15 |
| bri1 | 30.03 | 34.22 | 30.56 | 34.99 | 29.95 |
| c2h2d | 3.72 | 8.76 | 16.57 | 39.17 | 54.94 |
Expression profiles of wild type, Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δfst4Δfst4, Δc2h2Δc2h2, Δfst3Δfst3, Δgat1Δgat1, Δhom1Δhom1, and Δbri1Δbri1 strains were compared at the moment the wild-type formed aggregates (day 8) and fruiting bodies (day 12). Principal component analysis of the RNA profiles of 8-day-old colonies revealed a first and second component explaining 38% and 29% of the variation, respectively. Two distinct clusters were observed (Supplementary Fig. 4). The first cluster consisted of Δwc-1Δwc-1, Δwc-2Δwc-2, and Δhom2Δhom2 that are all affected in early stages of fruiting body development. The second cluster consisted of Δgat1Δgat1 and Δfst3Δfst3 that are affected in late stages of development. The other deletion strains did not cluster but rather showed a gradual change in expression. Principal component analysis of the RNA profiles of 12-day-old colonies revealed a first and second component explaining 72% and 7% of the variation, respectively. In this case, Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2and Δfst4Δfst4 clustered, whereas the other strains clustered with the wild-type (Supplementary Fig. 4).
The number of up- and down-regulated genes were between 86 and 1392 and 131 and 1463, respectively, when expression of the 8- and 12-day-old cultures of the deletion strains was compared with wild type (Supplementary Table 1). Enriched functional categories in the upregulated genes of 8-day-old colonies of Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2and Δfst4Δfst4 were mainly linked to carbohydrate metabolism, when compared to the 8-day-old wild-type colonies (Supplementary Tables 2 & 3). Downregulated functional groups were linked to energy transfer in these strains. Functional groups involved in energy transfer and carbohydrate metabolism were upregulated in 12-day-old colonies of Δwc-1Δwc-1 and Δwc-2Δwc-2 (Supplementary Tables 3 & 4), while downregulated genes were enriched for nucleosome, catalytic and oxidoreductase activity. The functional group ATPase activity was also overrepresented within the downregulated genes of Δwc-2Δwc-2. In 12-day-old colonies of Δhom2Δhom2 functional groups involved in translation, energy transfer, and carbohydrate metabolism were overrepresented in the upregulated genes, while functional groups involved in transport and energy transfer were overrepresented in the downregulated genes. In 12-days-old colonies of Δfst4Δfst4 functional groups involved in carbohydrate processes and energy transfer were enriched in the upregulated genes. Groups involved in oxidoreductase activity, metabolic process and nucleosome, amongst others, were enriched in the downregulated genes. Functional categories involved in energy transfer and carbohydrate metabolism were upregulated in 8-day-old colonies of Δc2h2Δc2h2, while processes involved in cytoplasm, ATP binding, nucleosome and peptidase activity were enriched in the downregulated genes. In 12-day-old colonies of Δc2h2Δc2h2 upregulated genes were enriched in functional groups related to energy transfer, carbohydrate metabolic process, and chitin catabolic process. Downregulated genes were enriched in groups involved in cell wall, catalytic activity, tryptophan and fatty acid synthesis. No functional groups were overrepresented in the upregulated genes in 8-day-old and 12-day-old colonies of Δhom1Δhom1 but functional groups related to chitinase activity, carbohydrate metabolism and energy transfer were enriched in the downregulated genes. 12-day-old Δhom1Δhom1 colonies showed overrepresentation of functional groups related to amino acid synthesis, oxidoreductase, and carbohydrate metabolism activity in the down regulated genes. The functional categories that were enriched in the upregulated genes of 8-day-old colonies of Δfst3Δfst3 and Δgat1Δgat1 were mainly involved in energy transfer, tryptophan metabolism, and cell wall processes. Genes involved in energy transfer and carbohydrate metabolism were enriched in the down-regulated genes. Groups involved in carbohydrate metabolism, methyltransferase, and leucine synthesis were enriched in the upregulated genes in 12-days-old colonies of Δfst3Δfst3. Downregulated genes were mainly enriched in cell wall processes and chitinase activity. Functional groups were upregulated for sphingolipid metabolism in 12-days-old colonies of Δgat1Δgat1. Downregulated genes were mainly enriched for translation and cell wall. No functional groups were overrepresented in the upregulated genes of 8-day-old and 12-day-old colonies of the Δbri1Δbri1 strain. In contrast, functional groups related to metabolic process, carbohydrate metabolism, transcription and cell wall were overrepresented in the downregulated genes of 8-day-old colonies. Downregulated genes in 12-day-old Δbri1Δbri1 colonies were mainly enriched for carbohydrate metabolism, hydrolase activity and transcription repressor activity.
Between 1 and 43 transcription factor genes were ≥2-fold up- or down-regulated in one of the deletion strains when compared to the wild-type (Fig. 3; Supplementary Table 5). Expression of hom1 and c2h2 had decreased in 12-day-old Δwc-1Δwc-1 colonies. In 12-day-old colonies of Δwc-2Δwc-2 hom2 expression was increased, while c2h2 was downregulated. Expression of c2h2, gat1, and hom1 was downregulated in 12-day-old Δhom2Δhom2 colonies. In 8-day-old Δfst4Δfst4 colonies expression of hom1 was downregulated and in 12-day-old colonies hom2 expression increased, while c2h2 and hom1 expression had decreased. In contrast, hom1 levels were upregulated in 8-day-old Δfst3Δfst3 colonies. Similarly, expression of hom1 but also c2h2 was increased in 8-day-old colonies of Δgat1Δgat1. In 8-day-old Δbri1Δbri1 colonies gat1 expression was increased, while hom2 was increased in 12-day-old colonies (Supplementary Fig. 5). Together, these data show that transcription factors involved in fruiting control other regulatory genes involved in this process.
Figure 3.
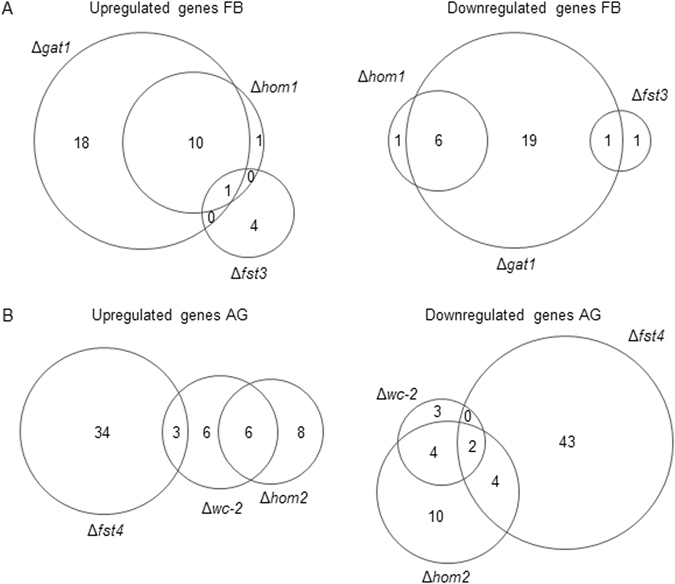
Venn diagrams showing overlapping differentially expressed transcription factor genes in Δfst3Δfst3Δhom1Δhom1and Δgat1Δgat1 (A) and Δwc-2Δwc-2, Δhom2Δhom2, and Δfst4Δfst4 (B) when compared to the wild-type aggregate and fruiting body stage, respectively.
Gene tea1 is involved in fruiting body development and represses vegetative growth
Transcription factor gene tea1 (protein ID 2519514) showed ≥2-fold decreased expression in 8-day-old colonies of Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δbri1Δbri1and Δfst4Δfst4 when compared to the 8-day-old aggregate forming colonies of wild-type, while it was upregulated ≥2-fold in 8-day-old colonies of Δfst3Δfst3, Δhom1Δhom1and Δgat1Δgat1. A ≥ 2-fold decreased or increased expression of tea1 was not observed in the deletion strains at day 12 when the wild-type had formed fruiting bodies. This is explained by reduced expression of tea1 in the wild-type colonies when compared to day 8. Gene c2h2d (proteinID 2703923) was also downregulated in Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2and Δfst4Δfst4 during aggregation, while it was upregulated ≥2-fold in Δgat1Δgat1 (Supplementary Fig. 5). Furthermore, expression of c2h2d was decreased >2-fold in 12-day-old colonies of Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δfst4Δfst4, Δfst3Δfst3and Δgat1Δgat1. Gene c2h2d has a predicted C2H2 DNA binding domain, while tea1 is a predicted TEA/ATTS transcription factor. Expression of tea1 and c2h2 in the wild-type strain peaked during primordia and fruiting body formation, respectively (Table 1).
Deletion constructs of c2h2d and tea1 were introduced in H4-8Δku80. PCR analysis confirmed that Δc2h2d and Δtea1 strains were obtained. These transformants were crossed with the compatible H4-8 wild-type strain and siblings were selected with an intact ku80 gene and a deleted tea1 or c2h2d gene. Compatible monokaryons were identified and then crossed to obtain the Δtea1Δtea1 and Δc2h2dΔc2h2d strains. Growth and mushroom formation of the Δc2h2dΔc2h2d strain were not affected (data not shown). In contrast, the Δtea1Δtea1 strain showed a 1.3-fold increase in biomass when compared to the wild-type. This increase was similar to that of Δhom2Δhom2 (Fig. 1). Transfer to the light did not induce irregular vegetative growth as observed in the wild-type. Moreover, newly formed light-exposed mycelium did not produce aerial hyphae (Fig. 4B). This resulted in a distinct border between the dark-grown mycelium and mycelium grown in the light. Fruiting body formation was almost completely abolished. The few spore-forming flask-like mushrooms were formed in clusters at random positions in the Δtea1Δtea1 colony, while a typical ring of fruiting bodies is formed in the wild-type (Fig. 4C,D). This and the fact that fruiting body and biomass formation was restored to wild-type levels in 2 independent complemented strains (data not shown) shows that tea1 represses vegetative growth and is involved in fruiting body formation.
Figure 4.
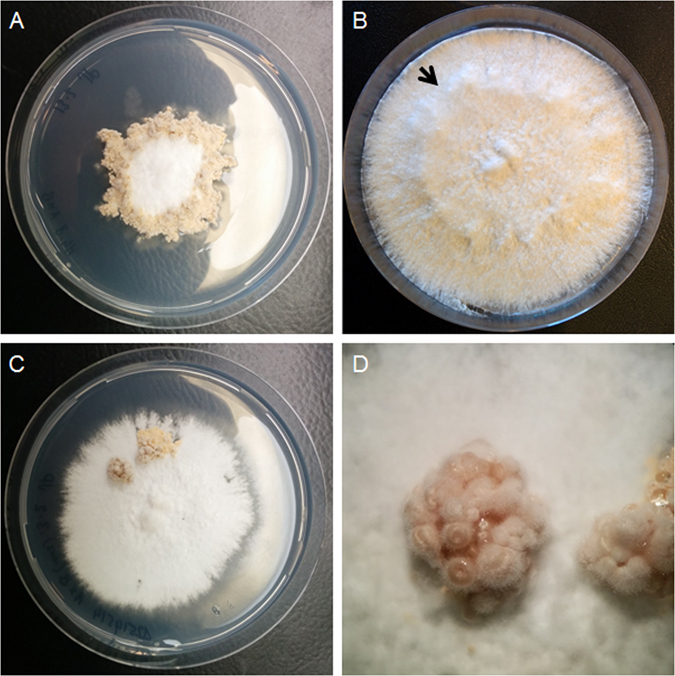
Aerial growth of dikaryotic colonies of H4-8 (A) and Δtea1 (B,C,D). Strain Δtea1Δtea1 produced less dense and high aerial hyphae when transferred to light (B) and only forms small clusters of fruiting bodies (C and D). Arrow indicates the transition from dense aerial hyphae production to thinner aerial hyphae production upon light induction.
Hom2con represses vegetative growth and induces fruiting body formation
Hom2 is a key gene involved in mushroom formation but it does not show differential expression during development (see above). Therefore, we searched for motifs that could mediate post-translational modification of this homeodomain protein. Hom2 was found to contain 4 predicted RRXS phosphorylation motifs. The serine codons within these motifs were replaced for alanine codons, resulting in hom2 con (name based on its phenotype; see below) and introduced in S. commune H4-8a and H4-8b. Two types of transformants were observed. 90% of the colonies had a growth speed similar to regenerating wild-type protoplasts, while 10% grew very slowly. Restriction PCR using RNA from colonies of the wild-type, normal growing transformants, and slow groing transformants showed that hom2 con was much higher expressed than the endogenous copy in the case of slow growing transformants (Fig. 5F). These transformants were selected for further experiments. Monokaryotic hom2 con transformants grew slower than wild type at 30 °C in the dark at high CO2. Clamp connections as observed in the wild-type dikaryon were absent (Fig. 5A,B). When colonies were transferred to fruiting conditions (low CO2, light, 25 °C) the monokaryotic hom2 con transformants started to form flask like fruiting bodies that did not expose their spore forming gills (Fig. 5D,E). Similar results were obtained when hom2 con was introduced in a Δhom2 monokaryon. These data show that Hom2con functions independently of the endogenous Hom2 and that it can induce fruiting in a sterile monokaryon. Monokaryotic hom2 con transformants crossed with the compatible wild type strain showed less severe phenotypes. However, they all grew slower than the wild type dikaryon and developed spore forming fruiting bodies immediately after transfer to the light at low CO2 conditions. In contrast, the wild-type continued vegetative growth for 1-2 days and then started to form fruiting bodies (Data not shown).
Figure 5.
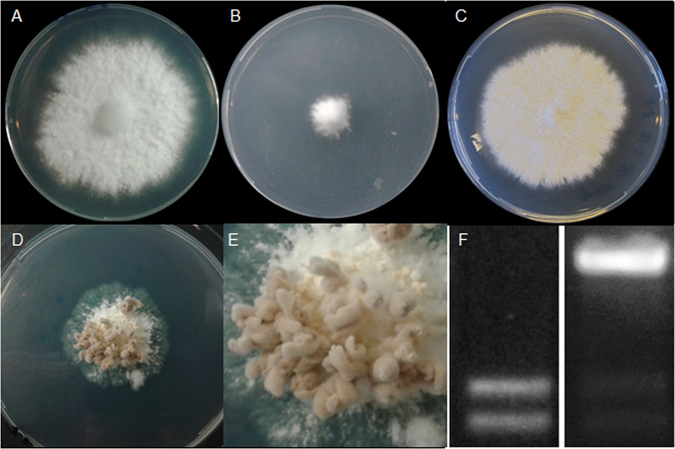
Six-day-old colonies of the wild type monokaryon (A,C) and a hom2 con strain (B,D) grown continuously at 30 °C in the dark at high CO2 (A,B) or transferred to 25 °C, low CO2, and light (C,D). (E) represents a close up of fruiting bodies of the hom2 con strain, while (F) shows relative hom2 and hom2 con expression in wild-type (left) and the hom2 con strain T4 (right).
Discussion
The transcription factor genes wc-2, hom2, fst4, bri1c2h2, fst3, gat1, and hom1 have been reported to be involved in fruiting body formation in S. commune 6–8. We here showed that the homeodomain domain protein Hom2, the zinc finger transcription factor Fst4, and the blue light complex transcription factor Wc-2 also repress vegetative growth, while the DNA binding BRIGHT domain protein Bri1 and the homeodomain protein Hom1 stimulate vegetative growth. We also showed that a newly identified transcription factor, called Tea1, stimulates mushroom formation, while repressing vegetative growth.
The Δbri1Δbri1 strain does not fruit after the standard growth period of 10 days7. However, we here showed that fully developed mushrooms had formed after 4 weeks. This shows that Bri1 is not required for fruiting. Delayed mushroom development may be the result of the reduced growth speed of Δbri1Δbri1. Lower biomass formation may well be explained by the fact that functional categories metabolic process, carbohydrate metabolism, catalytic activity, transcription, and cell wall were down-regulated in 8-day-old colonies of the deletion strain. 12-day-old colonies showed down-regulation of functional categories carbohydrate metabolism, hydrolase activity, and transcription repressor activity. As a consequence of reduced biomass formation, a quorum sensing pathway may become activated at a later moment delaying the switch to fruiting body formation12. Notably, Bri1 deletion has an effect on expression of tea1 and of gat1. The repression of tea1 may be a direct or an indirect effect due to its stimulatory effect on gat1 expression. Together, Bri1 stimulates vegetative growth and functions in mushroom formation by its effect on tea1 and gat1 expression (Fig. 6).
Figure 6.
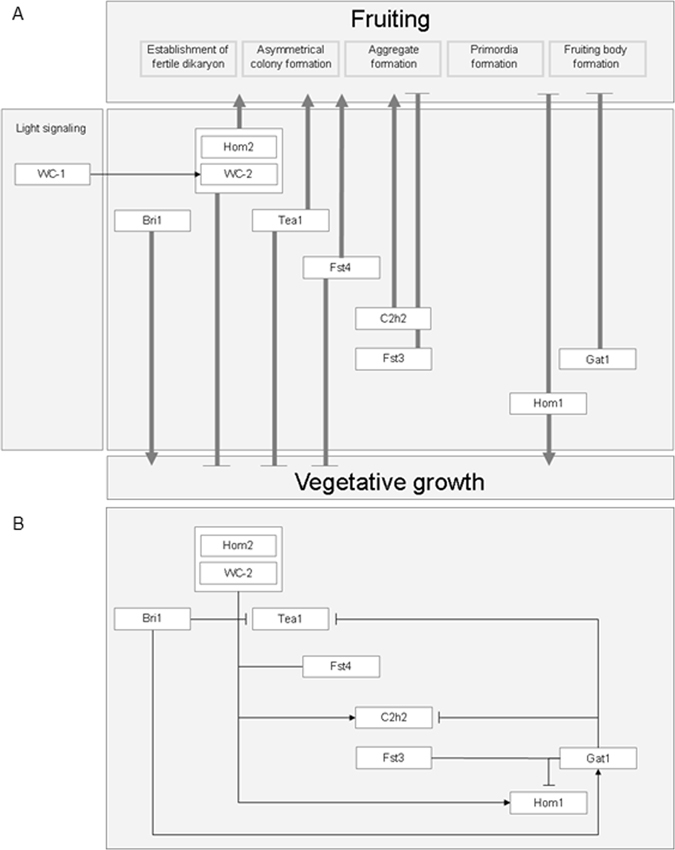
Model of regulation of vegetative growth and fruiting body formation in S. commune. Transcription factor genes control both vegetative growth and fruiting body development (A) and influence each other's expression levels (B).
Genes hom2 and wc-2 are involved in the switch from vegetative growth to fruiting. Inactivation of these genes abolishes early stages of fruiting body formation6, 7 but also increased the vegetative growth rate. Strain Δwc-2Δwc-2 formed more biomass on sucrose and pectin when compared to the wild-type, while Δhom2Δhom2 formed more biomass on glucose, sucrose, and pectin. Increased biomass formation was associated with an increased hyphal diameter and with a higher radial extension rate. In addition, upregulated genes were enriched in the functional class carbohydrate metabolism in 8-day-old colonies, and of functional groups involved in carbohydrate metabolism and energy transfer in 12-day old colonies. Deletion of wc-2 and hom2 resulted in a ≥2-fold downregulation of c2h2 in 12-day-old colonies. Downregulation was also observed in 8-day-old colonies, although the effect was less pronounced. Together, this confirms that Wc-2 and Hom2 stimulate c2h2 expression6, 7(Fig. 6).
Expression of hom2 and wc-2 is rather constant during development, suggesting post-translational regulation of these genes. In the case of Wc-2, this may be accomplished by the interaction with the blue light sensor Wc-16, while post-translational modification of Hom2 was shown to be mediated via its 4 predicted protein kinase motifs. Introduction of hom2 con that has its 4 protein kinase motifs eliminated resulted in severe phenotypes in sterile wild-type and Δhom2 monokaryons. The hom2 con strains grew very slowly when compared to their parental strains, indicating that hom2 in its non-phosphorylated state represses vegetative growth. Transformants even fruited when exposed to light and low CO2, thus resembling a dikaryon. This suggests that Hom2 is maintained in a phosphorylated state in the monokaryon resulting in fast vegetative growth and repression of fruiting. The fact that the Δhom2 monokaryon grows faster than the wild-type implies that a fraction of Hom2 in the monokaryon is not phosphorylated thus impacting vegetative growth to some extent. Data also indicate that the phosphorylation state of Hom2 is the only inhibitory factor that suppresses fruiting in monokaryons in conditions favorable for mushroom formation in the dikaryon. Moreover, data imply that fruiting body formation can be independent of the mating type genes. Indeed, frt1 of S. commune also initiates fruiting in a homokaryon when copies of the gene are introduced in homokaryons with a different frt1 allele13. The role of the mating type genes could be restricted to the establishment and maintenance of the dikaryotic state. Alternatively, different pathways exist that each can lead to fruiting body formation and that may at some point interconnect.
Gene fst4 is constitutively expressed during the S. commune life cycle. Like Hom2 and Wc-2 it is involved in the switch from vegetative growth to fruiting. Strain Δfst4Δfst4 grows irregular in the light like the wild-type but does not aggregate. It formed more biomass than the wild-type on xylose, sucrose, and pectin but not on glucose. Hyphae of Δfst4Δfst4 were more wide compared to wild-type but not as wide as Δhom2Δhom2. Together, these data indicate that Fst4 and Hom2 represent different parts of the repression pathway of vegetative growth. In liquid shaken cultures with glucose as carbon source these pathways may merge explaining why Δhom2Δhom2Δfst4Δfst4 formed more biomass than Δhom2Δhom2. Strain Δfst4Δfst4 showed enrichment of carbohydrate metabolism in the upregulated genes of 8-day-old and 12-day-old-colonies similar to that observed in Δwc-2Δwc-2 and Δhom2Δhom2. The fact that fst4 expression is not affected in Δwc-2Δwc-2 and Δhom2Δhom2 strengthens the hypothesis that Fst4 and Hom2 represent different pathways. Gene fst4 stimulates c2h2 like hom2 and wc-2 do. This indicates that Fst4, Hom2, and Wc-2 input are channeled into the fruiting pathway via c2h2 (Fig. 6).
Transcription factor gene tea1 was downregulated in Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δbri1Δbri1and Δfst4Δfst4 when compared to the aggregating wild-type (8-day-old colonies), while it was upregulated in Δfst3Δfst3, Δhom1Δhom1and Δgat1Δgat1. This indicates it is upregulated during early stages of development while it is repressed during late stages of mushroom formation. This agrees with the expression profile in the wild-type. The Δtea1Δtea1 strain formed more biomass on glucose when compared to the wild-type. Moreover, it was severely affected in mushroom formation. Local clusters of fully developed mushrooms were only occasionally formed in the deletion strain. This phenotype may be explained by a reduced sensitivity of a signaling pathway leading to a developmental switch from “off” to “on”.
Expression of c2h2 increased > 2-fold when RNA profiles of 5- and 8-day-old colonies were compared. The increased expression during the aggregation stage and the further increased expression in primordia and fruiting bodies agrees with the phenotype of Δc2h2Δc2h2 forming aggregates but not primordia and fruiting bodies7 (Fig. 6). C2H2 did not affect biomass formation implying that it is downstream of the switch between vegetative growth and mushroom development.
Deletion of fst3, gat1, or hom1 results in more, but smaller mushrooms7. In addition, Hom1 and Gat1 are involved in mushroom tissue formation7. Gene fst3 is constitutively expressed and its expression is not affected by any of the other transcriptional regulators. This suggests that Fst3 is subject to post-transcriptional regulation. The fact that a higher number of genes are differentially expressed in 8-day-old colonies (wild-type forming aggregates) when compared to 12-day-old colonies (wild-type forming fruiting bodies) (i.e. 882 and 217 genes, respectively) suggest that Fst3 exerts its effect already early in mushroom development. Upregulated functional groups were involved in energy transfer and cell wall processes in 8-day-old colonies, while downregulated functional groups were involved in carbohydrate metabolism. In 12-day-old colonies these groups were regulated in the opposite direction. Expression of gat1 was highest during fruiting body formation. It is repressed by Bri1 in 8-day-old colonies, while it is activated by Hom2 during fruiting body formation. Expression of hom1 gradually increases during progression of fruiting. Wc-1, Hom2, Fst4, and probably Wc-2, stimulate expression of hom1, while Fst3 and Gat1 have the opposite effect. The fact that Hom1 has both an effect on vegetative growth and on tissue formation in mushrooms suggests that Hom1 operates at two distinct stages of development (Fig. 6). Alternatively, the reduced size of the Δhom1Δhom1 fruiting bodies and the effect of Hom1 on tissue formation in mushrooms may be explained solely by the reduced size of the vegetative feeding mycelium.
The model of development of S. commune may apply to other mushroom forming basidiomycetes as well. Hom2, Hom1, Fst3, Fst4, C2H2, and Gat1 are basidiomycete-specific regulatory proteins14. Homologues of these genes were identified in L. bicolor and A. bisporus 8, 9. The homologues for fst4, fst3, c2h2, and hom1 were upregulated during sexual development in two A. bisporus varieties9, while homologues of hom2, fst4, c2h2, fst3, gat1 and hom1 showed similar expression expression profiles in L. bicolor showed similarity to S. commune 9. Expression patterns of c2h2, fst3, hom1 and gat1 were also found to be similar in C. cinerea 10. Moreover, it was recently shown that over-expression of the c2h2 homoloque of A. bisporus accelerated mushroom development in this mushroom forming fungus11, which agrees with the phenotype of the Δc2h2Δc2h2 phenotype of S. commune.
This is the first time a direct link has been shown between repression of vegetative growth and initiation of sexual reproduction. Previously, a link has been shown between vegetative growth and asexual development in Aspergillus 15. This link involves trimeric G-protein signaling. The activity of the Gα-subunit of Aspergillus is regulated by the FlbA protein16. Inactivation of this gene results in a strain that cannot initiate asexual development. Notably, S. commune has a homologue of flbA called thn. Inactivation of this gene results in a strain unable to form fruiting bodies17. This suggests that similar signaling pathways are involved in the decision to stop vegetative growth and to invest in reproduction in ascomycetes and basidiomycetes. A link between vegetative growth and development may also exist in Podospora anserina. Inactivation of the homeodomain gene pah1 resulted in increased production of male gametes and increased branching of vegetative hyphae, resulting in smaller but more dense colonies18, 19.
Methods
Culture conditions and strains
The compatible S. commune strains H4-8 (matA43matB41; FGSC 9210)8 and H4-8b (matA41matB43)20, their derived wild-type dikaryotic strain, as well as the deletion strains Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δfst4Δfst4, Δc2h2Δc2h2, Δfst3Δfst3, Δhom1Δhom1Δbri1Δbri1, Δgat1Δgat1 6–8, Δhom2Δhom2Δfst4Δfst4 and Δhom2Δhom2Δfst3Δfst3 were used in this study. The Δku80 H4-8 strain21 was used for gene inactivation. Strains were grown in the dark or in the light (1200 lux white LED light; Conrad Electronic, Hirschau, Germany) at 25 °C on minimal medium (MM) containing 1.5% agar and 2% glucose22. Cultures were inoculated with a point inoculum taken from the periphery of a 7-day-old colony. Liquid shaken cultures were inoculated with a mycelial homogenate23 and grown at 250 rpm in 250 ml Erlenmeyers containing 100 ml MM. To assess growth on other carbon sources, glucose was replaced by 4% xylose, 3.4% sucrose, or 1% pectin.
Gene inactivation and complementation
Deletion vectors for tea1 (Protein ID 2519514; http://genome.jgi-psf.org/Schco3) and c2h2d (Protein ID 2703923) were constructed using pDelcas that contains a nourseothricine and a phleomycin resistance cassette20. Upstream and downstream flanks of tea1 and c2h2d were cloned at either site of the nourseothricine resistance cassette. To this end, the flanks were amplified by PCR using Taq polymerase and H4-8 chromosomal DNA as template. The 906 bp upstream flank and the 946 bp downstream flank of tea1 were amplified using the primer combination Δ2519514ufw/Δ2519514urv and Δ2519514dfw/Δ2519514drv, respectively (Supplementary Table 6). Primer pair combinations ∆2703923ufw/∆2703923urv and ∆2703923dfw/∆2703923drv were used to amplify the 897 bp upstream and 975 bp downstream flank of c2h2d, respectively (Supplementary Table 6). The PCR products were cloned into pGEM-T Easy (Promega, Madison, USA). The upstream flanks were retrieved from the resulting constructs using Van91I and introduced into the Van91I site of pDelcas, resulting in pDel-2519514-UF and pDel-2703923-UF. The downstream flanks were retrieved from the pGEM-T Easy derived constructs using SfiI and introduced into the SfiI site of pDel_2519514-UF and pDel_2703923-UF. This resulted in the knock-out constructs pDelcas-2519514 and pDelcas-2703923.
To complement strain Δtea1 the coding region of tea1 was amplified by PCR using primer pair tea1fw1/tea1rv1 (Supplementary Table 6) that introduce AarI and BamHI sites, respectively. The AarI/BamHI fragment was introduced in NcoI/BamHI cut vector pRO15124 that has a pUC20 backbone containing 1 kb HindIII/NcoI promoter sequence of gpd1 gene from S. commune, a 350 bp BamHI/EcoRI terminator sequence from the S. commune sc3 gene, and a EcoRI fragment containing a phleomycin resistance cassette. This resulted in pTEA1comp.
Expression vector for Hom2con of S. commune
Plasmid pHom2 is a pGMTphleoB derivative containing the S. commune gene hom2 7. The 1832 bp HindIII/SbfI fragment of pHom2 containing hom2 coding and flanking sequences was cloned in pSP72 using the same sites, resulting in pHom2HF. In the next step, a 692 hom2 coding fragment was synthesized (Genscript, Piscataway, NJ, USA), in which the serine codons in the 4 RRXS motifs were replaced for alanine (hom2 con). As a consequence of the substitution in the second motif a unique PvuI site was removed in hom2 con. A fusion PCR was performed with the 692 bp fragment and pHom2HF using T7 and SP6 primers annealing at both sides of the multiple cloning site of pHom2HF. The fusion fragment resulted in a mixture of 1922 bp hom2 and hom2 con fragments. The wild-type fragment was digested with PvuI. The remaining 1922 bp band was cut with HindIII/SbfI and used to replace the corresponding fragment in pHom2. This resulted in plasmid pHom2* containing the hom2 con gene.
Transformation of S. commune
Constructs were introduced in H4-8 or H4-8Δku80 as described22 using 1·107 protoplasts. For gene deletions, protoplasts were incubated with 20 μg vector DNA and regenerated overnight without antibiotic. Selection of transformants took place for 4 days at 30 °C on MM plates containing 8 μg ml−1 nourseothricin. Transformants were transferred to a second selection plate containing 5 μg ml−1 phleomycin to distinguish between homologous and ectopic integrations. Gene deletion was confirmed by PCR using primers outside the flanks and inside the nourseothricin cassette. Primer pairs 2519514ufcfw/nourdelrev and 2519514dfcrv/sc3tersqf were used to screen for tea1 deletion (Supplementary Table 6), while primer pairs c2h2dufcfw/nourdelrev and c2h2ddfcrv/sc3tersqf were used to confirm c2h2d deletion (Supplementary Table 6).
For complementation or expression constructs, protoplasts were incubated with 5 μg vector DNA and regenerated overnight in the presence of 25 μg ml−1 phleomycin. Selection of transformants took place for 4 days at 30 °C on MM plates containing 25 μg ml−1 phleomycin.
Whole genome expression analysis
The wild-type dikaryon and strains Δwc-1Δwc-1, Δwc-2Δwc-2, Δhom2Δhom2, Δfst4Δfst4, Δc2h2Δc2h2, Δfst3Δfst3, Δhom1Δhom1, Δbri1Δbri1, and Δgat1Δgat1 were grown for 5 days in the dark at 22 °C, after which they were transferred to the light25. Biological duplicate whole cultures were harvested at the moment the wild-type dikaryon formed aggregates (day 8) or fruiting bodies (day 12). H4-8 colonies were also harvested at the moment they were transferred to the light (day 5) and when they had formed primordia (day 10). Mycelium was frozen in liquid nitrogen and homogenized using the TissueLyser II (Qiagen, Düsseldorf, Germany). RNA was extracted using TriZol (Life technologies, Carlsbad, USA) and purified using the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany). Quality of RNA was checked using the BioAnalyzer and sent to ServiceXS (Leiden, the Netherlands) for Illumina Next Generation Sequencing. RNAseq data have been deposited at NCBI under BioProject PRJNA323434.
RNA-Seq Analysis Pipeline
STAR aligner26 was used to align the 100 bp paired end reads to the S. commune v3.0 genome (http://genome.jgi-psf.org/Schco3/Schco3.home.html). The size of introns was limited to a maximum of 1500 bp based on the largest intron sizes in the genome annotation. Abundance estimation and differential expression were analysed by Cufflinks version 2.1.127, and Cuffdiff using a Benjamini Hochberg false discovery rate of 0.0528 excluding genes that had a number of fragments per kilobase of exon per million <1 in one or both of the strains or conditions. Enrichments of GO terms were analysed within sets of differentially expressed genes. Proteins annotated to contain a DNA-binding or regulatory protein domain were defined as transcription factors8.
Restriction PCR
RNA was extracted from 10-day-old colonies grown on MM plates on top of a polycarbonate membrane using Trizol (Thermo Scientific, Waltham, MA, USA). cDNA was made using the QuantiTect Reverse Transcription Kit (Qiagen, Düsseldorf, Germany). In the next step, cDNA was amplified by PCR with the intron spanning primers hom2rtfw and hom2rtrv (Supplementary Table 6). The amplified wild-type 1117 bp fragment was cut with PvuI resulting in fragments of 609 and 508 bp. The hom2 con fragment could not be cut by this enzyme, leaving the 1117 bp band intact.
Bioinformatics
RRXS domains were identified in the protein sequences of S. commune using a custom Python script. Enrichment analyses were performed with Python and R to analyze over- and under-representation of functional annotation terms in sets of genes using the Fisher Exact test. The Benjamini-Hochberg correction was used to correct for multiple testing using a p-value ≤ 0.05. Orthologues of S. commune hom2 in other basidiomycetes were identified by the presence of a homeobox PFAM domain (PF00046)29 and a reciprocal best blastp hit to hom2. These hits were manually curated using MycoCosm30 based on the available expression and homology data for that genome. The protein sequences were aligned using MAFFT version 7.12331 with the E-INS-i method. The alignment was visualized and manually curated using Jalview 2.8.232. Conserved RRXS sites were identified in the alignment. A phylogenetic tree was reconstructed using RaxML version 8.1.1633 with the PROTGAMMAWAG model with 100 rapid bootstrap partitions. The tree and domain structures were visualized using the ETE Toolkit version 2.2.
Biomass of colonies
Colonies were grown as liquid shaken cultures or on agar medium on a PC-membrane (diameter 76 mm, pore size 0.1 µm; Osmonics, GE Water Technologies) using biological triplicates. Mycelium of liquid cultures was separated from the medium using Miracloth filter (Merck Millipore, Billerica, USA). Mycelium was freeze-dried and weighed. Statistical analysis was done with an independent sample t-test (p-value ≤ 0.05) using IBM SPSS 20.
Microscopy
Strains were grown in Petri dishes with a diameter of 4.5 cm within a 125 µm layer of MM agar for 2 days at 30 °C. Petri dishes were covered with wet tissue, 2 layers of plastic foil, and 1 layer of aluminum foil. Morphology of hyphae was studied with an inverted microscope using a 400-fold magnification. Statistical analysis was done with a χ2 test (p-value ≤ 0.05).
Electronic supplementary material
Acknowledgements
The genome sequence data of Schizophyllum commune v3.0 together with annotation data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community.
Author Contributions
J.P., M.H., T.G. and L.L. performed, analysed, interpreted, and designed the experiments; H.W., L.L. and M.R. supervised the project; J.P., L.L., and H.W. wrote the manuscript.
Competing Interests
This research is supported by the Dutch Technology Foundation STW, which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs. The authors declare not to have a competing financial interest.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00483-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Han A. B. Wösten, Email: h.a.b.wosten@uu.nl
Luis G. Lugones, Email: l.g.lugones@uu.nl
References
- 1.Kües U, Liu Y. Fruiting body production in basidiomycetes. Appl. Microbiol. Biotechnol. 2000;54:141–152. doi: 10.1007/s002530000396. [DOI] [PubMed] [Google Scholar]
- 2.Kües U, Navarro-González M. How do Agaricomycetes shape their fruiting bodies? 1. Morphological aspects of development. Fungal Biol. Rev. 2015;29:63–97. doi: 10.1016/j.fbr.2015.05.001. [DOI] [Google Scholar]
- 3.Perkins JH. Morphogenesis in Schizophyllum commune. I. Effects of white light. Plant Physiol. 1969;44:1706–1711. doi: 10.1104/pp.44.12.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niederpruem DJ. Role of carbon dioxide in the control of fruiting of Schizophyllum commune. J. Bacteriol. 1963;85:1300–1308. doi: 10.1128/jb.85.6.1300-1308.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raudaskoski M, Viitanen H. Effects of aeration and light on fruit-body induction in Schizophyllum commune. Trans. Brit. Mycol. Soc. 1982;78:89–96. doi: 10.1016/S0007-1536(82)80080-6. [DOI] [Google Scholar]
- 6.Ohm RA, Aerts D, Wösten HAB, Lugones LG. The blue light receptor complex Wc-1/2 of Schizophyllum commune is involved in mushroom formation and protection against phototoxicity. Environ. Microbiol. 2013;15:943–955. doi: 10.1111/j.1462-2920.2012.02878.x. [DOI] [PubMed] [Google Scholar]
- 7.Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011;81:1433–1445. doi: 10.1111/j.1365-2958.2011.07776.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohm RA, et al. Genome sequence of the model mushroom Schizophyllum commune. Nature Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 9.Morin E. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA. 2012;109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plaza DF, Lin CW, van der Velden NSJ, Aebi M, Künzler M. Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development. BMC Genomics. 2014;15:492. doi: 10.1186/1471-2164-15-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelkmans JF, et al. The transcriptional regulator c2h2 accelerates mushroom formation in Agaricus bisporus. Appl. Microbiol. Biotechnol. 2016;100:7151–7159. doi: 10.1007/s00253-016-7574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wösten HAB, Willey JM. Surface active proteins enable microbial aerial hyphae to grow into the air. Microbiology. 2000;146:767–773. doi: 10.1099/00221287-146-4-767. [DOI] [PubMed] [Google Scholar]
- 13.Horton JS, Raper CA. The mushroom-inducing gene FRT1 of Schizophyllum commune encodes a putative nucleotide binding protein. Mol. Gen. Genet. 1995;247:358–366. doi: 10.1007/BF00293204. [DOI] [PubMed] [Google Scholar]
- 14.Todd RB, et al. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics. 2014;15:240. doi: 10.1186/1471-2164-15-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krijgsheld P, et al. Development in Aspergillus. Stud. Mycol. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu JH, Wieser J, Adams TH. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler TJ, Mitton MF. Scooter, a new active transposon in Schizophyllum commune, has disrupted two genes regulating signal transduction. Genetics. 2000;156:1585–1594. doi: 10.1093/genetics/156.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnaise S, Zickler D, Poisier C, Debuchy R. pah1: a homeobox gene involved in hyphal morphology and microconidiogenesis in the filamentous ascomycete Podospora anserina. Mol. Microbiol. 2001;39:54–64. doi: 10.1046/j.1365-2958.2001.02163.x. [DOI] [PubMed] [Google Scholar]
- 19.Coppin E, et al. Systematic deletion of homeobox genes in Podospora anserina uncovers their roles in shaping the fruiting body. PLoS ONE. 2012;7:e37488. doi: 10.1371/journal.pone.0037488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohm RA, et al. An efficient gene deletion procedure for the mushroom-forming basidiomycete Schizophyllum commune. World J. Microbiol. Biotechnol. 2010;26:1919–1923. doi: 10.1007/s11274-010-0356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong JF, Ohm RA, de Bekker C, Wösten HAB, Lugones LG. Inactivation of ku80 in the mushroom-forming fungus Schizophyllum commune increases the relative incidence of homologous recombination. FEMS Microbiol. Lett. 2010;310:91–95. doi: 10.1111/j.1574-6968.2010.02052.x. [DOI] [PubMed] [Google Scholar]
- 22.van Peer AF, de Bekker C, Vinck A, Wösten HAB, Lugones LG. Phleomycin increases transformation efficiency and promotes single integrations in Schizophyllum commune. Appl. Environ. Microbiol. 2009;75:1243–1247. doi: 10.1128/AEM.02162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wetter MA, Wösten HAB, Sietsma JH, Wessels JGH. Hydrophobin gene expression affects hyphal wall composition in Schizophyllum commune. Fungal Genet. Biol. 2000;31:99–104. doi: 10.1006/fgbi.2000.1231. [DOI] [PubMed] [Google Scholar]
- 24.Ohm, R. A. Regulation of mushroom formation in Schizophyllum commune. Ph. D. thesis, Utrecht Univerisity, Utrecht, the Netherlands (2010).
- 25.Gehrmann, et al. Schizophyllum commune has an extensive and functional alternative splicing repertoire. Sci. Rep. 2016;6:336–340. doi: 10.1038/srep33640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nature Prot. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-Seq. Nature Biotechnol. 2012;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. Pfam: The protein families database. Nucl. Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoriev IV, et al. MycoCosm Portal: Gearing up for 1000 fungal genomes. Nucl. Acids Res. 2014;42:699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


