Abstract
Diversity of antibodies and T cell receptors is generated by gene rearrangement dependent on RAG1 and RAG2, enzymes predicted to have been derived from a transposable element (TE) that invaded an immunoglobulin superfamily gene early in the evolution of jawed vertebrates. Now, Huang et al. report the discovery of ProtoRAG in the lower chordate Amphioxus, the long-anticipated TE related to the RAG transposon.
Immunoglobulin (or antibody) and T cell receptor germline gene segments rearrange to “mix and match” them to provide the first level of antigen receptor diversity. Furthermore, one of the loops in the receptors that recognizes antigen, CDR3, is generated by the rearrangement-induced, double-stranded DNA break and subsequent processing and repair (Figure 1). This process, which permits a large range of amino acid length in CDR3, is a novel mechanism that, by far, is the major generator of diversity (GOD) in these immune genes (Hsu, 2011). Genomes are polluted with transposable elements (TEs) considered to be “selfish DNA” and thought by most scientists to be of little functional consequence. However, this view has been changing with the increasing number of examples of TE involvement in gene modification and expression (Biémont, 2010). One of the best TE-taming examples has been the RAG transposon, which invaded an immunoglobulin superfamily (IgSF) exon 600 million years ago in an ancestor of the jawed vertebrates (gnathostomes) (Thompson, 1995). This invasion ignited the new form of DNA rearrangement in developing lymphocytes. In this paper, Huang et al. (2016) identify a RAG transposon in the lower chordate Amphioxus, complete with the two enzymes, RAG1-like(L) and RAG2L, repetitive elements at either end resembling regions of the recombination signal sequences (RSSs) found adjacent to rearranging gene segments (terminal-inverted repeats; TIRs), and transposon target-site duplications (TSDs) indicative of the translocation (see Figure 1 in Huang et al., 2016).
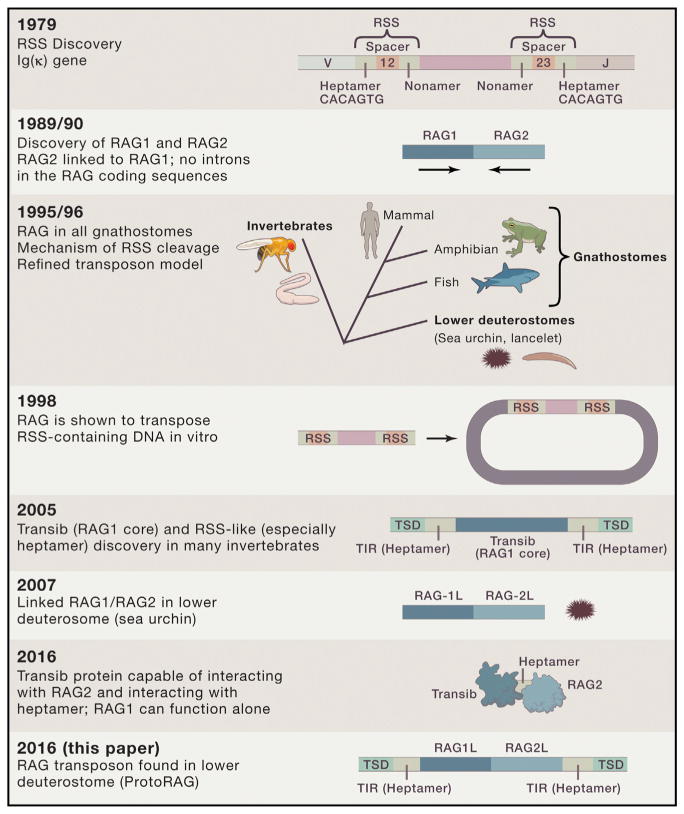
Figure 1. Historical Events Culminating in the Discovery of ProtoRag in Amphioxus.
The discoveries of RSS (by Tonegawa and colleagues), the RAG proteins (by Baltimore and colleagues), and the mechanism of RSS cleavage (by Oettinger/Gellert and colleagues) resulted in the theory that a TE invaded an IgSF gene, eventually leading to Ig/TCR gene rearrangement (by Thompson). Experiments showing that the RAGs are capable of DNA transposition (by Schatz/Gellert and colleagues), the presence of RAG in all gnathostomes (by Marchalonis and colleagues), and the presence of a RAG1 core and TIR in the invertebrates (by Kaptinov and Jurka) were consistent with the model. Discovery of linked RAG1L and RAG2L in lower deuterostomes (by Fugmann/Rast and colleagues), including a complete and active RAG transposon in Amphioxus (Huang et al., 2016), are consistent with the hypothesis.
Not long after antibody gene rearrangement was discovered, DNA sequencing revealed repetitive elements at the edge of the rearranging segments, having a conserved heptamer and nonamer separated by a spacer of either 12 or 23 base pairs (now called RSS, Figure 1; note, the history of VDJ rearrangement is reviewed in Fugmann, 2010 and crucial references are noted in the figure legend). It was suggested that these repetitive elements resembled transposon-like, inverted-repeat elements involved in DNA jumping in prokaryotes (Sakano et al., 1979). A decade later, one of the enzymes involved in rearrangement, RAG1, was discovered in a rearrangement screen of genomic DNA as well as cDNAs derived from developing B and T cells (Figure 1) (Schatz et al., 1989). The RAG1-induced rearrangement was very inefficient, and soon afterward a second enzyme, RAG2, was discovered after transfection of cells with genomic DNA that greatly enhanced the rearrangement; luckily, the two RAG enzymes were closely linked which made the experiment with genomic DNA possible. The synteny of RAG1 and RAG2 and their lack of introns were reminiscent of genes from prokaryotes involved in transposition (Figure 1). Later, two groups showed that RAGs could transpose linear DNA with RSS ends into DNA, providing a model for the original insertion into the IgSF gene (Figure 1). RAG1 and RAG2 were expressed in developing T and B lymphocytes in all gnathostomes, but apparently not lower deuterostomes or protostomes (Figure 1). However, with the rush of genome projects, a RAG1L and RAG2L gene set eventually was found in genomic DNA of the lower deuterostome sea urchin and a RAG1 core called “transib” and TIR were found in varying copies in the genomes of many organisms, including many invertebrates (Kapitonov and Jurka, 2005)(Figure 1).
The Huang et al. (2016) paper identifies a candidate RAG transposon, ProtoRAG, in the lower chordate Amphioxus (lancelet), with RAG 1L and RAG2L genes flanked by a TIR that has the conserved heptamer (Figure 1) and other repetitive elements specific to Amphioxus (including a lancelet-specific “nonamer”). This finding is similar to the situation in sea urchins, but no TIR were detected (Fugmann, 2010). Phylogenetic analysis showed that ProtoRAG was recently active in the germline. Amphioxus RAG1L can cleave DNA and generate hairpins in its TIR like its vertebrate relative, but cannot interact with mammalian RAG2 and RSS, perhaps due to the great phylogenetic distance. However, in previous experiments, Amphioxus RAG1L with N- and C-termini of mammalian RAG1 (Figure 1) complemented RAG1 knockout (KO) mice in being able to induce rearrangement of antigen receptor genes in vivo (Zhang et al., 2014). Mutation of residues involved in catalysis in mammalian RAG1, the so-called catalytic triad of negatively charged amino acids, abrogated Amphioxus RAG1L activity, and the DNA-binding protein HMGB1 enhanced the activity of the lancelet RAGs, like it does in vertebrates. RAG2L lacks the plant homeodomain (PHD) element involved in binding to active chromatin, which is likely a derived state since the domain is present in both sea urchin and vertebrate RAG2. Finally, TSD are found on both sides of the RAG/TIR elements (Figure 1), also consistent with recent activity of ProtoRAG in the germline. All in all, there is little doubt that a hallowed RAG transposon has been uncovered.
As mentioned, in the earliest publication on RAG, RAG1 was shown to induce low levels of rearrangement when transfected with a recombination substrate into fibroblasts (Schatz et al., 1989). This work has been repeated recently, and an invertebrate transib was also capable of RSS interaction (Figure 1) (Carmona et al., 2016). Study of the sea urchin system led to the hypothesis that a transib gene may have inserted near a RAG2 gene, a normal cellular gene. While RAG1 has features consistent with several prokaryotic cut-and-paste transposases like Tn10, RAG2 contains elements/domains found only in eukaryotes. Since RAG2 binds active chromatin (trimethylated histone H3K4), one hypothesis is that such binding helps recruit RAG1 to RSS sites in developing lymphocytes (Fugmann, 2010); eventually, the two enzymes became codependent in the rearrangement process. If this is true, then RAG2 only became part of the rag transposon after being shanghaied by a transib (RAG1core) transposon. Alternatively, RAG2, sans the PHD element that interacts with chromatin, was part of the original transposon, and it recruited the chromatin-interacting PHD element later. In summary, the RAG1 core (transib), as the major player as the catalyst of rearrangement, and the TIR/RSS heptamer were no doubt part of the original transposon, while RAG2’s origins and function(s) are still unclear. It will be interesting to determine whether the RAGs serve any cellular function in the lower deuterostomes: expression data published to date have not been revealing.
Since McClintock’s early work on transposition in maize, several other TEs have been shown to be domesticated (Biémont, 2010), and surely, there will be many more examples of such taming of TEs in eukaryotes. Archeologists search for evidence of Old Testament miracles such as the parting of the Red Sea and the tumbling of Jericho’s walls. With the discovery of ProtoRAG, molecular archeologists have given us a glimpse of the ancestral transposon that initiated immune generation of diversity.
References
- Biémont C. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona LM, Fugmann SD, Schatz DG. Genes Dev. 2016;30:909–917. doi: 10.1101/gad.278432.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD. Semin Immunol. 2010;22:10–16. doi: 10.1016/j.smim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E. Curr Opin Immunol. 2011;23:156–162. doi: 10.1016/j.coi.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Hüppi K, Heinrich G, Tonegawa S. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu K, Deng A, Fu X, Xu A, Liu X. Proc Natl Acad Sci USA. 2014;111:397–402. doi: 10.1073/pnas.1318843111. [DOI] [PMC free article] [PubMed] [Google Scholar]