Abstract
Traditional Chinese medicines (TCMs) have important therapeutic value in long-term clinical practice. However, because TCMs contain diverse ingredients and have complex effects on the human body, the molecular mechanisms of TCMs are poorly understood. In this work, we determined the gene expression profiles of cells in response to TCM components to investigate TCM activities at the molecular and cellular levels. MCF7 cells were separately treated with 102 different molecules from TCMs, and their gene expression profiles were compared with the Connectivity Map (CMAP). To demonstrate the reliability and utility of our approach, we used nitidine chloride (NC) from the root of Zanthoxylum nitidum, a topoisomerase I/II inhibitor and α-adrenoreceptor antagonist, as an example to study the molecular function of TCMs using CMAP data as references. We successfully applied this approach to the four ingredients in Danshen and analyzed the synergistic mechanism of TCM components. The results demonstrate that our newly generated TCM data and related methods are valuable in the analysis and discovery of the molecular actions of TCM components. This is the first work to establish gene expression profiles for the study of TCM components and serves as a template for general TCM research.
Introduction
Traditional Chinese medicine (TCM), a system of ancient medical practices that differs in methodology and philosophy from modern medicine, plays an important role in health maintenance for the peoples of Asia and is considered a complementary or alternative medical system in most Western countries1. Despite the important therapeutic value of TCMs, great challenges remain in understanding the scientific basis of TCMs at the molecular level and from a systemic perspective. The recent application of state-of-the-art technologies in chemical biology to characterize commonly used TCM formulae has provided the means to identify biological targets for the active ingredients in TCMs2, 3. However, as TCMs contain a large number of ingredients and many of the active ingredients of TCMs have effects on multiple diseases, the combinatorial rules and roles of most TCM formulae in complex diseases remain to be elucidated.
Recently, there has been growing interest in incorporating expression microarrays as an effective technology for drug development. Since the emergence of the Connectivity Map (CMAP), a novel pathway-independent approach employing gene expression profiles, numerous achievements have been made in the field of drug repurposing, target discovery and elucidating mechanisms of action4. The CMAP database is a collection of gene expression profiles from cultured human cell lines treated with drugs. Moreover, pattern-matching software was applied to mine these data and compare gene expression signatures to identify connections among small molecules, genes and diseases5, 6. The current version of the CMAP (build 02) contains 6100 expression profiles reflecting 1309 bioactive compounds (http://www.broadinstitute.org/cmap/). To date, the CMAP database has been employed in several studies of TCMs. In a recent study published in Cell, Liu et al.7 employed the CMAP database to identify candidate drugs for the treatment of obesity. Celastrol, a pentacyclic triterpene extracted from the roots of Tripterygium wilfordii (thunder god vine) plant, increases leptin sensitivity to suppress food intake and dramatically reduce body weight in obese mice. Wen et al. used the CMAP database8 to identify the model TCM formula Si-Wu-Tang (SWT), which is widely used for women’s health, as a nuclear factor erythroid 2-related factor 2 (Nrf2) activator and phytoestrogen. These studies demonstrate the feasibility of combining microarray-based gene expression profiles with CMAP mining to elucidate the mechanisms of action and discover the targets/pathways of TCM components.
However, compounds in the CMAP database mainly include US Food and Drug Administration (FDA)-approved and experimental drugs, most of which are not derived from TCMs, and research on the gene expression profiles of TCM molecules has been sparse. Thus, the current work seeks to establish the public and unified gene expression profiles of TCM components constructed according to the CMAP database. Gene expression profiles were produced from a human breast cancer epithelial cell line (MCF7) treated with 102 TCM ingredients to clarify the effects of TCM molecules on gene expression levels. The elucidation of the gene expression profiles, targets/pathways of small molecules, and mechanisms of activity of Chinese herbs and TCM formulae by combining the CMAP database and other bioinformatics methods will contribute to the efficacy of pharmacological prediction and drug discovery. As described below, we have employed gene expression profiles to mine molecular functions of TCM components and elucidate the synergistic mechanisms of TCM molecules, and the analytical results were validated with functional experiments.
Results
Generation of gene expression profiles for 102 TCM components
The schematic view of data construction and processing is presented in Fig. 1. All molecules were derived from TCMs and were mainly active ingredients in Chinese herbs and TCM formulae. MCF7 cells were then treated with the derived molecules and total RNA was extracted for microarray analysis. Finally, the gene expression profiles of TCM components were established for the study of TCMs. It is possible that some gene expression profiles of TCM ingredients have been reported in previous studies. However, we produced the gene expression profiles on a unified platform that was more conducive to the collective analysis of the different ingredients. In total, 102 components are provided in Table 1. The raw data of gene expression profiles of TCM components are available through the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), and the GEO series accession number is GSE85871. The gene expression profile data can be analyzed in combination with public database CMAP and other bioinformatics methods.
Figure 1.
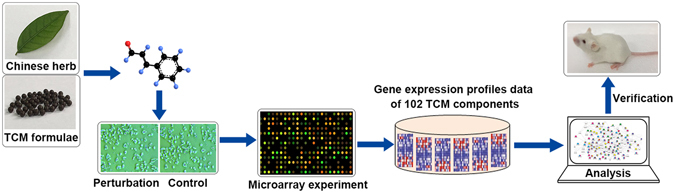
The schematic view of data construction and process.
Table 1.
The list of 102 molecules in TCMs.
| NO. | Molecules | Molecular formula | Concentration | NO. | Molecules | Molecular formula | Concentration |
|---|---|---|---|---|---|---|---|
| 1 | Glycyrrhizic acid | C42H62O16 | 10 μM | 52 | 1β-hydroxyalantolactone | C15H20O3 | 10 μM |
| 2 | Hydroxysafflor yellow A | C27H32O16 | 10 μM | 53 | Salvianic acid A sodium | C9H10O5 | 10 μM |
| 3 | Anhydroicaritin | C21H20O6 | 10 μM | 54 | Isoalantolactone | C15H20O2 | 10 μM |
| 4 | Hyperoside | C21H20O12 | 10 μM | 55 | Alantolactone | C15H20O2 | 10 μM |
| 5 | Hesperidin | C28H34O15 | 10 μM | 56 | Resibufogenin | C24H32O4 | 1 μM |
| 6 | Puerarin | C21H20O9 | 10 μM | 57 | Bufalin | C24H34O4 | 1 μM |
| 7 | Aconitine | C34H47NO11 | 10 μM | 58 | Arenobufagin | C24H32O6 | 1 μM |
| 8 | Stachydrine hydrochloride | C7H14ClNO2 | 10 μM | 59 | Cinobufagin | C26H34O6 | 1 μM |
| 9 | Ephedrine hydrochloride | C10H16ClNO | 10 μM | 60 | Bufotoxin | C40H60N4O10 | 1 μM |
| 10 | Berberine | C20H18NO4 + | 10 μM | 61 | Telocinobufagin | C24H34O5 | 1 μM |
| 11 | Ginkgolide B | C20H24O10 | 10 μM | 62 | Bufotaline | C26H36O6 | 1 μM |
| 12 | Bilobalide | C15H18O8 | 10 μM | 63 | Cinobufotalin | C26H34O7 | 1 μM |
| 13 | Andrographolide | C20H30O5 | 10 μM | 64 | Salidroside | C14H20O7 | 10 μM |
| 14 | Paeoniflorin | C23H28O11 | 10 μM | 65 | Daidzin | C21H20O9 | 10 μM |
| 15 | Tanshinone IIA | C19H18O3 | 10 μM | 66 | Schisantherin A | C30H32O9 | 10 μM |
| 16 | Lobetyolin | C20H28O8 | 10 μM | 67 | Schizandrin | C24H32O7 | 10 μM |
| 17 | Emodin | C15H10O5 | 10 μM | 68 | Oxymatrine | C15H24N2O2 | 10 μM |
| 18 | Ginsenoside Rg1 | C42H72O14 | 10 μM | 69 | Matrine | C15H24N2O | 10 μM |
| 19 | Ginsenoside Rb1 | C54H92O23 | 10 μM | 70 | Osthole | C15H16O3 | 10 μM |
| 20 | Astragaloside IV | C41H68O14 | 10 μM | 71 | Silybin | C25H22O10 | 10 μM |
| 21 | Saikosaponin A | C42H68O13 | 10 μM | 72 | Oleanic acid | C30H48O3 | 10 μM |
| 22 | Saikosaponin D | C42H68O13 | 10 μM | 73 | Phillyrin | C27H34O11 | 10 μM |
| 23 | Salvianolic acid B | C36H30O16 | 10 μM | 74 | Curculigoside | C22H26O11 | 10 μM |
| 24 | Chlorogenic acid | C16H18O9 | 10 μM | 75 | Scutellarein | C15H10O6 | 10 μM |
| 25 | Ferulic acid | C10H10O4 | 10 μM | 76 | β-ecdysterone | C27H44O7 | 10 μM |
| 26 | Gastrodin | C13H18O7 | 10 μM | 77 | Strychnine | C21H22N2O2 | 10 μM |
| 27 | Acteoside | C29H36O15 | 10 μM | 78 | Magnolol | C18H18O2 | 10 μM |
| 28 | Imperatorin | C16H14O4 | 10 μM | 79 | Honokiol | C18H18O2 | 10 μM |
| 29 | Artemisinin | C15H22O5 | 10 μM | 80 | Geniposide | C17H24O10 | 10 μM |
| 30 | Resveratrol | C14H12O3 | 10 μM | 81 | Gallic acid | C7H6O5 | 10 μM |
| 31 | Hyodeoxycholic acid | C24H40O4 | 10 μM | 82 | Notoginsenoside R1 | C47H80O18 | 10 μM |
| 32 | Deoxycholic acid | C24H40O4 | 10 μM | 83 | Liquiritin | C21H22O9 | 10 μM |
| 33 | Ursodeoxycholic acid | C24H40O4 | 10 μM | 84 | L-scopolamine | C17H21NO4 | 10 μM |
| 34 | Chenodeoxycholic acid | C24H40O4 | 10 μM | 85 | Gentiopicroside | C16H20O9 | 10 μM |
| 35 | Cholic acid | C24H40O5 | 10 μM | 86 | Benzoylhypaconitine | C31H43NO9 | 10 μM |
| 36 | Cinnamic acid | C9H8O2 | 10 μM | 87 | Benzoylaconitine | C32H45NO10 | 10 μM |
| 37 | Cinnamaldehyde | C9H8O | 10 μM | 88 | Tetrahydropalmatine | C21H25NO4 | 10 μM |
| 38 | Muscone | C16H30O | 10 μM | 89 | Hypaconitine | C34H45NO11 | 10 μM |
| 39 | Isoborneol | C10H18O | 10 μM | 90 | Glycyrrhizic acid | C42H62O16 | 10 μM |
| 40 | Borneol | C10H18O | 10 μM | 91 | 6-gingerol | C17H26O4 | 10 μM |
| 41 | Benzyl benzoate | C14H12O2 | 10 μM | 92 | Macrozamin | C13H24N2O11 | 10 μM |
| 42 | Ginsenoside Rb3 | C53H90O22 | 10 μM | 93 | Sennoside A | C42H38O20 | 10 μM |
| 43 | Ginsenoside Rc | C53H90O22 | 10 μM | 94 | Oridonin | C20H28O6 | 1 μM |
| 44 | Ginsenoside Rb2 | C53H90O22 | 10 μM | 95 | Dioscin | C45H72O16 | 1 μM |
| 45 | Ginsenoside Re | C48H82O18 | 10 μM | 96 | (+) 2-(1-hydroxyl-4-oxocyclohexyl) ethyl caffeate | C17H20O6 | 10 μM |
| 46 | Ginsenoside Rd | C48H82O18 | 10 μM | 97 | Bruceine D | C20H26O9 | 10 μM |
| 47 | Nitidine chloride | C21H18ClNO4 | 10 μM | 98 | Narciclasine | C14H13NO7 | 10 μM |
| 48 | Protocatechuic aldehyde | C7H6O3 | 10 μM | 99 | Santonin | C15H18O3 | 10 μM |
| 49 | Britanin | C19H26O7 | 10 μM | 100 | Ainsliadimer A | C30H34O7 | 1 μM |
| 50 | Japonicone A | C32H40O7 | 10 μM | 101 | Chelerythrine | C21H18NO4 + | 1 μM |
| 51 | Bacopaside I | C46H74O20S | 10 μM | 102 | Sanguinarine | C20H14NO4 + | 1 μM |
Analysis of TCM component activities
We first performed a comparison analysis between the gene expression profiles in response to TCM components and the CMAP database to discover their molecular functions. Nitidine chloride (NC, Fig. 2a) is a natural phytochemical alkaloid and a major active compound isolated from the well-known traditional Chinese medicinal herb Zanthoxylum nitidum (Roxb.) DC. Previous studies have reported that NC is a potential anti-tumour drug via the modulation of multiple targets/pathways9–11. In the present study, the gene expression profiles of NC-treated MCF7 cells were selected to search the CMAP database. The query signature consisted of 752 genes (173 up-regulated and 579 down-regulated; Supplementary Data 1) that were simultaneously submitted to the CMAP database for analysis.
Figure 2.
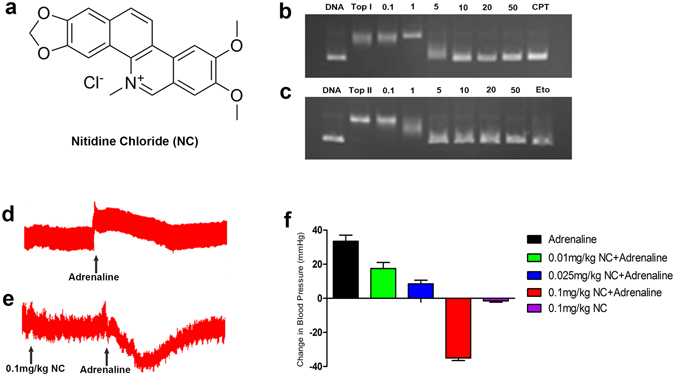
The results of validation experiments. (a) Chemical structure of NC. (b) Effect of NC on TopI mediated DNA relaxation indifferent concentration. Lane 1, supercoiled pBR322 plasmid DNA; lane 2, DNA + TopI; lane 3–8, DNA + TopI + NC (0.1, 1, 5, 10, 20, 50 μM); lane 9, DNA + TopI + 50 μM camptothecin. (c) Inhibition of TopII relaxation activity. Lane 1, supercoiled pBR322 plasmid DNA; lane 2, DNA + TopII; lane 3–8, DNA + TopII + NC (0.1, 1, 5, 10, 20, 50 μM); lane 9, DNA + TopII + 50 μM etoposide. (d) The effect of adrenaline (5 μg/kg) on arterial blood pressure. (e) NC (0.1 mg/kg) caused reversal of the adrenaline pressor response. (f) The effect of NC (0.01, 0.025, 0.1 mg/kg) on the arterial blood pressure response to adrenaline.
The similarity between the gene expression profiles of the query signature and a CMAP instance was measured using the connectivity score (from −1 to 1). A highly positive connectivity score indicates inducement of the expression of the query signature by the corresponding drug. The CMAP yielded highly positive connectivity scores for NC-treated MCF7 cells. In the detailed results of CMAP analysis, the top 10 instances of positive correlations are presented in Table 2. The results revealed a total of five compounds in the top ten instances, including irinotecan, phenoxybenzamine, hycanthone, camptothecin, and daunorubicin. Among these five compounds, irinotecan and camptothecin are topoisomerase I inhibitors, and daunorubicin is a topoisomerase II inhibitor, as reported in previous studies12–14. Furthermore, phenoxybenzamine is a known α-adrenoreceptor antagonist in the treatment of hypertension, but there have been no reports that NC acts as an α-adrenoreceptor antagonist. An extension of this finding would be to hypothesize that NC might perform activities based on same or similar activities as the compounds with the highest positive connectivity scores.
Table 2.
Top CMAP analysis of nitidine chloride in detailed results.
| Rank | Batch | Cmap name | Dose | Cell | Score |
|---|---|---|---|---|---|
| 1 | 1090 | irinotecan | 100 µM | MCF7 | 1 |
| 2 | 1082 | irinotecan | 100 µM | MCF7 | 0.917 |
| 3 | 726 | phenoxybenzamine | 12 µM | MCF7 | 0.895 |
| 4 | 758 | phenoxybenzamine | 12 µM | MCF7 | 0.871 |
| 5 | 1091 | irinotecan | 100 µM | PC3 | 0.792 |
| 6 | 708 | hycanthone | 11 µM | MCF7 | 0.781 |
| 7 | 687 | camptothecin | 11 µM | MCF7 | 0.747 |
| 8 | 1084 | daunorubicin | 1 µM | MCF7 | 0.746 |
| 9 | 755 | phenoxybenzamine | 12 µM | MCF7 | 0.741 |
| 10 | 1088 | daunorubicin | 1 µM | MCF7 | 0.727 |
Topoisomerase I/II are main targets for antitumor drugs, and some studies have reported that NC inhibits topoisomerase activities15–17. However, few reports have conducted experiments on the simultaneous inhibition of topoisomerase I/II activities by NC. To validate the topoisomerase I/II inhibitory activity of NC, we evaluated the effect of NC on the stabilization of the cleavable complex that forms in the presence of topoisomerase I/II and DNA. As illustrated in Fig. 2b and c, NC was active against the topoisomerase I/II-mediated relaxation of supercoiled DNA. In addition, the effect of NC completely inhibited topoisomerase I at a concentration of 10 μM and completely inhibited the cleavage activity of topoisomerase II at 5 μM.
Furthermore, to validate the function of NC as an α-adrenoreceptor antagonist, we performed the classic adrenaline reversal experiment. α-Adrenoreceptor-blocking agents have the ability to cause a reversal of the adrenaline pressor response18. Adrenaline administration to animals results in an increased mean arterial blood pressure given that its α-receptor agonist properties predominate. However, when an α-adrenoreceptor antagonist is present, the β-receptor agonist property of adrenaline plays a leading role and causes a decrease in arterial pressure or reversal of the pressor response. As shown in Fig. 2d and e, after rats were injected with adrenaline (5 μg/kg), the arterial blood pressure increased abruptly. However, after pre-treatment with an injection of NC (0.1 mg/kg) followed by an injection of adrenaline (5 μg/kg), the arterial blood pressure significantly decreased. In addition, NC dose-dependently reduced the adrenaline pressor response (Fig. 2f). NC (0.1 mg/kg) caused a significant reversal of adrenaline pressor response with a reduction to normal levels (35 ± 1.4 mmHg).
In summary, most molecules have more than one effect, especially TCM molecules. The results of CMAP analysis showed that topoisomerase I/II inhibitor and α-adrenoreceptor antagonist produced high positive connectivity scores. After topoisomerase I/II inhibitory activity assays and adrenaline reversal experiments, NC was validated as an effective topoisomerase I/II inhibitor and candidate α-adrenoreceptor antagonist.
Synergistic mechanism of TCM components
TCMs consist of numerous ingredients, and the therapeutic effect of TCMs mainly originates from the synergistic effect of these multiple components19. Synergy is one of the fundamental advantages of multicomponent therapeutics, indicating that combinational effects are greater than the sum of the individual effects20, 21. However, the mechanisms of synergistic action remain poorly understood. We attempted to conduct a systematic analysis to explore the rationality of the synergistic effects of the principal compounds in Danshen (Salvia miltiorrhiza roots). Danshen is one of the most versatile TCMs based on its properties of improving microcirculation, causing coronary vasodilatation, suppressing the formation of thromboxane, inhibiting platelet adhesion and aggregation, and protecting against myocardial ischemia, among other effects22, 23. Therefore, Danshen has been used to treat cardiovascular diseases, including coronary artery disease, hypercholesterolemia, hypertension, arrhythmias, and other cardiovascular diseases, for hundreds of years23.
In this work, we selected the four main active components (tanshinone IIA, salvianic acid A sodium, protocatechuic aldehyde and salvianolic acid B) in Danshen to elucidate their synergistic effect in the treatment of cardiovascular diseases. In this study, the differential expression levels of genes in MCF7 cells treated with each compound and four mixtures (Supplementary Data 2) were selected and analyzed in the CMAP database. Only drugs associated with cardiovascular diseases with positive connectivity scores and p < 0.01 were summarized (Table 3). The CMAP analysis results of four mixtures suggested that 11 drugs were related to cardiovascular diseases, including cardiovascular agents, cardiotonic agents, vasodilator agents, anti-arrhythmia agents, antihypertensive agents and calcium channel blockers. Most of these drugs ranked at or near the top, which indicated significant positive enrichment. However, the CMAP analysis results of single compounds indicated that fewer drugs were associated with cardiovascular diseases and received a lower ranking. The CMAP analysis results indicated that four mixtures possessed more positive effects on the therapeutic efficacy of cardiovascular diseases than any other individual components.
Table 3.
Results of CMAP analysis.
| Rank | Cmap name | Enrichment | p | Therapeutic Use | |
|---|---|---|---|---|---|
| Four mixtures | 1 | phenoxybenzamine | 0.981 | 0 | Antihypertensive Agents, Vasodilator Agents |
| 2 | lanatoside C | 0.981 | 0 | Cardiovascular Agents | |
| 3 | digitoxigenin | 0.978 | 0 | Cardiotonic Agents | |
| 5 | digoxigenin | 0.967 | 0 | Cardiotonic Agents | |
| 7 | ouabain | 0.962 | 0 | Cardiotonic Agents | |
| 8 | digoxin | 0.959 | 0 | Anti-Arrhythmia Agents, Cardiotonic Agents | |
| 28 | proscillaridin | 0.968 | 0.00004 | Cardiotonic Agents | |
| 60 | beta-escin | 0.663 | 0.00427 | Cardiovascular Agents | |
| 62 | bepridil | 0.778 | 0.00464 | Anti-Arrhythmia Agents, Vasodilator Agents | |
| 73 | strophanthidin | 0.738 | 0.00927 | Cardiovascular Agents | |
| 75 | tetrandrine | 0.733 | 0.00989 | Calcium Channel Blockers | |
| Tanshinone IIA | 15 | phenoxybenzamine | 0.85 | 0.00072 | Antihypertensive Agents, Vasodilator Agents |
| 17 | lanatoside C | 0.738 | 0.00085 | Cardiovascular Agents | |
| 23 | beta-escin | 0.711 | 0.00167 | Cardiovascular Agents | |
| 44 | ouabain | 0.733 | 0.00989 | Cardiotonic Agents | |
| Salvianic acid A sodium | 16 | timolol | 0.761 | 0.00621 | Anti-Arrhythmia Agents |
| 17 | labetalol | 0.761 | 0.00623 | Antihypertensive Agents | |
| 18 | fenofibrate | 0.847 | 0.00693 | Hypolipidemic Agents | |
| 20 | propranolol | 0.749 | 0.0076 | Anti-Arrhythmia Agents, Antihypertensive Agents, Vasodilator Agents | |
| protocatechuic aldehyde | 12 | pronetalol | 0.771 | 0.00539 | Antihypertensive Agents |
| 23 | guanabenz | 0.676 | 0.00927 | Antihypertensive Agents | |
| Salvianolic acid B | 15 | benfluorex | 0.796 | 0.00338 | Hypolipidemic Agents |
We also applied the algorithm of random walk with restart (RWR) to elucidate the synergistic effects of multi-components of Danshen. RWR is a widely accepted algorithm that globally scores each gene in the entire network by computing the effects of seed genes24–26. Therefore, we computed the cardiovascular effect scores of the mixture of the four components and each single component separately. In addition, to verify whether their effect scores regarding cardiovascular disease were notable, we computed their Z-scores and compared them with random counterparts. The results are listed in Table 4. The mixture of the four components obtained a high effect score of 0.72, which was much higher than that of any single component. An absolute Z-score greater than 3 is generally deemed as a threshold, which suggests a statistically significant deviation between the actual value and the random values. Thus, the Z-score of 4.958 for the mixture of the four components was much higher than that of any single component, which suggested that the synergy among the four components is significantly associated with the effects of Danshen on cardiovascular disease. Therefore, the RWR algorithm showed that the synergistic effect of the mixture of the four components on multiple target genes outperforms the effects of single components.
Table 4.
Effect scores and Z-scores of four mixtures and its single component to cardiovascular disease.
| Components | Target number | Effect scores | Z-scores |
|---|---|---|---|
| Four mixtures | 966 | 0.720 | 4.958 |
| Tanshinone IIA | 549 | 0.396 | 2.319 |
| Salvianic acid A sodium | 326 | 0.231 | 1.121 |
| protocatechuic aldehyde | 354 | 0.266 | 2.274 |
| Salvianolic acid B | 170 | 0.131 | 1.627 |
In addition, cardioprotective effect assays were conducted on single compounds and four mixtures to validate the synergistic mechanisms of components. As showed in Fig. 3, hypoxia/reoxygenation (H/R)-induced cell injury significantly reduced cell viability. At concentration of 10 µM, all single compounds and four mixtures could notably protect H9c2 cells from H/R-induced cell injury. The cell viability of four mixtures was higher than that of any single compound. Thereby, four mixtures can exert more cardioprotective effect than single compounds, which sheds light on the synergistic therapeutic mechanisms of TCM components.
Figure 3.
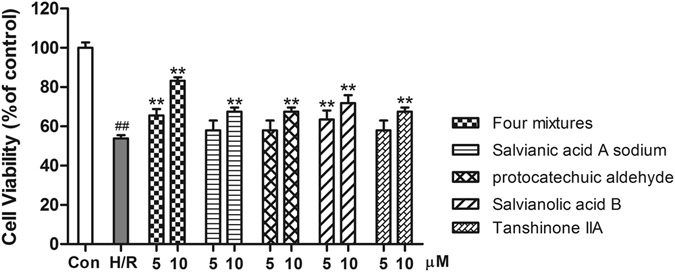
Effects of single compounds and four mixtures on H/R-induced H9c2 cell injury. Cell viability by CCK-8 assay. ## P < 0.01 vs. Con; **P < 0.01 vs. H/R. Data are shown as the mean ± SD of three independent experiments.
Discussion
With the application of TCMs in various diseases receiving increasing attention worldwide, more and more efforts have been made to elucidate the mechanisms of TCMs. TCMs are highly diverse, with abundant ingredients, making TCMs a potential repository of molecules for drug discovery. Thus, developing an effective method to investigate the molecular mechanisms of TCMs is necessary. Since the emergence of microarray techniques, gene expression profiling has been widely used in the field of TCMs, and various achievements have been attained using this methodology. Lee et al.27 used gene expression profiles combined with the CMAP database to elucidate the mechanism of the Chinese herbal medicine berberine, which inhibits global protein synthesis and basal AKT activity and induces endoplasmic reticulum (ER) stress and autophagy. Li et al.28 adopted gene expression profiles to elucidate the multi-compound, multi-target and multi-pathway mechanism of action of a TCM formula, QiShenYiQi, on myocardial infarction. Therefore, gene expression profiles can provide new insight into the mechanisms of TCMs at the molecular and gene levels.
In this study, we established the gene expression profiles of cells in response to 102 TCM molecules, and this information will likely accelerate progress in understanding the molecular mechanisms of TCMs. Then, we used gene expression profiles combined with the CMAP database to explore the molecular functions of NC. After validation experiments, NC was identified as a topoisomerase I/II inhibitor and a potential new α-adrenoreceptor antagonist. This result indicates that some compounds in TCMs have more than one function and the approach is efficient. In addition, we analyzed the gene expression profiles of four active ingredients in Danshen and elucidated the theory of synergistic action in TCM multicomponent therapeutics. These results demonstrate the reliability and utility of our gene expression profile data. This approach provides an integrative platform to simultaneously analyze a large number of genes associated with TCM ingredients and offers convenience to researchers.
We only selected 102 molecules from TCMs, and these molecules only represent a small part of the numerous compounds in TCMs. In subsequent studies, we will further expand the number of small molecules. Such data will provide more possibilities for research on the molecular mechanisms of TCMs. For example, the theory of detoxification in TCMs is that it occurs mainly through herbal ingredient interactions, which can likely be elucidated by analyzing the gene expression profiles of molecules at the gene level. In the future, the gene expression profiles of cells in response to TCM components can help researchers to perform their own bioinformatics analyses to clarify the mechanisms of action of TCMs in real time. In short, this is just the beginning, and additional outcomes will depend on the use of the data.
Methods
The establishment of gene expression profiles of TCM components
The selection of components in TCMs
We selected 102 small molecules that are commonly found in Chinese herbs and TCM formulae, such as Radix Salviae Miltiorrhizae, Rhizoma Coptidis, and Shexiang Baoxin Pill. Most of these 102 compounds are the quality control components of TCMs from the China Pharmacopoeia and are selected to represent a broad range of activities and diverse structures.
Cell lines
The gene expression profile data for 102 molecules were produced for MCF7 cells. MCF7 cells are commonly used in the worldwide laboratories as a reference cell line, have clear biological characteristics, remain stable after prolonged culture, and can be cultured in microplates. In addition, MCF7 is the one of the main cell lines used in the CMAP database4. Furthermore, the MCF7 cell line was procured from American Type Culture Collection (ATCC) and cultured in MEM/EBSS (Hyclone) supplemented with 10% foetal bovine serum, 1 mmol/L sodium pyruvate, 0.1 mmol/L MEM non-essential amino acids, 100 unit/mL penicillin, and 100 mg/mL streptomycin in an incubator containing 5% CO2 at 37 °C.
Small molecule-treated cells
The gene expression profiles can be affected by the concentration and duration of compound treatment. According to the CMAP database, the concentration of small molecules was set at a single dose of 10 μM, which is also internationally recognized as a reasonable concentration for high-throughput screening4. Simultaneously, the cell’s survival rate was investigated upon treatment with compounds at a concentration of 10 μM using the MTT assay. If the cell survival rate was <40%, the number of cells could not meet the needs of microarray analysis. Therefore, the concentration of compounds was decreased to 1 μM until the cell survival rate was >40%. For each compound, the duration of the treatment was 12 h and two biological replicates were performed. Full details of small molecules and treatment conditions are provided in Table 1.
RNA isolation and quality
After pre-treatment, MCF7 cells were harvested and total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, US) according to the manufacturer’s instructions. To control the quality and purity of isolated total RNA, formaldehyde agarose gel electrophoresis and spectrophotometry (NanoDrop, Wilmington, DE, USA) were performed. Moreover, DMSO-treated cells were selected as a control.
Microarray analysis
The gene expression profiles were assessed using microarray technology with Affymetrix Human Genome U133A 2.0 (Santa Clara, CA, US), which was used in numerous studies, covering 18,400 transcripts and including 14,500 characterized human genes29–31. Total RNA was purified using a QIAGEN RNeasy Kit (GmBH, Germany) according to the manufacturer’s protocols, and biological duplicates were employed for each cell line. Then, total RNA was used to generate double-stranded cDNA and biotin-labelled cRNA. Following fragmentation, cRNA products were hybridized to an Affymetrix Human Genome U133A 2.0. GeneChip array, and hybridized arrays were washed and stained using a GeneChip® Hybridization, Wash and Stain Kit (Affymetrix). Finally, the fluorescent signals were measured with the GeneChip® Scanner 3000 (Affymetrix).
The data from this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO series accession number: GSE85871). Raw data (CEL files) were normalized by MAS 5.0 algorithm, Gene Spring Software 11.0 (Agilent Technologies, Santa Clara, CA, US). Subsequently, quality control (QC) analysis was performed on the expression data, including overview of QC analysis, quality on-chip analysis, comparative analysis among multiple samples, PCA, and RNA degradation analysis, by using the affy package in R language. The results indicated that all data met the requirements for bioinformatics analysis.
Similarity search against the CMAP
In the investigation of the function of a small molecule, a similarity search against the CMAP was performed. For each treatment of one compound (one treated versus the corresponding DMSO pair), Fold Change was used to filter the differential expression probes which was calculated as follows: first of all, average normalized expression values were calculated for two replications for each small molecule and DMSO; second, Fold Change was represented by average normalized expression value of treatment divided by the average normalized expression value corresponding to DMSO. Then, differential expression probes were selected according to fold change (e.g., FC ≥2 or ≤0.5), the criteria used for filtering the differential expression probes were consistent among the small molecules. The gene-expression signature of the compound was represented by two sets (‘up-’ and ‘down-’ probe sets, saved as. grp files and required as the inputs for CMAP), which was made up by the significant up/down regulation probes respectively. The query in the CMAP was performed as a “quick query” in the query section of http://portals.broadinstitute.org/cmap/.
Topoisomerase I/II inhibitory activity assay
Topoisomerase I/II inhibitory activity assays were conducted according to the procedure described in a previous study32. Compound concentrations, pBR322 plasmid DNA (0.25 μg) and 1 unit of TopI (TaKaRa Biotechnology Co., Ltd., Dalian) were combined in a final volume of 20 μL buffer (35 mM pH 8.0 Tris-HCl, 72 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol, 5 mM spermidine, 0.1% bovine serum albumin). Then, the mixed reaction buffers were incubated for 15 min at 37 °C and stopped by the addition of 2 μL of 10× loading buffer. The samples were analyzed by electrophoresis on a 0.8% agarose gel in TAE (Tris-acetate-EDTA) for 1 h and then stained with 0.5 μg/mL of ethidium bromide for 30 min. Finally, the DNA band was visualized using UV light and photographed with a G:BOX gel imaging system (Gene Co., Ltd., Hong Kong).
The DNA TopIIα inhibitory activity of the compounds was measured using a Topoisomerase IIα Drug Screening Kit (TopoGEN, Inc.). Compound concentrations, pBR322 plasmid DNA (0.25 μg) and 0.75 unit of TopII were combined in a final volume of 20 μL buffer (50 mM pH 8.0 Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol, 30 μg/Ml bovine serum albumin, 2 mM ATP). Then, the following experimental procedures were performed according to Topoisomerase I inhibitory activity assay.
Adrenaline reversal experiments
Male Sprague-Dawley rats (300–350 g) were obtained from the Slac Laboratory Animal Co., Ltd. (Shanghai, China), and given free access to tap water and food pellets. All animal experiments were carried out under standard conditions according to the guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Committee on the Ethics of Animal Experiments of the Second Military Medical University, China. The animals were anesthetized with urethane (1 g/kg) with minimal suffering. Mean blood pressure was continuously monitored from a cannulated carotid artery using a pressure transducer to a polygraph (Alcott Biotech Co., Ltd. Shanghai). All care was taken that animals could breathe normally. Adrenaline and NC were administered through a catheter inserted into the tail vein. Experiments were performed only after completion of the operative procedures to permit arterial blood pressure to stabilize.
Animals were randomly divided into five groups. Group A rats received a single injection of adrenaline (5 μg/kg). Group B, C, and D rats received an injection of NC (0.01, 0.025, and 0.1 mg/kg, respectively) after a 2-min injection of adrenaline (5 μg/kg). Group E rats received a single injection of NC (0.1 mg/kg). After the experiments, all animals were sacrificed by tail vein air injection.
RWR-based evaluation of compounds’ effect
Data preparation
In total, 301 distinct genes associated with cardiovascular diseases were collected by searching the key word “Cardiovascular Disease” in a plugin of Cytoscope, DisGeNet. In addition, Version 10 of the STRING database was employed as a resource of the PPI network. We extracted interactions with confidence scores greater than 0.9 or target-related edges with maximum scores. Thus, a PPI network was constructed, and its greatest weighted component had 10,270 nodes and 176,739 edges.
RWR algorithm
RWR can globally score seed genes’ effects on each gene in the entire network and can be denoted as follows:
| 1 |
where P is the column-normalized adjacency matrix of the network, χ 0 is the initial vector that indicates the seed nodes’ strength, and χ t denotes a probability vector in which the ith element holds the chance of the walker being at v i node at step t. Parameter r indicates a restart probability indicating the likelihood that the walker will return to seed set at step t. In practice, 0.3 is an optimal value. A steady state of χ t will be reached after performing the eq. (1) iteratively with sufficient time, which can disclose to which extent each node is affected by seed nodes.
In this paper, disease genes and differentially expressed genes under the treatment of the molecules are regarded as seed nodes separately. To calculate disease effect, the corresponding component of the disease gene in the initial vector is χ 0(v) = 1. To compute drug effect, if node v is a drug target, we define its corresponding component in the initial vector χ 0 as χ 0(v) = 0.01. After running the RWR with these initial vectors, we obtain the drug and disease effect vectors χ drug and χ disease, respectively. Then, we compute the inner product between the effect vectors of drug and disease to measure how the drug-affected network and disease-affected network overlap. The equation is
| 2 |
To measure the statistical significance of the score s, we randomly generate 1000 counterparts that have the same number of drug targets and calculate s scores. Suppose and Δs r are the mean and standard deviation of these random counterparts’ scores, respectively. Then, the z-score can quantify the score difference among the original seed set and counterparts as follows:
| 3 |
Typically if the z-score is greater than 3, the drug can be considered as exhibiting statistically stronger effects than random cases.
Cardioprotective effect assay
Cardioprotective effect assays were performed by determing the effects of single compounds and four mixtures against H/R-induced H9c2 cells injury. Rat H9c2 cardiomyocyte cell line was obtained from Chinese Academy of Sciences Cell Bank (Shanghai, China) and maintained in DMEM supplemented with 10% foetal bovine serum at 37 °C in CO2 incubation. To mimic the ischemic injury in vitro, H9c2 cells were placed in a humidifed chamber containing the cells with 95% N2 and 5% CO2 for 4 h and maintained in serum-free and glucose-free DMEM. Then, the cells were transferred to normal conditions for 20 h and cultured in routine culture medium to achieve reoxygenation. The single compounds and four mixtures (1:1:1:1) were added 1 h before the hypoxia period. Cell viability was determined by Cell Counting Kit-8 assay (CCK-8; Dojindo, Kumamoto, Japan).
Electronic supplementary material
Acknowledgements
The work was supported by Professor of Chang Jiang Scholars Program, NSFC (81230090, 81520108030, 81573318, 31271409, 81373301, 1302658, 61372194, 81260672), Shanghai Engineering Research Center for the Preparation of Bioactive Natural Products (10DZ2251300), the Scientific Foundation of Shanghai China (12401900801, 13401900103, 13401900101), National Major Project of China (2011ZX09307-002-03), special Fund for strategic pilot technology Chinese Academy of Sciences (XDA08020104) and the National Key Technology R&D Program of China (2012BAI29B06).
Author Contributions
C.L. performed the majority of experiments and data analysis, as well as prepared the manuscript; X.W. participated in data analysis, and drafted the manuscript; X.W., J.Z. and H.Z. participated in data analysis; J.S., S.L. and R.L. performed part of the experiment; X.L., H.L. and W.Z. designed the experiments and prepared the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Chao Lv and Xueting Wu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00535-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Honglin Li, Email: hlli@ecust.edu.cn.
Xuan Li, Email: lixuan@sibs.ac.cn.
Weidong Zhang, Email: wdzhangy@hotmail.com.
References
- 1.Efferth T, Li PCH, Konkimalla VSB, Kaina B. From traditional Chinese medicine to rational cancer therapy. Trends. Mol. Med. 2007;13:353–361. doi: 10.1016/j.molmed.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, et al. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. PNAS. 2008;105:4826–4831. doi: 10.1073/pnas.0712365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam W, et al. The Four-Herb Chinese Medicine PHY906 Reduces Chemotherapy-Induced Gastrointestinal Toxicity. Sci. Transl. Med. 2010;2:45ra59. doi: 10.1126/scitranslmed.3001270. [DOI] [PubMed] [Google Scholar]
- 4.Qu XA, Rajpal DK. Applications of Connectivity Map in Drug Discovery and Development. Drug Discov. Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 6.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, et al. Treatment of Obesity with Celastrol. Cell. 2015;161:999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen Z, et al. Discovery of Molecular Mechanisms of Traditional Chinese Medicinal Formula Si-Wu-Tang Using Gene Expression Microarray and Connectivity Map. PLoS One. 2011;6:e18278. doi: 10.1371/journal.pone.0018278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J, et al. Nitidine chloride inhibits hepatic cancer growth via modulation of multiple signaling pathways. BMC Cancer. 2013;14:1–11. doi: 10.1186/1471-2407-14-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Z, et al. Nitidine chloride induces apoptosis and inhibits tumor cell proliferation via suppressing ERK signaling pathway in renal cancer. Food Chem. Toxicol. 2014;66:210–216. doi: 10.1016/j.fct.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 11.Pan X, et al. Nitidine Chloride inhibits breast cancer cells migration and invasion by suppressing c-Src/FAK associated signaling pathway. Cancer Lett. 2011;313:181–191. doi: 10.1016/j.canlet.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Ando M, et al. Phase I study of sequentially administered topoisomerase I inhibitor (irinotecan) and topoisomerase II inhibitor (etoposide) for metastatic non.small-cell lung cancer. Brit. J. Cancer. 1997;76:1494–1499. doi: 10.1038/bjc.1997.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin Induces Protein-linked DNA Breaks via Mammalian DNA Topoisomerase I. J. Biol. Chem. 1985;260:4873–4878. [PubMed] [Google Scholar]
- 14.Bodley A, et al. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res. 1989;49:5969–5978. [PubMed] [Google Scholar]
- 15.Gatto B, et al. Identification of topoisomerase I as the cytotoxic target of the protoberberine alkaloid coralyne. Cancer Res. 1996;56:2795–2800. [PubMed] [Google Scholar]
- 16.Makhey D, et al. Coralyne and Related Compounds as Mammalian Topoisomerase I and Topoisomerase II Poisons. Bioorgan. Med. Chem. 1996;4:781–791. doi: 10.1016/0968-0896(96)00054-5. [DOI] [PubMed] [Google Scholar]
- 17.Prado S, et al. Synthesis and cytotoxic activity of benzo[c][1,7] and[1,8]phenanthrolines analogues of nitidine and fagaronine. Bioorgan. Med. Chem. 2004;12:3943–3953. doi: 10.1016/j.bmc.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 18.Graham RM, Oates HF, Stoker LM, Stokes GS. Alpha blocking action of the antihypertensive agent, prazosin. J. Pharmacol. Exp. Ther. 1977;201:747–752. [PubMed] [Google Scholar]
- 19.Zhang F, et al. Chemical profile- and pharmacokinetics-based investigation of the synergistic property of Platycodonis Radix in Traditional Chinese Medicine formula Shengxian Decoction. J. Ethnopharmacol. 2014;152:497–507. doi: 10.1016/j.jep.2014.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, et al. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. PNAS. 2008;105:4826–4831. doi: 10.1073/pnas.0712365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Zhang B, Zhang N. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst. Biol. 2011;5:1–13. doi: 10.1186/1752-0509-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng TO. Danshen: a versatile Chinese herbal drug for the treatment of coronary heart disease. Int J Cardiol. 2006;113:437–438. doi: 10.1016/j.ijcard.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Cheng TO. Cardiovascular effects of Danshen. Int. J. Cardiol. 2007;121:9–22. doi: 10.1016/j.ijcard.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Fang H, et al. Analysis of Cynandione A’s Anti-Ischemic Stroke Effects from Pathways and Protein-Protein Interactome. PLoS One. 2015;10:e0124632. doi: 10.1371/journal.pone.0124632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang H, et al. Bioinformatics Analysis for the Antirheumatic Effects of Huang-Lian-Jie-Du-Tang from a Network Perspective. Evid-Based Compl. Alt. 2012;2013:391–392. doi: 10.1155/2013/245357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler S, Bauer S, Horn D, Robinson PN. Walking the Interactome for Prioritization of Candidate Disease Genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KH, et al. A gene expression signature-based approach reveals the mechanisms of action of the Chinese herbal medicine berberine. Sci. Rep. 2014;4:6394–6394. doi: 10.1038/srep06394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, et al. A Network Pharmacology Study of Chinese Medicine QiShenYiQi to Reveal Its Underlying Multi-Compound, Multi-Target, Multi-Pathway Mode of Action. PLoS One. 2014;9:e95004. doi: 10.1371/journal.pone.0095004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang H, et al. Transcriptome analysis of early organogenesis in human embryos. Dev. Cell. 2010;19:174–184. doi: 10.1016/j.devcel.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, et al. PML/RARalpha Targets Promoter Regions Containing PU.1 Consensus and RARE Half Sites in Acute Promyelocytic Leukemia. Cancer Cell. 2010;17:186–197. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, et al. Gene expression-based classification and regulatory networks of pediatric acute lymphoblastic leukemia. Blood. 2009;114:4486–4493. doi: 10.1182/blood-2009-04-218123. [DOI] [PubMed] [Google Scholar]
- 32.Dong G, et al. New Tricks for an Old Natural Product: Discovery of Highly Potent Evodiamine Derivatives as Novel Antitumor Agents by Systemic Structure-Activity Relationship Analysis and Biological Evaluations. J. Med. Chem. 2012;55:7593–7613. doi: 10.1021/jm300605m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


