Abstract
Background
p14ARF, a tumor suppressor protein, encoded by the p16 tumor suppressor gene, has been reported to be associated with the clinicopathological features of lung cancer. However, the evaluated outcomes were inconsistent and remained inconclusive. In this study, we conducted a meta-analysis to clarify the significance of p14ARF expression in lung cancer pathogenesis.
Materials and methods
Electronic databases, PubMed, Web of Knowledge, Embase, and CNKI, were retrieved to collect relevant articles with inclusion and exclusion criteria. Using Stata 12.0 software, 95% confidence intervals (CIs) and odds ratios (ORs) were calculated.
Results
A total of 15 eligible case–control studies that evaluated the relationship between p14ARF expression and lung cancer were included in the meta-analysis. The results demonstrated that there were significant associations between p14ARF expression and the risk of non-small-cell lung cancer (NSCLC), lung adenocarcinoma, and lung squamous carcinoma (for NSCLC, OR =11.02, 95% CI =5.30–22.92; for lung adenocarcinoma, OR =7.28, 95% CI =3.92–13.50; and for lung squamous carcinoma, OR =14.40, 95% CI =2.83–73.24). In the stratified analysis based on race, significant associations between p14ARF expression and lung cancer risk were found in Chinese population and Caucasians (for Chinese population, OR = 7.02, 95% CI =4.48–11.00 and for Caucasians, OR =4.19, 95% CI =1.42–12.38). Furthermore, the expression of p14ARF was significantly associated with the TNM-stage of lung cancer in Chinese population (OR =2.07, 95% CI =1.38–3.10).
Conclusion
p14ARF expression was significantly associated with the risk of lung cancer. In addition, the data of the meta-analysis showed that there was a significant correlation between p14ARF expression and the TNM-stage of lung cancer in Chinese population.
Keywords: p14ARF, expression, lung cancer, meta-analysis
Introduction
Lung cancer is one of the leading causes of cancer-related death around the world.1 The major histological classes are small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC), which includes lung adenocarcinoma, lung squamous carcinoma, and large cell lung cancer. NSCLC has been hardly cured with the current standard therapies, and the 5-year survival has been reported to be only 15%.2 It has been reported that smoking is a major risk factor for lung adenocarcinoma and lung squamous carcinoma.3 Although the pathological mechanism is not clear, smoking induces genetic and epigenetic abnormalities that lead to lung cancer. These genetic variations are associated with the occurrence and progression of lung cancer. On the basis of these gene mutations, many molecular targeted therapies were studied to improve the survival quality and survival rate of lung cancer patients. For instance, the molecular targeted therapy that was directed against receptor tyrosine kinases (RTKs) had a good response in the lung cancer patients who had certain gene alternations such as epidermal growth factor receptor (EGFR), anaplastic lymphomakinase (ALK), and ROS proto-oncogene 1 (ROS1).4 In addition, other gene alterations that were found in the BRAF, ERBB2, NTRK1, and MET genes also became the therapeutic targets of lung cancer.5 Gefitinib was an EGFR tyrosine kinase inhibitor and was often used in the treatment of lung cancer.6 The therapeutic outcomes indicated that lung tumor had high response rates and EGFR was a key predictive biomarker for gefitinib. Many studies have also found that EGFR mutations were one of the most frequent driver mutations in lung cancer.7,8 At the same time, researchers made efforts to find more molecular targets and developed more advanced molecular targeted agents. Although second-generation or third-generation inhibitors have been developed to cure lung cancer, more genetic alternations were still needed.
The alternate reading frame (ARF) of CDKN2A locus encodes the p14ARF protein, while the CDKN2A locus encodes the p16INK4a protein. The expression of p14ARF leads to the activation of p53 by sequestering Mdm2.9 P53 could protect cells against excessive growth through induction of the p14ARF protein. Therefore, p14ARF plays a crucial role in tumor suppression.10 It was reported that promoter methylation and gene mutations of p14ARF gene reduced the expression of p14ARF and had a significant association with the risk of lung cancer.11,12 Furthermore, studies have found that there was a significant association between p14ARF expression and risk of cancers, such as breast cancer, liver cancer, ovarian cancer, and laryngeal cancer.13–16 Although some researchers performed studies to explore the relationship between p14ARF expression and lung cancer risk, the conclusions remained unclear due to many factors, such as sample size, ethnicity, and disease subtype. Hence, the aim of this meta-analysis was to assess the potential value of p14ARF expression in lung cancer risk and clinicopathological features of lung cancer.
Materials and methods
Literature search
PubMed, Embase, Web of Knowledge, and CNKI were searched to retrieve relevant articles that evaluated the association between p14ARF expression and lung cancer. Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines were applied to perform the literature retrieval. The following terms were used in the retrieval: “lung cancer”, “lung carcinoma”, “Lung Neoplasms”, “non small cell lung cancer”, “NSCLC”, “Small-Cell Lung Cancer”, “SCLC” “p14”, “p14ARF”, “ARF”, “gene expression”, and “expression”. Additional eligible studies were identified based on references cited, reviews, and meta-analysis. The searching data were up to December 2016.
Study selection and inclusion and exclusion criteria
Two investigators independently searched and collected eligible articles. Articles that evaluated the relationship between p14ARF expression and lung cancer risk and possessed enough data were included in this meta-analysis. Furthermore, these articles must meet the following criteria: 1) articles that evaluated the association between p14ARF expression and lung cancer, 2) articles that possessed enough data for p14ARF expression and lung cancer, 3) case–control studies or cohort studies, 4) articles that have been published online, and 5) articles published in English or Chinese. If the articles did not meet the following criteria, they were excluded: 1) meta-analysis and reviews, 2) articles that lacked the necessary data of p14ARF expression and lung cancer, and 3) the data of articles were same as those of other articles.17
Data extraction and quality assessment
All data were independently extracted from the eligible publications by two reviewers. The extracted information was as follows: name of authors, publication year, data of p14ARF expression, disease type, methods of detection, sample type, clinical features, country, and ethnicity. Disagreements were resolved via discussion between the two reviewers. In addition, the Newcastle–Ottawa quality assessment scale was applied to assess the methodological quality of included articles.
Statistical analysis
Stata 12.0 (Stata Corporation, College Station, TX, USA) was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs), which quantitatively determined the association between P14ARF expression and clinicopathological features of lung cancer. Q-test and I2 statistic were used to evaluate the heterogeneity among studies.18 P<0.05 or I2>50 suggested that there was a significant heterogeneity among studies. Furthermore, if significant heterogeneity existed, the random-effects model was applied. Otherwise, the fixed-effects model was used.19,20 The funnel plot was used to evaluate the publication bias. The publication bias was quantitatively assessed according to Begg’s test and Egger’s test, while P<0.05 indicated a significant publication bias.21,22 Sensitivity analysis was performed to detect whether one single study had a significant impact on the overall estimate.
Results
Identification of eligible studies
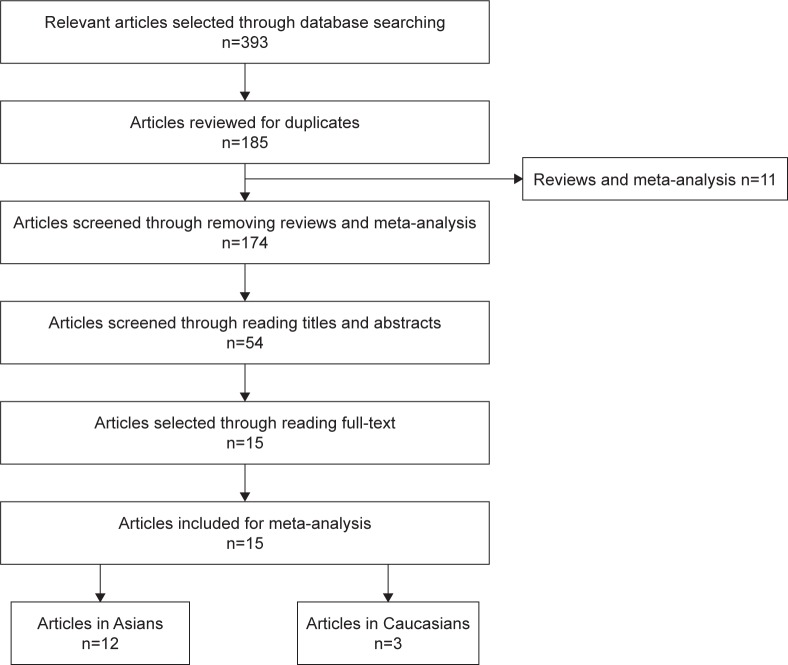
A total of 393 articles were acquired in the initial retrieval from the databases of PubMed, Embase, Web of Knowledge, and CNKI. Of these 393 articles, 208 articles overlapped with other studies, and 185 articles remained after removal of these repeated articles. Nine reviews and 2 meta-analyses were removed, which left 174 articles for title and abstract evaluation. Fifty-four articles remained after reading the titles and abstracts of relevant articles. Finally, 15 articles were left after exclusion of 39 articles according to the full-text evaluation.23–37 These articles included the data of p14ARF expression in control group and case group, method of p14ARF expression detection, race and source of patients, type of sample, evaluation criterion of staining results, and other clinical information of lung cancer patients. Of the 15 articles, 12 articles evaluated Asians patients and 3 articles assessed Caucasian patients. Moreover, 11 studies evaluated the association of p14ARF expression with differentiation of lung tumor; 9 studies assessed the relationship between p14ARF expression and TNM-stage of lung tumor; and 10 studies explored the correlation between p14ARF expression and lymph node metastasis of lung cancer. Immunohistochemistry (IHC) and Western blotting (WB) were used to detect the expression of p14ARF protein in the included studies (Figure 1; Table 1). According to the results of retrieval, this was the first meta-analysis to assess the correlation between p14ARF expression and lung cancer risk.
Figure 1.
Flow diagram of eligible articles.
Table 1.
Main characteristics of included studies
| First author | Year | Country | Ethnicity | Disease | Control
|
Case
|
Methods | Control type | Case type | Reference | Score | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||||||||
| Chen et al | 2012 | China | Asians | LC | 53 | 18 | 37 | 34 | IHC | Adjacent tissue | Tumor tissue | 23 | 8 |
| Yang et al | 2008 | China | Asians | LC | 19 | 1 | 26 | 34 | IHC | Normal tissue | Tumor tissue | 24 | 8 |
| Yang et al | 2008 | China | Asians | NSCLC | 14 | 1 | 54 | 44 | IHC | Normal tissue | Tumor tissue | 25 | 6 |
| Yang et al | 2008 | China | Asians | NSCLC | 15 | 3 | 54 | 44 | IHC | Adjacent tissue | Tumor tissue | 25 | 8 |
| Zhao et al | 2008 | China | Asians | NSCLC | 19 | 1 | 41 | 21 | IHC | Adjacent tissue | Tumor tissue | 26 | 6 |
| Mascaux et al | 2008 | Belgium | Caucasians | LC | 16 | 4 | 26 | 14 | IHC | Normal tissue | Tumor tissue | 27 | 6 |
| Tian et al | 2006 | China | Asians | LC | 38 | 2 | 28 | 12 | IHC | Adjacent tissue | Tumor tissue | 28 | 8 |
| Tian et al | 2005 | China | Asians | NSCLC | 28 | 0 | 20 | 8 | WB | Adjacent tissue | Tumor tissue | 29 | 8 |
| Tian et al | 2004 | China | Asians | LC | 38 | 2 | 58 | 22 | IHC | Adjacent tissue | Tumor tissue | 30 | 8 |
| Hu et al | 2004 | China | Asians | NSCLC | 20 | 0 | 30 | 73 | IHC | Benign tissue | Tumor tissue | 31 | 6 |
| Chaussade et al | 2001 | France | Caucasians | LC | 20 | 0 | 12 | 8 | IHC | Normal tissue | Tumor tissue | 32 | 6 |
| Cortot et al | 2014 | France | Caucasians | LA | NT | NT | 70 | 18 | IHC | NT | Tumor tissue | 33 | – |
| Li et al | 2005 | China | Asians | NSCLC | NT | NT | 30 | 73 | IHC | NT | Tumor tissue | 34 | – |
| Li et al | 2004 | China | Asians | SCLC | NT | NT | 9 | 15 | IHC | NT | Tumor tissue | 35 | – |
| Chen et al | 2003 | China | Asians | NSCLC | NT | NT | 25 | 14 | IHC | NT | Tumor tissue | 36 | – |
| Xue et al | 2002 | Japan | Asians | LA | NT | NT | 4 | 46 | IHC | NT | Tumor tissue | 37 | – |
Abbreviations: IHC, immunohistochemistry; LA, lung adenocarcinomas; LC, lung cancer; NT, not stated; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; WB, Western blotting.
Quality evaluation
On the basis of evaluation of the methodology quality, six studies got a score of 8 and were considered to be of high quality. Five studies scored 6 and were considered to be of moderate quality. In addition, five studies were not added in the quality evaluation because these studies did not have a control group.
Quantitative synthesis
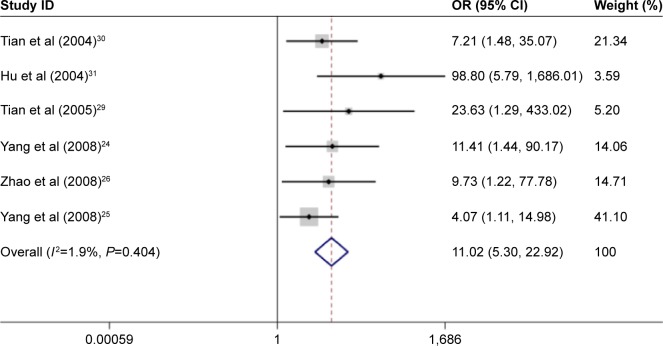
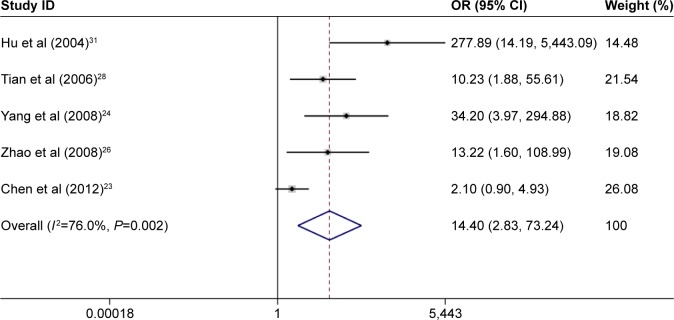
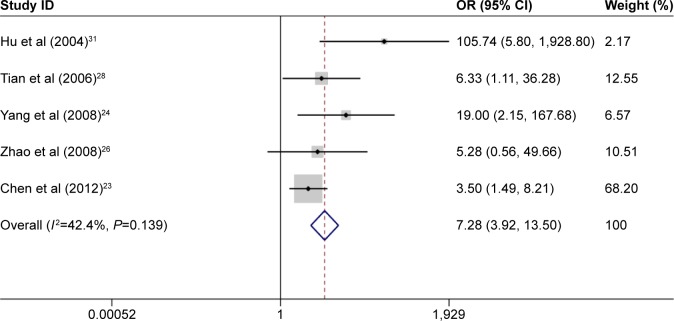
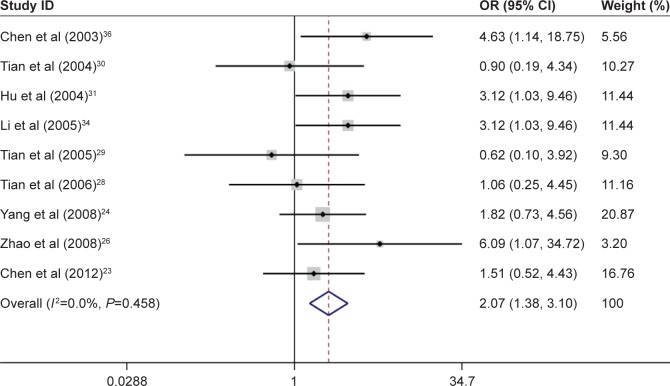
A meta-analysis that included 11 studies was performed to evaluate the association between p14ARF expression and lung cancer risk. The result indicated a significant association between p14ARF expression and lung cancer risk (OR =6.55, 95% CI =4.33–9.90). The frequency of p14ARF-negative expression in the lung cancer group was higher than that in the control group (case group vs control group, p14ARF-negative expression vs p14ARF positive expression: 81.35% vs 11.43%). In the subgroup analysis based on disease subtype, there was a significant association of p14ARF expression with NSCLC, lung adenocarcinoma, and squamous cell carcinoma (for NSCLC, OR =11.02, 95% CI =5.30–22.92; for lung adenocarcinoma, OR =7.28, 95% CI =3.92–13.50; and for lung squamous carcinoma; OR =14.40, 95% CI =2.83–73.24). The sample of control group in the included studies consisted of adjacent tissue and normal tissue. Hence, the subgroup analysis by sample type was conducted to explore the relationship between p14ARF expression and lung cancer, and the results showed a significant association of p14ARF expression with lung cancer risk (for normal tissue, OR =7.97, 95% CI =3.41–18.64 and for adjacent tissue, OR =4.76, 95% CI =2.88–7.84). Furthermore, stratified analysis based on ethnicity demonstrated significant relationships between p14ARF expression and lung cancer risk in Asians and Caucasians (for Asians, OR =7.02, 95% CI =4.48–11.00 and for Caucasians, OR =4.19, 95% CI =1.42–12.38). In the analysis of clinicopathological features, a significant correlation between p14ARF expression and TNM-stage of lung cancer was found in Chinese population (OR =2.07, 95% CI =1.38–3.10). Heterogeneity was not highly significant in the meta-analysis. If heterogeneity existed, the random-effects model and subgroup analysis were performed (Figures 2–5; Table 2).
Figure 2.
Meta-analysis for p14ARF expression and non-small-cell lung cancer risk.
Abbreviations: CI, confidence interval; OR, odds ratio.
Figure 3.
Meta-analysis for p14ARF expression and lung squamous carcinoma risk.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; OR, odds ratio.
Figure 4.
Meta-analysis for p14ARF expression and lung adenocarcinoma risk.
Abbreviations: CI, confidence interval; OR, odds ratio.
Figure 5.
Meta-analysis for p14ARF expression and the TNM-stage of lung cancer.
Abbreviations: CI, confidence interval; OR, odds ratio.
Table 2.
Meta-analysis results of p14ARF expression in lung cancer
| Variables | Studies (n) | OR | 95% CI | P-value | Heterogeneity
|
|
|---|---|---|---|---|---|---|
| I2 (%) | P-value | |||||
| Overall | 11 | 6.55 | 4.33–9.90 | <0.05 | 40.10 | 0.08 |
| Asians | 9 | 7.02 | 4.48–11.00 | <0.05 | 40.30 | 0.10 |
| Caucasians | 2 | 4.19 | 1.42–12.38 | <0.05 | 62.20 | 0.10 |
| Adjacent tissue | 6 | 4.76 | 2.88–7.84 | <0.05 | 0.00 | 0.43 |
| Normal tissue | 4 | 7.97 | 3.41–18.64 | <0.05 | 50.20 | 0.11 |
| Adenocarcinoma | 5 | 7.28 | 3.92–13.50 | <0.05 | 42.40 | 0.14 |
| Squamous cell carcinoma | 5 | 14.4 | 2.83–73.24 | <0.05 | 76.00 | 0.002 |
| NSCLC | 6 | 11.02 | 5.30–22.92 | <0.05 | 1.90 | 0.40 |
| Gender | 6 | 0.79 | 0.32–1.96 | >0.05 | 65.60 | 0.01 |
| Differentiation | 11 | 0.57 | 0.26–1.23 | >0.05 | 67.30 | 0.001 |
| TNM-stage | 9 | 2.07 | 1.38–3.10 | <0.05 | 0.00 | 0.46 |
| Lymph node metastasis | 10 | 0.93 | 0.66–1.30 | >0.05 | 40.40 | 0.09 |
Abbreviations: CI, confidence interval; OR, odds ratio; NSCLC, non-small-cell lung cancer.
Sensitivity analysis and publication bias
The results of funnel plot showed that moderate publication bias was found in this meta-analysis (for lung cancer risk: Begg’s test, P=0.001, Egger’s test, P=0.000 and for TNM-stage: Begg’s test, P=0.677, Egger’s test, P=0.705). Therefore, meta-regression was conducted to explore the causes of publication bias, and the results showed that ethnicity and sample type were not the main cause of moderate publication bias. However, publication bias decreased after subgroup analysis was conducted based on disease type. Thus, we speculated that disease type may be one of the causes of the publication bias. Sensitivity analysis was also conducted using Stata 12.0 software, and the result showed that the overall ORs did not have a significant change. At the same time, no significant publication bias was found in the analysis of association between p14ARF expression and TNM-stage, differentiation, and lymph node metastasis of lung cancer (Figure 6).
Figure 6.
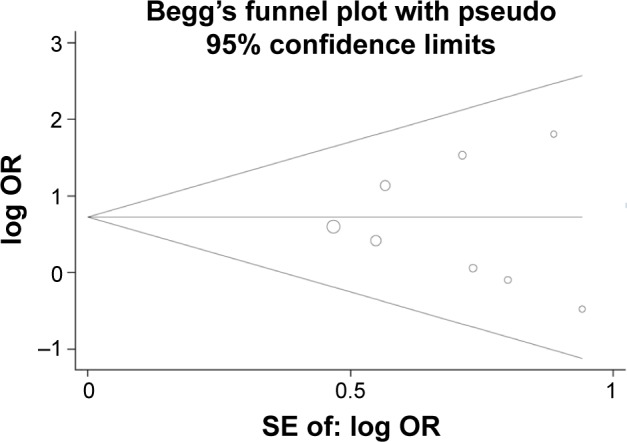
Begg’s funnel plot of association between p14ARF expression and the TNM-stage of lung cancer.
Abbreviations: OR, odds ratio; SE, standard error.
Discussion
p14ARF, a tumor suppressor protein, has been reported to be a regulator in the cell cycle arrest and apoptosis.38 Although p14ARF and p16INK4A share the common exon 2 and exon 3 of INK4A gene, the two proteins have unrelated structure and function.39 It has been reported that p14ARF played a key role in DNA damage and the regulation of cellular or viral oncogenes.40 Under normal conditions, the signal pathway of p53-MDM2-p14ARF maintained growth and development of cells. If the gene of the pathway was downregulated or upregulated, some diseases could be found in the body. P53 gene is a common gene that is related to the oncogenesis. Recent studies have found that MDM2 could act as a principal cellular regulator of p53 in response to the stress signals triggered by cellular oncogenes.41 For example, the study by Jin et al found that single nucleotide polymorphisms (SNPs) of MDM2 were significantly associated with the risk of salivary gland carcinoma.42 MDM2 could bind the N-terminal end p53 protein to block the transcriptional activation function. Moreover, MDM2 inhibited the expression of p53 protein through its ubiquitin ligase activity.43 p14ARF restrained the ubiquitin E3 ligase activity of MDM2 to antagonize its regulation to p53 protein, and p14ARF sequestered MDM2 to nucleus, which prevented the contact between MDM2 and p53, resulting in apoptosis and cell cycle arrest.44 In addition, p14ARF also acted as a sensor of excessive proliferation and inhibited the function of oncoproteins such as Myc and E2F1.45,46 Several changes in the tumor suppressor gene were found in the development of lung cancer. Hence, p14ARF, playing a central role that inhibited tumor transformation, might have an important role in the occurrence and development of lung tumor.
The results of the meta-analysis indicated that there was a significant association of p14ARF expression with lung cancer risk. To further investigate the relationship between p14ARF expression and lung cancer, subgroup analysis based on lung cancer subtype was conducted to explore the association of p14ARF expression with NSCLC, lung squamous carcinoma, and lung adenocarcinoma. According to the results of the subgroup analysis, significant correlations between p14ARF expression and NSCLC, lung squamous carcinoma, and lung adenocarcinoma risk were found. On the basis of sample type of the included studies, we performed stratified analysis, and the results suggested that significant associations were found both in normal tissue and in adjacent tissue. Furthermore, the TNM-stage of lung cancer was significantly associated with p14ARF expression in Chinese population. This result showed that the frequency of p14ARF-negative expression in stages III–IV of lung cancer was higher than the frequency in stages I–II of lung cancer. Previous studies also found consistent results.26,31,34,36 In these included studies, five studies indicated contrasting results.23,25,28–30 However, there were no significant associations of p14ARF expression with differentiation and lymph node metastasis of lung tumor. Most of the included studies indicated consistent results.23–26,28–31,34–37 Overall, the p14ARF-negative expression significantly correlated with the development and progression of lung cancer.
In this meta-analysis, although heterogeneity among studies was not very significant, the subgroup analysis and random-effects model were applied to decrease the heterogeneity. Because moderate publication bias was found from the results of Begg’s test and Egger’s test, meta-regression was conducted, and no significant cause was found. Hence, we speculated that other causes might lead to the publication bias. However, no other clinical information was extracted because of limited clinical information in the included studies. To further demonstrate these associations, more studies that include clinical information should be conducted.
Although the results showed that p14ARF expression has an important role in lung cancer development, several limitations were found in this meta-analysis. First, little clinical information was extracted so that a relevant meta-analysis could not be performed, and this might be one of the reasons of publication bias. Second, moderate publication bias existed in the meta-analysis, which affected the accuracy of results. Third, the patients of included studies were mostly from Chinese population. Hence, the results mostly suggested that p14ARF expression was significantly associated with the risk of lung cancer in Chinese population.
Conclusion
This meta-analysis demonstrated that there was a significant association between p14ARF expression and lung cancer risk. Furthermore, p14ARF expression was significantly associated with the TNM-stage of lung cancer in Chinese population. However, due to limitations in this study, future studies that are derived from different ethnicities and have enough clinical information are needed to clarify these conclusions.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 3.Samet JM, Avila-Tang E, Boffetta P, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15(18):5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med. 2013;188(7):770–775. doi: 10.1164/rccm.201305-0843PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciardiello F, Tortora G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res. 2001;7(10):2958–2970. [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 9.Lomazzi M, Moroni MC, Jensen MR, Frittoli E, Helin K. Suppression of the p53- or pRB-mediated G1 checkpoint is required for E2F-induced S-phase entry. Nat Genet. 2002;31(2):190–194. doi: 10.1038/ng891. [DOI] [PubMed] [Google Scholar]
- 10.Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127(10):2239–2247. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- 11.Timofeeva MN, Hung RJ, Rafnar T, et al. Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21(22):4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Yang L, Dai W, Ye B. Role of p14ARF and p15INK4B promoter methylation in patients with lung cancer: a systematic meta-analysis. Onco Targets Ther. 2016;9:6977–6985. doi: 10.2147/OTT.S117161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Ding S, Zhong Q, Li G, Zhang Y, Huang XC. Significance of MMP11 and P14(ARF) expressions in clinical outcomes of patients with laryngeal cancer. Int J Clin Exp Med. 2015;8(9):15581–15590. [PMC free article] [PubMed] [Google Scholar]
- 14.Cabral VD, Cerski MR, Sa BI, Kliemann LM. p14 expression differences in ovarian benign, borderline and malignant epithelial tumors. J Ovarian Res. 2016;9(1):69. doi: 10.1186/s13048-016-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pare R, Shin JS, Lee CS. Increased expression of senescence markers p14(ARF) and p16(INK4a) in breast cancer is associated with an increased risk of disease recurrence and poor survival outcome. Histopathology. 2016;69(3):479–491. doi: 10.1111/his.12948. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T, Nakao K. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J Gastroenterol. 2004;39(4):355–361. doi: 10.1007/s00535-003-1302-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhen Y, Chunlei G, Wenzhi S, et al. Clinicopathologic significance of legumain overexpression in cancer: a systematic review and meta-analysis. Sci Rep. 2015;5:16599. doi: 10.1038/srep16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 21.Song F, Gilbody S. Bias in meta-analysis detected by a simple, graphical test. Increase in studies of publication bias coincided with increasing use of meta-analysis. BMJ. 1998;316(7129):471. [PMC free article] [PubMed] [Google Scholar]
- 22.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Pang JW, Wang W, Xiao L, Zu GF. Clinicopathological significance and expression of E2F-1, c-myc and p14ARF protein in squamous cell carcinoma and adenocarcinoma of lung. Chin J Cancer Prev Treat. 2012;19:115–118. [Google Scholar]
- 24.Yang ZH, Yong HR, Jin KW, Gao Q, Zhang L. The expression of p14arf in lung cancer. Chinese Journal of Ethnomedicine & Ethnopharmacy. 2008;(07):4–6. [Google Scholar]
- 25.Yang TD, Wu JF, Li XJ, Fan WY, Wang YL. Relationship between the expression of p14ARF, mtp53 protein and clinical pathological parameters in non-small cell lung cancer. J Xinxiang Med Coll. 2008;25:336–339. [Google Scholar]
- 26.Zhao ZH, Wang SF, Cong DG, Yu L, Wang J. Correlation of the expression of pokemon with p14ARF and bcl-2 and their effects on prognosis of non small cell lung cancer. Tumor. 2008;28:121–124. [Google Scholar]
- 27.Mascaux C, Bex F, Martin B, et al. The role of NPM, p14arf and MDM2 in precursors of bronchial squamous cell carcinoma. Eur Respir J. 2008;32(3):678–686. doi: 10.1183/09031936.00008408. [DOI] [PubMed] [Google Scholar]
- 28.Tian KH, Lin LS, Jia YT, Guo XJ, Zhang L. Promoter methylation status and protein expression of p14ARF gene in squamous cell carcinoma and adenocarcinoma of the lung. Chin J Lung Cancer. 2006;9(1):40–44. doi: 10.3779/j.issn.1009-3419.2006.01.11. [DOI] [PubMed] [Google Scholar]
- 29.Tian KH, Shen Y, Luo YR, Zhang L. Expression of p14ARF gene in non-small cell lung cancer. Med J Qilu. 2005;20:389–393. [Google Scholar]
- 30.Tian KH, Zhao HR, Zhao F, Zhang L, Qu HC. Expression of p14ARF, p16INK4a, and p53 gene in non-small cell lung cancer. Chin J Clin Oncol. 2004;31:376–383. [Google Scholar]
- 31.Hu YD, Li JG, Ye FM, et al. Study on the expressions and clinical significances of cell cycle control protein p14ARF in non-small cell lung cancer. Sichuan Med J. 2004;25:1191–1193. [Google Scholar]
- 32.Chaussade L, Eymin B, Brambilla E, Gazzeri S. Expression of p15 and p15.5 products in neuroendocrine lung tumours: relationship with p15(INK4b) methylation status. Oncogene. 2001;20(45):6587–6596. doi: 10.1038/sj.onc.1204798. [DOI] [PubMed] [Google Scholar]
- 33.Cortot AB, Younes M, Martel-Planche G, et al. Mutation of TP53 and alteration of p14(arf) expression in EGFR- and KRAS-mutated lung adenocarcinomas. Clin Lung Cancer. 2014;15(2):124–130. doi: 10.1016/j.cllc.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Li JG, Hu YD, Ye MF, et al. The co-expression and clinical significance of p14ARF and p16INK4a proteins in non-small cell lung cancer. Chin J Clin Oncol. 2005;32:1021–1024. [Google Scholar]
- 35.Li JG, Ye FM, Hu YD. Expressions of p14ARF and p16INK4a proteins in small cell lung cancer and the clinical significance. Acta Acad Med Militaris Tertia. 2004;26:1098–1101. [Google Scholar]
- 36.Chen QK, Ding JA, Gao W. Immunohistochemical analysis of P14ARF protein expression in non-small cell lung cancer: its prognostic signifi-cance. Chin J Lung Cancer. 2003;6(4):283–285. doi: 10.3779/j.issn.1009-3419.2003.04.10. [DOI] [PubMed] [Google Scholar]
- 37.Xue Q, Sano T, Kashiwabara K, Saito M, Oyama T, Nakajima T. Aberrant expression of pRb, p16, p14ARF, MDM2, p21 and p53 in stage I adenocarcinomas of the lung. Pathol Int. 2002;52(2):103–109. doi: 10.1046/j.1440-1827.2002.01321.x. [DOI] [PubMed] [Google Scholar]
- 38.Muer A, Overkamp T, Gillissen B, et al. p14(ARF)-induced apoptosis in p53 protein-deficient cells is mediated by BH3-only protein-independent derepression of Bak protein through down-regulation of Mcl-1 and Bcl-xL proteins. J Biol Chem. 2012;287(21):17343–17352. doi: 10.1074/jbc.M111.314898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12(15):2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103(17):6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 42.Jin L, Xu L, Song X, Wei Q, Sturgis EM, Li G. Genetic variation in MDM2 and p14ARF and susceptibility to salivary gland carcinoma. PLoS One. 2012;7(11):e49361. doi: 10.1371/journal.pone.0049361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 44.Johansson HJ, El-Andaloussi S, Holm T, et al. Characterization of a novel cytotoxic cell-penetrating peptide derived from p14ARF protein. Mol Ther. 2008;16(1):115–123. doi: 10.1038/sj.mt.6300346. [DOI] [PubMed] [Google Scholar]
- 45.Eymin B, Karayan L, Seite P, et al. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene. 2001;20(9):1033–1041. doi: 10.1038/sj.onc.1204220. [DOI] [PubMed] [Google Scholar]
- 46.Martelli F, Hamilton T, Silver, et al. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci U S A. 2001;98(8):4455–4460. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]