Abstract
Molecular diagnostics are increasingly performed routinely in the diagnosis and management of patients with melanoma due to the development of novel therapies that target specific genetic mutations. The development of next-generation sequencing (NGS) technologies has enabled to sequence multiple cancer-driving genes in a single assay, with improved sensitivity in mutation detection. The main objective of this study was the design and implementation of a melanoma-specific sequencing panel, and the identification of the spectrum of somatic mutations in a series of primary melanoma samples. A custom panel was designed to cover the coding regions of 35 melanoma-related genes. Panel average coverage was 2,575.5 reads per amplicon, with 92,8% of targeted bases covered ≥500×. Deep coverage enabled sensitive discovery of mutations in as low as 0.5% mutant allele frequency. Eighty-five percent (85/100) of the melanomas had at least one somatic mutation. The most prevalent mutated genes were BRAF (50%;50/199), NRAS (15%;15/100), PREX2 (14%;14/100), GRIN2A (13%;13/100), and ERBB4 (12%;12/100). Turn-around-time and costs for NGS-based analysis was reduced in comparison to conventional molecular approaches. The results of this study demonstrate the cost-effectiveness and feasibility of a custom-designed targeted NGS panel, and suggest the implementation of targeted NGS into daily routine practice.
Introduction
Malignant melanoma is the most aggressive form of skin cancer, with a poor prognosis for patients with metastatic disease. Melanomas are currently classified based on clinical and histologic characteristics of the primary tumors; in addition, it has been described that distinct patterns of genetic alterations contribute to the development of the different subtypes of primary melanoma. It is well known that superficial spreading melanoma (SSM) and nodular melanoma (NM) are associated with BRAF or NRAS mutations; acral lentiginous melanoma (ALM), lentigo maligna (LM), and mucosal melanoma are more often associated with KIT mutations; and ocular melanomas are not associated with these common oncogenes, but rather with GNAQ or GNA11 alterations1, 2. A recent integrative analysis of cutaneous melanoma from The Cancer Genome Atlas (TCGA) has established a new genomic classification into four subtypes, based on the identification of the most prevalent mutated genes [BRAF, RAS, NF1 and triple-wild type (wt) subtypes]3.
The advances on melanoma molecular pathogenesis have opened a new insight for the management of advanced melanoma due to the development of novel therapies that target causative genetic events, and improve disease free survival and overall survival4. The selective BRAF kinase inhibitors (Vemurafenib and dabrafenib) are effective in BRAF mutant melanoma; MEK inhibitors (trametinib and cobimetinib) show efficacy against both BRAF- and KRAS/NRAS-driven tumors; KIT inhibitors (imatinib, dasatinib, sunitinib and nilotinib) have demonstrated clinical responses in melanoma arising from acral, mucosal, and chronic sun-damaged cutaneous sites; and additionally, there are novel therapeutic monoclonal antibodies targeted against immunosuppressive molecules such as CTLA4, PD-1 and PD-L1.
Therefore, molecular diagnostics are increasingly performed routinely in the diagnosis and management of patients with melanoma. Conventional molecular analyses for detecting cancer somatic alterations have relied on methods such as Sanger sequencing and real-time quantitative polymerase chain reaction (RQ-PCR). These approaches have the limitation that are performed separately for each gene and therefore consume a high turn-around time. Moreover, Sanger sequencing has a relatively low sensitivity, and sometimes it can be challenging to detect somatic mutations, especially when tumor material is mixed with normal tissue. In this context, the development of next-generation sequencing (NGS) technologies has enabled to massively analyze millions of DNA segments in parallel, thus allowing to sequence multiple cancer-driving genes in a single assay, with improved sensitivity in mutation detection. One of the developed NGS methodologies is the new Ion Torrent sequencing platform, based on the detection of hydrogen ions released on each cycle of DNA polymerization. It has been described to be cost and time effective5, and its applicability in formalin-fixed and paraffin-embedded (FFPE) specimens with small amounts of DNA has been proved in several reports6, 7. The use of a multi-gene screening panel may potentially allow a more personalized approach to cancer therapy by identifying less common but potentially actionable mutations.
In the present study, we have used Ion Torrent sequencing technology with the Personal Genome Machine (PGM) and a custom AmpliSeq Panel including 35 genes. The main objective was the design and implementation of a melanoma-specific sequencing panel, based on the inclusion of relevant melanoma-genes with clinical diagnostic, prognostic or treatment value. Moreover, we aimed to describe the mutation profile in a series of primary melanoma samples in order to provide new insights into the molecular subclassification of melanoma. The translation of these study results may provide further understanding of the molecular alterations that lead to the development of melanoma, and therefore may contribute to the improvement of a personalized medicine.
Results
Next-generation sequencing quality metrics and sensitivity assay
Quality control analysis revealed excellent performance of the panel (Fig. 1). Of note, the average total reads per sample was 1,305,617 with an average coverage of 2,575.5 reads per amplicon (Figure S1). Moreover, 97.8% of targeted bases covered ≥100x and 92.8% ≥500x. The coverage uniformity of amplicon sequencing was very high with an average of 91.9%. In addition, the sensitivity assay performed with serial dilutions in two independent experiments confirmed a high sensibility of the IonTorrent PGM (Table 1). BRAF mutations could be detected at an allele frequency as low as 0.05% with a rate of false reads of 0.1%. Therefore adjusting the sensitivity to our error-rate, the limit of detection was 0.5% (Figure S2).
Figure 1.
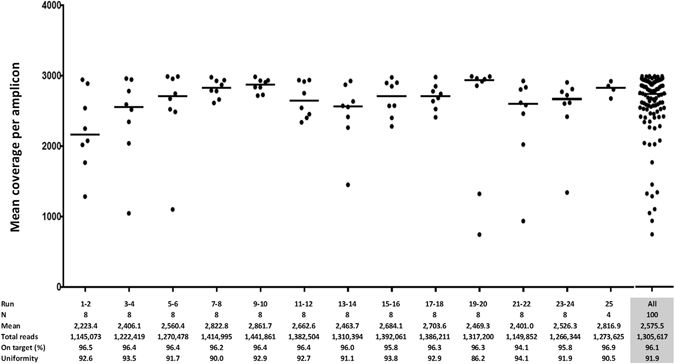
Quality metrics for 25 runs including mean coverage, total reads, reads on target and uniformity.
Table 1.
Sequencing results of serially diluted DNA isolated from two adenocarcinoma cell lines (HT-29 y Caco-2) with known variants in BRAF and MC1R genes.
| HT-29 in Caco-2 (variant frequency) | BRAF gene, p.V600E (heterozygous) | MC1R gene, p.R160W (heterozygous) | ||||||
|---|---|---|---|---|---|---|---|---|
| Allele frequency (%) | Coverage (x) | Allele frequency (%) | Coverage (x) | |||||
| Exp 1 | Exp 2 | Exp 1 | Exp 2 | Exp 1 | Exp 2 | Exp 1 | Exp 2 | |
| Undiluted (50%) | 57.12 | 52.85 | 2027 | 2182 | 61.42 | 64.48 | 1894 | 2329 |
| 1:1 (25%) | 26.22 | 25.69 | 4366 | 5780 | 32.66 | 32.93 | 2103 | 2708 |
| 1:3 (12.5%) | 13.13 | 12.85 | 5620 | 3507 | 14.84 | 15.53 | 2182 | 2524 |
| 1:24 (2%) | 1.82 | 1.75 | 3720 | 3523 | 1.95 | 1.21 | 1894 | 1981 |
| 1:49 (1%) | 1.12 | 0.68 | 4266 | 3927 | 0.80 | 0.87 | 2967 | 2631 |
| 1:99 (0.5%) | 0.65 | 0.64 | 3977 | 3430 | 0.57 | 0.34 | 2624 | 1458 |
| 1:999 (0.05%) | 0.04 | 0.10 | 4993 | 2945 | 0.03 | 0.19 | 3065 | 3138 |
Exp: Experiment.
Mutations detected by Next-generation sequencing in melanoma
Mutation prevalence
Sequencing identified on average 56.6 total variants and 8.9 exonic variants per sample, that were subsequently filtered in order to exclude variants without impact on protein function. After filtering, a total of 135 different pathogenic variants were finally reported in all the samples (Table S1). In the entire cohort, 94% (94/100) had at least one pathogenic variant, and 51% (51/100) had ≥3. Without considering MC1R polymorphisms, 85% (85/100) of the melanomas had at least one somatic mutation.
The most prevalent mutated genes were BRAF (50%; 50/100), NRAS (15%; 15/100), PREX2 (14%; 14/100), GRIN2A (13%; 13/100), and ERBB4 (12%; 12/100). All the mutated genes are represented in Fig. 2. Regarding BRAF gene, the most frequent mutation was p.V600E (80%; 40/50), followed by p.V600K (10%; 5/50), p.K601E (4%; 2/50), p.L597R (2%; 1/50), p.L584F (2%; 1/50), and p.G464R (2%; 1/50). The most frequent affected codon in NRAS was Q61 (87,6%; 14/16), followed by G12 (6,3%; 1/16) y E62 (6,3%; 1/16). KRAS oncogene mutations were identified in 3% (3/100), and the most prevalent mutation was p.Q61R (75% ; 2/3); KIT mutations were detected in 5% (5/100) of the samples, and two of them showed the hotspot mutation p.L576P. In regard to MAP2K1 and MAP2K2, the overall prevalence was 3% (3/100). PTEN mutations were present in 9% (9/100) of the melanomas, with a recurrent point mutation in two of the samples (p.F278L). PIK3CA mutations were not identified in our cohort. NF1 mutations were detected in 8% (8/100) of the melanomas; and nonsense mutations were the most prevalent alteration [62.5% (5/8)], including one recurrent mutation present in two samples (p.R1362*). The overall prevalence of variations in PREX2 and GRIN2A genes, was 28% (28/100) and 21% (21/100), with a total of 31 and 27 different variants dispersed throughout the entire gene, respectively. After filtering out, the prevalence of pathogenic variants was 14% (14/100) for PREX2 gene and 13% (13/100) for GRIN2A gene. ERBB4 variants were also identified in 15% (15/100) of the samples, and 80% (12/15) of them were predicted as pathogenic. Hotspot mutations were detected in RAC1 (p.P29S and p.P69L) and PPP6C (p.D193Y, p.P209L, and p.R264C) in 8% and 6% of the samples, respectively. None of the melanomas harbored the hotspot activating mutation p.S722F in TRRAP, although we identified one sample with another TRRAP pathogenic variant (p.P814S).
Figure 2.

Frequency of somatic gene mutations. Each column represents 1 sample and each row represents 1 gene. The column on the left indicates the percentage of samples with specific gene mutation.
Although paired tumor-normal samples were not collected, melanoma related polymorphisms were analyzed. MC1R polymorphisms were detected in 66% (66/100) of the patients; 4% (4/100) showed polymorphisms in CDK4; and 2% (2/100) in MITF. The variant p.V60L in MC1R was the most prevalent polymorphism [36% (36/100)], followed by p.V92M [17% (17/100)] and p.D294H [9% (9/100)]. Regarding CDK4 gene, two of the samples showed the polymorphism p.R24H, one p.R24C and one p.L22R. Moreover, the MITF p.E318K variant was detected in two additional samples.
Associations between mutations and clinicopathological characteristics
The prevalence of gene mutations varied by melanoma location, as most of the KIT mutated melanomas were distributed in chronically sun-damaged skin and acral melanomas, and were significantly associated with ALM [p = 0.004; OR = 4.4 (1.6–12.1)]. Conversely, BRAF mutations were associated with histological subtypes other than ALM [p = 0.013; OR = 0.5 (0.2–0.8)]. In adition, GRIN2A and RAC1 mutations were also associated with chronically sun-exposed melanomas [p = 0.011; OR = 5.9 (1.5–23.2) and p = 0.026; OR = 3.2 (1.1–9.2), respectively]. Interestingly, NF1 mutations were more prevalent in melanomas with increased Breslow thickness [p = 0.025; OR = 2.16 (1.10–4.26)], ulceration [p = 0.003; OR = 26.89 (3.11–232.10)], and fast-growing melanomas8 [p = 0.004; OR = 12.16 (2.26–65.37)]. Furthermore, PREX2 and GRIN2A mutations were significantly more prevalent in melanomas with mitosis [p = 0.042; OR = 4.0 (1.0–15.4) and p = 0.023; OR = 6.2 (1.3–29.5), respectively]. All significant associations with clinicopathological characteristics are shown in Table 2. No correlation between mutations and clinical outcome was found.
Table 2.
Associations of mutations with clinicopathological characteristics.
| Variables | Mutational status | Total N | OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|
| Wildtype N (%) | Mutated N (%) | |||||
| BRAF | 50 (50%) | 50 (50%) | ||||
| Histological subtype | ||||||
| LMM | 5 (55.6) | 4 (44.4) | 9 | 0.5 | 0.2–0.8 | 0.013 |
| SSM | 29 (41.2) | 40 (54.8) | 69 | |||
| NM | 8 (57.1) | 6 (42.9) | 14 | |||
| ALM | 8 (100) | 0 (0) | 8 | |||
| KIT | 95 (95%) | 5 (5%) | ||||
| Histological subtype | ||||||
| LMM | 9 (100) | 0 (0) | 9 | |||
| SSM | 68 (98.5) | 1 (1.5) | 69 | 4.4 | 1.6–12.1 | 0.004 |
| NM | 13 (92.9) | 1 (7.1) | 14 | |||
| ALM | 5 (62.5) | 3 (37.5) | 8 | |||
| Ulceration | ||||||
| No | 73 (98.6) | 1 (1.4) | 74 | 13.3 | 1.4–124.9 | 0.024 |
| Yes | 22 (84.6) | 4 (15.4) | 26 | |||
| Tumor stage | ||||||
| I–II | 79 (97.5) | 2 (2.5) | 81 | 7.4 | 1.1–47.9 | 0.036 |
| III–IV | 16 (84.2) | 3 (15.8) | 19 | |||
| PREX2 | 86 (86%) | 14 (14%) | ||||
| Mitosis | ||||||
| <1 | 45 (93.8) | 3 (6.3) | 48 | 4.0 | 1.0–15.4 | 0.042 |
| ≥1 | 41 (78.8) | 11 (21.2) | 52 | |||
| GRIN2A | 87 (87%) | 13 (13%) | ||||
| Sun related site | ||||||
| Non exposed | 20 (95.2) | 1 (4.8) | 21 | |||
| Ocasionally exposed | 56 (88.9) | 7 (11.1) | 63 | 3.2 | 1.1–9.2 | 0.026 |
| Usually exposed | 11 (68.8) | 5 (31.3) | 16 | |||
| Mitosis | ||||||
| <1 | 46 (95.8) | 2 (4.2) | 48 | 6.2 | 1.3–29.5 | 0.023 |
| ≥1 | 41 (78.8) | 11 (21.2) | 52 | |||
| RAC1 | 92 (92%) | 8 (8%) | ||||
| Sun related site | ||||||
| Non exposed | 21 (100) | 0 (0) | 21 | |||
| Ocasionally exposed | 59 (93.7) | 4 (6.3) | 63 | 5.9 | 1.5–23.2 | 0.011 |
| Usually exposed | 12 (75) | 4 (25) | 16 | |||
| NF1 | 92 (92%) | 8 (8%) | ||||
| Breslow | ||||||
| ≤1 | 45 (95.7) | 2 (4.3) | 47 | |||
| 1–2 | 21 (100) | 0 (0) | 21 | 2.2 | 1.1–4.3 | 0.025 |
| 2–4 | 15 (78.9) | 4 (21.1) | 19 | |||
| >4 | 10 (76.9) | 3 (23.1) | 13 | |||
| Ulceration | ||||||
| No | 72 (97.3) | 2 (2.7) | 74 | 26.9 | 3.1–232.1 | 0.003 |
| Yes | 19 (73.1) | 7 (26.9) | 26 | |||
| Growth rate | ||||||
| SGM | 74 (97.3) | 2 (2.7) | 76 | 12.2 | 2.3–65.4 | 0.004 |
| FGM | 17 (70.8) | 7 (29.2) | 19 | |||
LMM: Lentigo maligna melanoma; SSM: Superficial spreading melanoma; NM: Nodular melanoma; ALM: Acral lentiginous melanoma; SGM: Slow growing melanoma; FGM: Fast growing melanoma.
Concurrent molecular alterations in melanomas
Several genes harbored concurrent mutations among the studied genes (Fig. 3).We observed mutual exclusivity between BRAF hotspot mutations (p.V600E, p.V600K and p.K601E) and NRAS/KRAS mutations in almost all samples; however, one sample concurrently harbored BRAF p.V600E and NRAS p.Q61R mutations. Moreover, we found concurrent mutations in BRAF non-hotspot mutations (p.L584F and p.L597R) with NRAS (p.Q61L and p.G12S). Strikingly, one sample showed two concurrent NRAS mutations (p.Q61R and p.E62Q), another sample showed concurrent mutations on KRAS (p.Q61R and p.A146V), and another one two concurrent mutations in KIT (p.Y553S and p.Y578C). The clinicopathological features of patients with concurrent mutations are listed in Table 3.
Figure 3.
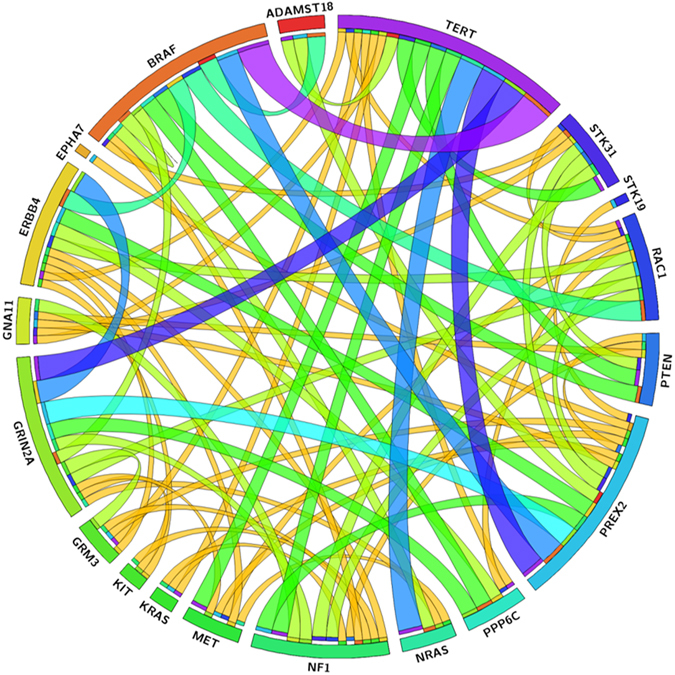
Circos diagram. Associations between the more prevalent genes.
Table 3.
Clinicopathological characteristics of the samples with concurrent mutations in the four genomic subtypes.
| No. | Gender | Age | Breslow (mm) | Anatomic site | Sun related site | Histological subtype | Gene | Amino acid change | Allele frequency (%) |
|---|---|---|---|---|---|---|---|---|---|
| 2 | M | 79 | 2.7 | Trunk | Occasional exposed | SSM | BRAF | p.Gly464Arg | 6.8 |
| NF1 | p.Arg711Cys | 4.7 | |||||||
| 28 | F | 58 | 0.7 | Lower extremities | Occasional exposed | SSM | NRAS | p.Glu62Gln | 8.0 |
| NRAS | p.Gln61Arg | 8.0 | |||||||
| 58 | M | 69 | 0.4 | Trunk | Occasional exposed | SSM | BRAF | p.Leu597Arg | 4.1 |
| NRAS | p.Gly12Ser | 5.0 | |||||||
| 62 | M | 66 | 4.2 | Head and neck | Usually exposed | LMM | KRAS | p.Gly12Val | 80.8 |
| NF1 | p.Trp1512* | 30.8 | |||||||
| 68 | M | 54 | 0.7 | Head and neck | Usually exposed | SSM | BRAF | p.Val600Glu | 25.4 |
| NF1 | p.Lys1844Th | 6.3 | |||||||
| 71 | F | 85 | 3 | Acral | Non exposed | ALM | KIT | p.Tyr553Ser | 31.8 |
| KIT | p.Tyr578Cys | 43.9 | |||||||
| 80 | F | 73 | 0.3 | Upper extremities | Occasional exposed | SSM | BRAF | p.Val600Glu | 2.0 |
| NRAS | p.Gln61Arg | 3.0 | |||||||
| 84 | F | 57 | 2 | Upper extremities | Occasional exposed | SSM | BRAF | p.Leu584Phe | 23.1 |
| NRAS | p.Gln61Leu | 24.0 | |||||||
| 107 | M | 59 | 6 | Head and neck | Usually exposed | NM | KRAS | p.Gln61Arg | 5.3 |
| KRAS | p.Ala146Val | 10.6 | |||||||
| NF1 | p.Gln1070* | 13.5 |
M: Male; F: Female; SSM: Superficial spreading melanoma; LM: Lentigo Maligna; NM: Nodular melanoma; ALM: Acral lentiginous melanoma.
To verify the variant calling accuracy, BRAF, NRAS, KRAS and KIT mutated samples were subsequently analyzed by HRM, Sanger and RQ-PCR. BRAF mutations were detected by HRM and Sanger in 54% (27/50) of the melanomas; the average allele frequency of those wt samples was 4.4% with an average variant coverage of 229.5 reads. NRAS and KRAS mutations were confirmed by RQ-PCR in all the samples, and KIT mutations were confirmed in 80% (4/5) of the samples. Above mentioned concurrent mutations of NRAS, KRAS and KIT in the same sample were also confirmed by direct sequencing.
Ninety percent of the samples were correctly classified in four genomic subtypes3 (45 BRAF, 10 RAS, 4 NF1 and 31 triple-wt); however apart from the already mentioned, concurrent mutations were also identified between NF1 and BRAF, and also between NF1 and KRAS. Considering BRAF, RAS and NF1 subtypes together, a total of 31% (31/100) of our samples were triple-wt, and 9% (3/31) and 6% (2/31) of them harbored KIT and PTEN mutations, respectively (Figure S3).
TERT promoter mutations
The prevalence of TERT promoter mutations was 33% (33/100), The most frequent somatic changes were −146C > T and −124C > T that were detected in 45% (15/33) and 30% (10/33) of the samples, respectively. Other recurrent changes identified were −124/−125CC > TT (6%; 2/33), −138/−139 CC > TT (15%; 5/33) and −57 A > C (3%; 1/33). We found mutual exclusivity between all the variants. TERT promoter mutations were observed in 32% (16/50) of BRAF, 47% (7/15) of RAS and 50% (4/8) of NF1 subtypes. Mutations were significantly associated with female gender [p = 0.021; OR = 2.76 (1.16–6.54)], increased Breslow thickness [p = 0.001; OR = 2.07 (1.37–3.13)], ulcerated melanomas [p = 0.010; OR = 3.37 (1.33–8.56)], presence of mitosis [p = 0.001; OR = 4.63 (1.82–11.78)], absence of regression [p = 0.039; OR = 0.45 (0.21–0.96)], and fast-growing melanomas [p = 0.015; OR = 3.25 (1.25–8.42)]. Results of the univariate analysis are represented in Figure S4. In the multivariate analysis female gender [p = 0.001; OR = 6.50 (2.06–20.49)], increased Breslow thickness [p = 0.002; OR = 2.50 (1.40–4.50)], nodular histological subtype [p = 0,043; OR = 0,45 (0.21–0.97)], and the presence of mitosis [p = 0.014; OR = 4.48 (1.35–14.78)] were independently associated with TERT promoter mutations.
Turn-around time and cost comparison
In order to compare NGS with conventional molecular testing under routine laboratory conditions, we calculated the turn-around time and costs for three commonly melanoma-related genes (BRAF, NRAS, KIT) in eight samples. Starting from FFPE samples, we were able to isolate DNA, prepare libraries and sequence eight samples within two sequencing runs on 318v2 chips in approximately three working days. The costs of consumables and laboratory personal for NGS was 415.5€ per sample. Grouping turn-around time analysis for BRAF (exon 15), NRAS (exons 2, 3 and 4) and KIT (exons 9, 11, 13 and 17) genes, the conventional methods resulted in approximately five working days. As expected, the mean turn-around time for NGS-based analysis was lower in comparison to routine methods. The total cost for conventional methods was more expensive in comparison to NGS (Table 4).
Table 4.
Turn-around time and cost comparison between Next-generation sequencing and conventional molecular analysis.
| Analysis | System | Time duration (min) | Costs** (€) | |
|---|---|---|---|---|
| NGS analysis (8 samples; 35 genes) | ||||
| Quantification and sample dilution | Qubit | 60 | 4 | |
| Library preparation | Veriti Thermal Cycler | 310 | 1819.8 | |
| Emulsion PCR | One Touch | 315* | 238.8 | |
| Enrichment | One Touch ES | 45 | 47.2 | |
| Sequencing (two 318 chip v2) | PGM System | 360 (x2) | 1214 | |
| Data processing and analysis | Ion Reporter | 110 | 0 | |
| Laboratory personal costs | NA | 288 | ||
| Total | 1,560 (=26 h) | 3323.8 | ||
| Working days | 3 | NA | ||
| Analysis | Method | |||
| Conventional molecular analysis (8 samples; 3 genes) | ||||
| Quantification | Nanodrop | 30 | 0 | |
| BRAF | Exon 15 | RQ-PCR HRM+ SS | 510 (=81/2 h) | 460.2 |
| NRAS | Exon 2/3/4 | RQ-PCR (hibridization probes)+ SS | 420 (=7 h) | 1,225.6 |
| KIT | Exon 9/11/13/17 | Conventional PCR + SS | 1,230 (201/2 h) | 1,555.4 |
| Laboratory personal costs | NA | 480 | ||
| Total | 2,160 (=36 1/2 h) | 3,723.2 | ||
| Working days | 5 | NA | ||
NGS: Next-generation sequencing; PGM: Personal Genome Machine PCR: polymerase chain reaction; RQ-PCR: real-time quantitative polymerase chain reaction; SS: Sanger sequencing; HRM: High-resolution melting; NA: Non applicable.
*The duration of the second emulsion PCR required is not included as it is done at the same time as the first sequencing run. **Costs: Cost of consumables and laboratory personal (as cost calculated from the time that the technician/physician is required for the analysis).
Discussion
In this study we analyzed the spectrum of mutations in 35 melanoma-related genes using a targeted NGS approach on 100 melanoma samples. This in-depth analysis provides important insights into the molecular alterations that contribute to the development and progression of melanoma. In this regard, NGS analysis using commercial pan-cancer panel has been performed in a large cohort of melanoma samples9; however as it has been pointed out the majority of genes analyzed in this commercialized cancer panels are not relevant in melanoma pathogenesis, and other recently described genes are not included10. Furthermore, in our analysis the deep coverage enabled sensitive discovery of mutations in as low as 0.5% mutant allele frequency, which may be important in FFPE specimens where tumor content may be low and DNA may be degraded. The applicability of FFPE in NGS was previously demonstrated in a study conducted by Chen et al.11, that sequenced a pair of matched fresh-frozen and FFPE tumor samples and found high concordances between both types of samples. This is relevant as in routine practice FFPE specimens are usually available and fresh-frozen samples are difficult to achieve, especially in melanoma. In addition, as described above and consistent with other reports12, sample processing in our hands took approximately three working days, and therefore in comparison to conventional molecular approaches, with NGS the overall time for sample mutation detection is reduced.
Recent WES studies have demonstrated that melanoma has one of the highest rates of somatic mutations among all cancers13. In our cohort, we found that the great majority of the analyzed samples (85%) showed at least one somatic mutation among the 35 genes of our customized panel. Moreover, the prevalence of mutations detected in the study among the different melanoma-related genes is in agreement with previous reports. In our primary melanoma series, BRAF mutations were the most prevalent (50%) followed by TERT promoter mutations (33%). The prevalence of mutations in RAS family was also similar to other studies, as NRAS mutations were present in 15% of the samples and KRAS in 3%. As previously described, mutations were detected at different frequencies across all melanoma subtypes; BRAF and NRAS were more frequent in SSM and NM, KIT mutations were the most prevalent in ALM, and TERT promoter mutations were significantly associated with NM2, 14.
Consistent with previous reports, we found mutual exclusivity between somatic mutations in BRAF and NRAS/KRAS in 97% (97/100) of the samples15. Interestingly, we also identified concurrent point mutations on the KRAS (Q61R and A146V) and NRAS (E62Q and Q61R) in two of the samples. To our knowledge these concurrent mutations have not been described before, as previously reported cases show coexisting mutations in codon 1216. It has been suggested that these co-mutations may cooperate to activate the MAPK pathway9, and therefore the identification could have prognostic and therapeutic implications.
Moreover, we aim to analyze the prevalence of mutations in new recurrently mutated genes in melanoma. Such is the case of PREX2 (a guanine nucleotide exchange factor and a PTEN regulating protein) and GRIN2A (a ionotropic glutamate receptor), both recently described in WES studies with mutations distributed along the length of the entire gene, and in frequencies around 14% and 33%, respectively17, 18. In our cohort, PREX2 frequency is consistent with previous reports; however GRIN2A prevalence was lower than previously described. In this respect, it is worth mentioning that all variants with a benign in silico prediction were excluded. Additionally, we have analyzed the ERBB4 gene, that was initially found to be somatically mutated in 19% of patients with cutaneous melanoma19, although subsequent studies have reported lower prevalences, in coexistence with BRAF or NRAS mutations20. The prevalence of pathogenic variants in our series was 12%, of whom 50% (6/12) were concurrent with BRAF/NRAS. Combination strategies for targeted therapies with BRAF inhibitors and ERBB family kinases inhibitors have been pointed out as a promising therapeutic option in the future. In addition, among the newly identified cancer genes are RAC1 and PPP6C. The RAC1 p.P29S mutation was present in 7% of the samples and as previously described, it was significantly associated with sun-exposed melanomas21. This activating mutation may have clinical implications as it has been associated to aggressive melanoma features22, and it has been described to regulate PD-L1 expression23 and to confer resistance to BRAF inhibitors24. Hotspot mutations in PPP6C have been found with an overall prevalence of 8%22, 25, significantly associated with NRAS-mutated melanomas26. We have identified an overall prevalence of 6%, with the p.R264C PPP6C hotspot in 4% of our samples, mostly in concurrence with BRAF mutations rather than NRAS mutations. Although several studies have highlighted the relevance of these recently described genes, clinical and therapeutic impact of such genes will need to be determined in future studies.
As recently described by TCGA3, our panel provides a genomic classification of melanoma with four different subtypes. It should be noted that we have a higher prevalence in the triple-wt subtype. This could be explained in part due to the lower prevalence in NF1 gene, as we did not performed WES and therefore our panel did not covered the entire CDS. An alternative explanation of the different prevalence is that our cohort only included primary melanomas.
Considering the recent development of targeted therapies, it is of great importance to identify molecular alterations that contribute to the appearance of resistances in a high percentage of the patients27. In this regard, our panel design covers genes that have been related to such mechanisms of resistance, as MAP2K1, MAP2K2, MITF, PTEN, PIK3CA, NF1, RAC1 and HOXD8. Therefore, application of this melanoma-specific panel could be of interest in the management of metastatic patients, not only for the detection of actionable mutations, but also to identify those patients who may more likely benefit from those treatments.
In summary, the cost-effectiveness (reduced DNA input amount, increase in sensitivity, low turn-around time, and simultaneous analysis of multiple cancer-driving genes) of our new NGS approach based on Ampliseq libraries and Ion PGM sequencing, strongly suggest its implementation in routine diagnostics. In this study we have successfully performed NGS in FFPE melanoma and we have found a wide variety of somatic mutations that may contribute to the pathogenesis of melanoma. Since there are multiple mutations in individual tumors, and each tumor has a specific genetic profile, characterization of the molecular alterations of individual samples seems to be necessary in order to develop a personalized medicine. In addition, application of this panel may also provide further information about the genetic mechanisms of resistance to available therapies, so it would be especially valuable for clinicians in the management of patients with metastatic disease.
Materials and Methods
Patients and tumor samples
One hundred melanoma FFPE tumor samples were retrieved from the tissue archives of the Departments of Dermatology and Pathology of the University Hospital La Fe, University General Hospital of Valencia, La Plana Hospital of Villarreal, and Instituto Valenciano de Oncología, Spain. This study was approved by the Institutional Review Board at University Hospital La Fe, University General Hospital of Valencia, La Plana Hospital of Villarreal, and Instituto Valenciano de Oncología, and was carried out in accordance with the approved guidelines. Writen inform consent from all patients were obtained. Main clinicopathological features of the patients are summarized in Table 5.
Table 5.
Clinicopathological characteristics of the patients included in the study.
| Variables | n = 100 |
|---|---|
| Epidemiological features | |
| Age at diagnosis [median, (range)] | 65.5 (21–90) |
| Age at diagnosis (%) | |
| <40 | 6 |
| 40–65 | 44 |
| >65 | 50 |
| Sex (%) | |
| Male | 53 |
| Female | 47 |
| Clinical melanoma features | |
| Anatomic site (%) | |
| Head/neck | 16 |
| Upper extremities | 16 |
| Trunk | 42 |
| Lower extremities | 18 |
| Acral | 8 |
| Sun related site (%) | |
| Non exposed | 21 |
| Occasionally exposed | 63 |
| Usually exposed | 16 |
| Pathological melanoma features | |
| Histological subtype (%) | |
| LMM | 10 |
| SSM | 68 |
| NM | 14 |
| AML | 8 |
| Breslow thickness (mean (±SD)) | 1.9 ± 2.1 |
| Breslow thickness (%) | |
| <1 mm | 47 |
| 1–2 mm | 21 |
| 2–4 mm | 19 |
| >4 mm | 13 |
| Ulceration (%) | |
| No | 74 |
| Yes | 26 |
| Regression (%) | |
| No | 52 |
| <50% | 41 |
| >50% | 7 |
| Mitoses/mm2 (%) | |
| <1 | 48 |
| >1 | 52 |
| Growth rate (%) | |
| SGM | 76 |
| FGM | 24 |
| Tumor stage (%) | |
| Localized (I–II) | 75 |
| Locoregional and metastatic disease (III–IV) | 25 |
| Months follow-up [median, (range)] | 32 (6–121) |
| Clinical outcome (%) | |
| Stable disease | 81 |
| Relapse (locoregional vs distant metastasis) | 8 |
| Exitus | 11 |
DNA preparation
Genomic DNA was isolated from two 10-μm thick FFPE sections using Deparaffinization Solution and the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. This isolation kit contains Uracil-DNA-Glycosylase (UNG) treatment that prevents formalin-fixation induced artifacts, which may lead to false-positive mutation callings28. DNA concentration was quantified by Qubit dsDNA HS Assay Kit (ThermoFisher Scientific). Genomic DNA with at least 10 ng/μL was subjected to library preparation.
Integrative molecular analysis by Next-generation sequencing
Melanoma-specific panel design
A custom panel was designed using Ion AmpliSeq Designer platform (www.ampliseq.com) to cover the coding regions of 35 melanoma-related genes (NRAS, ERBB4, HOXD8, ALK, MITF, BPA1, PIK3CA, KIT, TERT, EPHA7, STK19, BRAF, MET, GRM3, RAC1, STK31, TRRRAP, PREX2, CDKN2A, GNAQ, TAF1L, PPP6C, PTEN, HRAS, KRAS, CDK4, MAP2K1, MC1R, GRIN2A, ADAMST18, NF1, GNA11, MAP2K2, STK11, IRS4). The selection of the genes to be included in this panel was based on the relatively high frequency of mutations over other whole-exome sequencing (WES) studies, as well as their possible diagnostic, prognostic and therapeutic implication. In five genes, sequencing of the complete coding sequence (CDS) was performed, whereas in the other 30 genes specific targeted exonic regions were studied, focusing on areas where hotspot mutations are known to occur. The panel covered 23,702 base pairs (bp) with an average coverage of 97.8%. A total of 515 amplicons were designed with an amplicon range of 125–175 bp. All panel details are summarized in Table S2.
Ion Torrent Library preparation
Library preparation was performed using the Ion Ampliseq Library kits 2.0 (Thermo Fisher Scientific). Multiplex PCR amplification of 10 ng of DNA was performed using the custom Ion AmpliSeq Primer Pool and the Ion AmpliSeq Hifi Mix. (Thermo Fisher Scientific) according to manufacturer’s protocol. Primer sequences were partially digested using FuPa Reagent, and Ion Torrent adapters and Ion Xpress Barcodes were ligated with DNA ligase. Following adapter’s ligation, amplicons were purified with Agencourt® AMPure® XP (Beckam Coulter), and subsequently quantification of the amplified library was performed using Ion Library Equalizer Kit (ThermoFisher Scientific) according to manufacturer’s protocol.
Emulsion PCR and DNA sequencing
The library pool was clonally amplified in an emulsion PCR reaction using Ion Sphere Particles (ISPs) on the One Touch 2 Instrument, and subsequently template-positive ISPs were enriched on the Ion One Touch ES (Thermo Fisher Scientific) as described by the manufacturer. Enriched template-positive ISPs were subjected to sequencing on the Ion Torrent PGM on a 318v2 Chip (four samples per chip) (Thermo Fisher Scientific).
Sensitivity assay
Sensitivity was assessed by sequencing serially diluted DNA isolated from two cell lines: DNA isolated from HT-29 (ACC-299), a human colorectal adenocarcinoma cell line with a known heterozygous mutation in BRAF (p.V600E) and a polymorphism in MC1R (p.R160W), was diluted into DNA obtained from another colorectal adenocarcinoma cell line Caco-2 (ACC-169), known to be wild-type for the mentioned variations, in ratios of 1:1, 1:3, 1:24, 1:49, 1:99, 1:999 resulting in 25%, 12.5%, 2%, 1%, 0.5% and 0.05% dilutions of the mutated allele, respectively. To asses reproducibility sequencing runs of the diluted DNA were performed in two independent experiments.
Variant Calling and experimental validation
Data from sequencing runs were transferred to the Torrent Server, and Ion Torrent Suite Software was used to generate initial variant calling. Filtered variants were annotated using Ion Reporter software. Exclusion of non-exonic variants and synonymous mutations was carried out. Subsequently, several steps were used in order to filter out variants with low read numbers: a minimum depth of total coverage ≥500 reads, an each variant coverage of ≥20 reads, and P-value < 0.01. Moreover, mutations were visually examined using Integrative Genomics Viewer (IGV) software. In addition, detected missense mutations in BRAF, NRAS, KRAS and KIT were subsequently confirmed by different methods. BRAF mutations were validated by High Resolution Melting (HRM) as previously described29, and Sanger’s sequencing; NRAS and KRAS mutations were confirmed by RQ-PCR using AmoyDx® KRAS/NRAS Mutations Detection Kit, according to manufacturer’s instructions; and KIT mutations were validated by direct sequencing.
Prediction tools analysis
Databases such as COSMIC, TCGA and dbSNP were used to assess recurrent known mutations and to exclude reported germline polymorphisms. Furthermore, in all the variants of unknown significance we aimed to identify those ones likely to impact protein function using five prediction tools such as Provean, SIFT (Sorting Intolerant From Tolerant), PolyPhen-2 (Polymorphism Phenotyping v2), SNPS&GO, and Condel, which use algorithms that predict the effect of amino acid substitution on the protein structure and function. We excluded the variants predicted as “benign” by at least three of the five applied prediction tools.
Detection of TERT promoter mutations by direct sequencing
Mutational status of the TERT promoter region (from position −27 to −286 from ATG start site) was determined by PCR and Sanger sequencing. A 260 bp region was amplified using a pair of primers previously described30. The PCR was performed in a conventional thermal cycler using the following cycling conditions: initial heating at 95 °C for 5 min followed by 40 cycles of 45 s denaturation at 95 °C, 45 s annealing at 59 °C, 36 s extension at 72 °C, and, finally, 72 °C for 10 min. PCR product was subsequently sequenced on the ABIprism 3130 (Applied Biosystems).
Statistical analysis
Quantitative variables were summarized by their mean and standard deviation, and categorical variables by relative and absolute frequencies. The relationship between mutations and clinicopathological features was evaluated using logistic regression analysis with estimation of OR and 95% CI. P values < 0.05 were considered statistically significant. Computations were performed using the SPSSv21.statistical package (Chicago, IL).
Electronic supplementary material
Acknowledgements
We would like to thank to the Genomic Unit (Health Research Institute La Fe) for having participate in the development of the study, and the Biostatistics Unit (Health Research Institute La Fe) for having contribute to the analysis of the results. This study was supported in part by the IISLaFe for having granted the projects 2014/0370 and 18/2015; the ISCIII for having granted the projects PIE13/00046 and PI16/01559; Conselleria d’ Educació, Investigació, Cultura i Esport for having granted the project GV/2016/064.
Author Contributions
B.U., R.M., S.P. and R.B. participated in designing the study. B.U., B.E., M.R., R.B., E.N. and R.B. provided patient specimens and collected clinical data, V.A. and M.L.L. conducted histopathological examinations. B.U., R.M., G.P., I.J., S.P. conducted experiments and participated in data analysis. B.U., R.M., S.P. and R.B. wrote the paper. All authors reviewed and approved the final version of the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Blanca de Unamuno Bustos and Rosa Murria Estal contributed equally to this work.
Sarai Palanca Suela and Rafael Botella Estrada jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00606-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 2.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–1155. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson IR, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niezgoda A, Niezgoda P, Czajkowski R. Novel Approaches to Treatment of Advanced Melanoma: A Review on Targeted Therapy and Immunotherapy. Biomed Res Int. 2015;2015:851387. Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 6.Singh RR, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Tsongalis GJ, et al. Routine use of the Ion Torrent AmpliSeq™ Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 8.Martorell-Calatayud A, et al. Defining fast-growing melanomas: reappraisal of epidemiological, clinical, and histological features. Melanoma Res. 2011;21:131–138. doi: 10.1097/CMR.0b013e328342f312. [DOI] [PubMed] [Google Scholar]
- 9.Siroy AE, et al. Beyond BRAF(V600): clinical mutation panel testing by next-generation sequencing in advanced melanoma. J Invest Dermatol. 2015;135:508–515. doi: 10.1038/jid.2014.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griewank KG, Schadendorf D. Panel sequencing melanomas. J Invest Dermatol. 2015;135:335–336. doi: 10.1038/jid.2014.420. [DOI] [PubMed] [Google Scholar]
- 11.Chen K, et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem. 2015;61:544–553. doi: 10.1373/clinchem.2014.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartels S, Schipper E, Kreipe HH, Lehmann U. Comprehensive Molecular Profiling of Archival Bone Marrow Trephines Using a Commercially Available Leukemia Panel and Semiconductor-Based Targeted Resequencing. PLoS One. 2015;29:10. doi: 10.1371/journal.pone.0133930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pópulo H, et al. TERT promoter mutations in skin cancer: the effects of sun exposure and X-irradiation. J Invest Dermatol. 2014;134:2251–2257. doi: 10.1038/jid.2014.163. [DOI] [PubMed] [Google Scholar]
- 15.Jakob JA, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jönsson, M. et al. Experiences from treatment-predictive KRAS testing; high mutation frequency in rectal cancers from females and concurrent mutations in the same tumor. BMC Clin Pathol9, doi:10.1186/1472-6890-9-8 (2009). [DOI] [PMC free article] [PubMed]
- 17.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prickett TD, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manca A, et al. Mutations in ERBB4 may have a minor role in melanoma pathogenesis. J Invest Dermatol. 2013;133:1685–1687. doi: 10.1038/jid.2013.27. [DOI] [PubMed] [Google Scholar]
- 21.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mar VJ, et al. Clinical and pathological associations of the activating RAC1 P29S mutation in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2014;27:1117–1125. doi: 10.1111/pcmr.12295. [DOI] [PubMed] [Google Scholar]
- 23.Vu HL, Rosenbaum S, Purwin TJ, Davies MA, Aplin AE. RAC1 P29S regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res. 2015;28:590–598. doi: 10.1111/pcmr.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson IR, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J, et al. A meta-analysis of somatic mutations from next generation sequencing of 241 melanomas: a road map for the study of genes with potential clinical relevance. Mol Cancer Ther. 2014;13:1918–1928. doi: 10.1158/1535-7163.MCT-13-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil-DNA glycosylase. Oncotarget. 2012;3:546–558. doi: 10.18632/oncotarget.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 30.Heidenreich B, et al. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat Commun. 2014;5:3401. doi: 10.1038/ncomms4401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


