Abstract
Genetically encoded fluorescent Zn2+ indicators (GEZIs) are highly attractive research tools for studying Zn2+ homeostasis and signaling in mammalian cells. Most current GEZIs are based on Förster resonance energy transfer (FRET) between a select pair of fluorescent proteins (FPs) fused with Zn2+-binding motifs. One drawback of such FRET-based GEZIs is their broad spectral profile bandwidths, creating challenges when monitoring multiple targets or parameters. To address this issue, we have engineered a group of intensiometric GEZIs based on single teal and red FPs that can be utilized to monitor subcellular Zn2+ diffusion and glucose-induced Zn2+ secretion in pancreatic INS-1E β-cells. These GEZIs offer the simplicity of intensiometric measurements, compatibility in multicolor imaging, large dynamic ranges, and relatively small molecular sizes, making them valuable additions to the molecular toolbox for imaging Zn2+.
Graphical Abstract
INTRODUCTION
Zinc ion (Zn2+) is one of the most abundant transition metal ions in living organisms and plays critical roles in maintaining and regulating protein structure, function, and dynamics1. As an essential element, Zn2+ can exert its biological functions as an enzyme cofactor1, signaling mediator2, and regulator of fundamental cellular processes, such as gene transcription, apoptosis, adaptive immunity, and neurotransmission3. Approximately 3,200 proteins (~10% of the human proteome) require Zn2+ to function properly4. Zn2+ deficiency has been linked to compromised immunity, impaired cognition, diarrhea, and abnormal brain development5; whereas Zn2+ overabundance potentially leads to cytotoxicity6. Moreover, the interplay between Zn2+ and other cellular components has implications in the pathogenesis of a variety of diseases, such as type I diabetes7, cancer8, and neurodegeneration9. Cells have evolved delicate systems to maintain Zn2+ homeostasis, including Zn2+-buffering metallothioneins and Zn2+-storing vesicles or granules10,11.
Fluorescent Zn2+ indicators have played pivotal roles in understanding the biological roles of Zn2+. The past two decades have witnessed the development of a large array of small molecule-based fluorescent Zn2+ indicators12–15. With diverse colors, response mechanisms, kinetics, and binding affinities, these synthetic Zn2+ indicators have proven to be invaluable tools16,17. While the development of novel synthetic Zn2+ indicators is still an attractive research field, a new focus on genetically encoded fluorescent Zn2+ indicators (GEZIs) has emerged. These genetically encoded indicators generally bestow a number of favorable features, such as high affinity and specificity obtained from naturally evolved Zn2+-binding domains, a capacity for precise subcellular localization, and the convenient choice of stable or transient transfection. In the past few years, these GEZIs, notably the eCALWY18 and ZapCY19 families, have been instrumental for elucidating steady-state Zn2+ concentrations and intracellular Zn2+ dynamics, such as those between the cytoplasm, mitochondria, endoplasmic reticulum (ER), and Golgi apparatus in a variety of cell lines. These GEZIs are typically based on Förster resonance energy transfer (FRET) between a cyan fluorescent protein (CFP) and a yellow fluorescent protein (YFP) linked by Zn2+-binding motifs16,20,21, resulting in ratiometric signal outputs in response to Zn2+ concentration changes. Recently, Merkx and co-workers created a de novo Cys2His2 Zn2+-binding pocket on the surfaces of CFP and YFP and directly fused the two to derive an alternative FRET-based Zn2+ sensor22. These FRET-based GEZIs respond to Zn2+ ratiometrically. Because FRET ratios are less dependent of protein expression levels than the intensities of intensiometric biosensors, these FRET-based GEZIs have been quite useful for (semi-)quantitative analysis of Zn2+ concentrations in live cells. Despite the progress, these FRET-based indicators have relatively large molecular sizes, modest dynamic ranges (i.e. changes of FRET ratios for ratiometric sensors), and broad fluorescence excitation and mission profiles that increase the technical difficulty to perform multicolor or/and multiplex imaging experiments. More recently, FRET-based GEZIs derived from fluorescent proteins spectrally compatible with CFP and YFP have been reported and used for dual-parameter imaging, but these indicators still suffer from significant spectral bleed-through and/or much reduced dynamic range22–24.
Our initial inspiration for this work was the tremendous success of developing single-FP based Ca2+ indicators, such as GCaMP25, Pericam26, and GECOs27. Compared to FRET-based GEZIs, common single-FP based Ca2+ sensors have a larger dynamic range (i.e. changes of fluorescent intensities for intensiometric sensors), an increased signal-to-noise ratio, and a narrower spectral profile better suited for multiparameter imaging. Although early studies created artificial Zn2+-binding sites28 or inserted zinc fingers29 into single fluorescent proteins, these constructs were not subjected to much optimization and only showed modest intensity changes and Zn2+ affinities. Herein, we report our efforts in designing, engineering, and characterizing several fluorescent GEZIs based on single-FP domains. We fused Zn2+-binding motifs with either teal or red FPs, followed by directed evolution to optimize their fluorescence responses to Zn2+. In particular, we have derived two green fluorescent Zn2+-turn-off sensors, ZnGreen1 and ZnGreen2, showing 26.3-fold and 8.7-fold Zn2+-induced fluorescence decreases, respectively. Additionally, we developed one red fluorescent turn-on sensor, ZnRed, showing a 3.8-fold fluorescence enhancement by Zn2+. To our knowledge, the dynamic range of ZnGreen1 is the largest among known FP-based probes excluding those well-optimized calcium indicators. The use of these sensors to image Zn2+ in cultured mammalian cells has also been demonstrated. This work may serve as the foundation for developing additional single-FP based GEZIs, which may complement existing FRET-based GEZIs and expand the fluorescent toolbox to accelerate our understanding of Zn2+ biology.
EXPERIMENTAL SECTION
General Materials and Methods
All chemicals were purchased from Sigma Aldrich (St. Louis, MO) or Fisher Scientific (Hampton, NH) and used directly. Glucose for cell stimulation during mammalian cell imaging was purchased as a 45% solution from Corning Cellgro (Manassas, VA). Synthetic DNA oligonucleotides and the Zap1 gene block were purchase from Integrated DNA Technologies (San Diego, CA). Restriction endonucleases were purchased from New England Biolabs (Ipswich, MA) or Thermo Scientific Fermentas (Vilnius, Lithuania). PCR and restriction digested DNA products were purified using Syd Laboratories Gel Extraction columns (Malden, MA). DNA sequences were analyzed by Retrogen (San Diego, CA). Absorbance and fluorescence spectroscopy data were collected on a monochromator-based BioTek Synergy Mx Microplate Reader (BioTek, Winooski, VT).
Engineering of Zn2+ Sensors and in vitro Characterization
The detailed procedure to engineer and characterize ZnGreen and ZnRed sensors is presented in the Supporting Information S-2.
Construction of Mammalian Expression Plasmids
To construct mammalian expression vectors, the full-length genes of the ZnGreen1, ZnGreen2, and ZnRed sensors were amplified from their corresponding E. coli expression vectors with oligo pairs pCMV-ZnGreen1-F and pCMV-ZnGreen1-R, pCMV-ZnGreen2-F and pCMV-ZnGreen2-R, and pCMV-ZnRed-F and pCMV-ZnRed-R, respectively. The amplified PCR products were digested with Hind III and Xho I and ligated into a pre-digested pcDNA3-pnGFP-WPRE plasmid30. To construct vectors with nuclear localization, oligos pNuc-ZnRed-F and pNuc-ZnRed-R were used to amplify the ZnRed gene from plasmid pTorPE-ZnRed. PCR products were then digested with Nhe I and Xho I, and ligated into a predigested phsGFP-Nuc plasmid 31 to create pNuc-ZnRed, which contains three copies of the nuclear localization signal (NLS) sequence (DPKKKRKV) at its C-terminus. To localize ZnGreen1 sensor to the extracellular cell surface, a modified pDisplay vector (Life Technologies, Carlsbad, CA) was first amplified with primers pDisplay-Vector-F and pDisplay-Vector-R. The ZnGreen1 gene was then amplified with primers ZnGreen1-Display-F and ZnGreen1-Display-R, and inserted into the pDisplay vector by the Gibson Assembly Cloning method32 to create pDisplay-ZnGreen1. Plasmid identities were verified by DNA sequencing. Sensor localizations were confirmed by fluorescence microscopy of transfected cells.
Mammalian Cell Culture, Transfection, and Imaging
Human Embryonic Kidney (HEK) 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Rat insulinoma INS-1E cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 Medium supplemented with 10% FBS, 10 mM HEPES, 2 mM L-glutamine, 1 mM sodium pyrurate and 0.05 mM 2-mercaptoethanol. All cells were incubated at 37°C with 5% CO2 in humidified air. HEK 293T cells were split into 35 mm Petri dishes with glass bottom coverslips pretreated with poly-lysine and transfected at 70% confluency with 3 μg of plasmid DNA and 10 μg of PEI (polyethylenimine, linear, 25 kDa) for two hours. Transfected HEK293T cells were then cultured in complete DMEM medium for an additional 48-hour. INS-1E cells were split and seeded similarly to HEK293T cells. At 50% confluency, INS-1E cells were transfected with jetPRIME transfection reagent (Polyplus-transfection SA, France) according to the manufacturer’s instructions. INS-1E cells were incubated in the transfection mixture for 3 hours and further cultured in complete RPMI 1640 medium for another 3 days before imaging. Cells were imaged in modified Krebs-Hepes-bicarbonate buffer (KHB) containing 140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 10 mM Hepes, 2 mM NaHCO3 and 3 mM glucose. Before imaging, KHB buffer was pre-equilibrated with 95:5 air:CO2. Transfected cells were washed three times with KHB buffer and were imaged on a Leica SP5 confocal fluorescence microscope with a 40× objective lens. The Leica SP5 microscope was equipped with an argon-ion laser with 488 nm to excite ZnGreen, and a HeNe laser with 543 nm to excite ZnRed. Fluorescence emission was collected with a highly sensitive prism spectral detector, and 495–540 nm and 580–650 nm were utilized to collect fluorescence emission for ZnGreen1 and ZnRed, respectively.
To test sensor responses to Zn2+, time-lapse fluorescence imaging was performed with images acquired every minute and stimulating chemicals added between acquisitions. The Zn2+ chelator TPEN (N,N,N′,N′-tetrakis(2-pyridylmethyl)ethane-1,2-diamine) and pyrithione ionophore were freshly prepared from DMSO stock solutions and diluted with KHB buffer to desired concentrations. ZnCl2 was prepared as a stock solution in slightly acidic pure water and was diluted in KHB buffer upon usage. 50 μM Zn2+ and 5 μM pyrithione was used to load cells with Zn2+ and 200 μM TPEN was used to deprive free Zn2+.
Dual color imaging was performed on the SP5 confocal microscope. Before imaging, cells were washed three times with and then maintained in KHB buffer. ZnGreen1 was excited with a 488 nm argon-ion laser, and the green fluorescence emission was collected between 495–540 nm with a PMT (photomultiplier tube) detector. ZnRed was excited with a 543 nm HeNe laser, and the red fluorescence emission was collected between 560–700 nm with a hybrid detector (HyD). Time-lapse series was acquired at 1 min interval and fluorescence emissions from both channels were scanned sequentially with the “between lines” mode. Following a few initial image acquisitions, Zn2+/ pyrithione/KHB (50 μM Zn2+ and 5 μM pyrithione) solution was added and the time series continued for another 10 min.
To stimulate zinc release in INS-1E cells, transfected cells expressing pDisplay-ZnGreen1 were first incubated with 10 μM EDTA/KHB solution for 15 min to remove pre-bound metals. Cells were then washed three times with and then maintained in KHB buffer to remove EDTA before imaging. Cells were imaged on the confocal SP5 microscope. ZnGreen1 was excited with a 488 nm argon-ion laser and green fluorescent emission was collected between 495–600 nm with a hybrid detector (HyD). Following the initial four image acquisitions, INS-1E cells were stimulated with 35 mM glucose/KHB solution and time-lapse fluorescence imaging continued for another 20 min.
All images and videos were processed and rendered with ImageJ.
RESULTS AND DISCUSSIONS
Development of Green Fluorescent Zn2+ Indicators
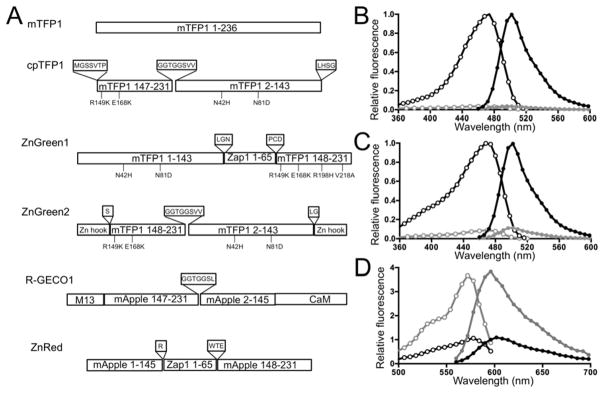
The first and second zinc fingers of Saccharomyces cerevisiae transcription factor, Zap1, is an ideal proteinaceous, Zn2+-binding domain for constructing GEZIs due to their small size, high Zn2+-binding affinity, and Zn2+-induced conformational change, and it has been successfully exploited to derive FRET-based GEZIs.19,33 In a similar strategy to the insertion of a Ca2+-binding calmodulin domain into a FP, as in “Camgaroo” indicators29, we reasoned that inserting Zap1 zinc fingers into FPs would be a promising strategy to create GEZIs. For the fluorescent element, we selected monomeric teal fluorescent protein (mTFP1) because of its high fluorescence brightness, excellent photostability, and efficient chromophore maturation at 37°C34. Directly inserting Zap1 into the β-barrel nearest the phenolate chromophore of mTFP1 would disrupt the chromophore folding and subsequently complicate attempts to rescue fluorescence loss caused by this insertion. Thus, to make the β-barrel more tolerable to modifications, we first created an efficiently folded, highly fluorescent circularly permutated version of mTFP1. We fused the original N- and C-terminal ends with a GGTGGS hexapeptide linker and split mTFP1 near the phenolate chromophore between residues 144 and 145 (Figure 1A). Because the resultant recombinant cpFP (circularly permuted FP) was essentially non-fluorescent, we then used degenerate codons to extend both the new N- and C-termini in order to improve the fluorescence. Screening the resultant library yielded a moderately improved fluorescent clone, cpTFP0.5, which harbored extra N- and C- terminal extensions (Supplemental Information Figure S1). We next performed five successive rounds of directed evolution by using error-prone PCR-generated libraries and screened the resultant bacterial colonies after over-night incubation at 37°C for improved fluorescence brightness. This effort led to cpTFP1, which harbors five additional mutations (N42H, N81D, S146P, R149K, and E168K; residues numbered according to the numbering of mTFP1 in PDB 2HQK; Figure S1) and shows bright fluorescence with fast chromophore maturation (Figure 1A). Next, we connected the N- and C-termini of cpTFP1 with Zap1 zinc fingers, and converted cpTFP1 back to the wild-type topology to re-establish its original N- and C-termini (ZnGreen0.1). ZnGreen0.1 expression in Escherichia coli yielded very dim fluorescence with poor Zn2+-dependent fluorescence decrease (20–30%). As such, we conducted systematic optimization of the two peptide linkers flanking the Zap1 domain and screened for mutants exhibiting larger Zn2+-dependent responses. The two linkers are important for tuning the responses, because of their structural proximity to the chromophore to participate in the intricate protonation network of mTFP1 and their direct roles in transducing conformational changes from fused domains to the chromophore environment, as suggested by similar work in genetically encoded Ca2+26,27,35 and glutamate36 indicators. To optimize the two linkers in ZnGreen0.1, we built nine gene libraries that harbored randomized sequences in the two linkers (Table S1). The libraries were used to transform E. coli and fluorescent colonies were then cultured in 96-well plates to harvest crude protein extracts for responses to Zn2+ compared to EDTA, a potent Zn2+ chelator. The dynamic range, expressed as the fluorescence ratio of EDTA- to Zn2+-treated crude protein extracts, was scored for each variant, with variants showing dynamic ranges at the extremes being processed for further analysis. These efforts led to ZnGreen1, a variant showing 26.3-fold fluorescence decrease upon Zn2+ binding (Figure 1B), which has a significantly greater absolute dynamic range than those of current FRET-based GEZIs18 and is comparable to commonly used small molecule-based Zn2+ probes15. Most clones in the libraries were not or only slightly responsive to Zn2+. Among the variants showing response to Zn2+, both turn-on and turn-off type variants were observed. Those fluorescent turn-on mutants were not further pursued due to their relatively small dynamic ranges. We randomly selected a Zn2+-irresponsive mutant and named it ZnGreen-N as a potential control probe in future experiments. ZnGreen-N is identical to ZnGreen1, except for two residues in one of the two linkers (Figure S1).
Figure 1. Engineering of single-FP based GEZIs.
(A) Domain arrangements of various constructs. Important linker sequences are shown in individual boxes, and mutations within the FP scaffold are also indicated. The full amino acid sequences of these constructs are available in Figures S1 and S2. (B–D) Fluorescence excitation (open circle) and emission (filled circle) spectra of ZnGreen1 (B), ZnGreen2 (C) and ZnRed (D), in the presence (gray line, treated with 100 μM ZnCl2) and absence (black line, treated with 100 μM EDTA) of Zn2+.
Because of the successful examples of genetically encoded Ca2+ indicators, such as GCaMP37 and GECO27 families, we also created single-FP based GEZIs based on cpFPs. We chose a minimal zinc hook peptide from Pyrococcus furiosus Rad50, which is known to undergo Zn2+-mediated homodimerization38. We fused one copy of this peptide to each of the two termini of cpTFP1 and performed optimization on both linkers based on Zn2+-induced responses using a strategy similar to that of ZnGreen1 screening (Table S2). We eventually derived ZnGreen2, with an 8.7-fold fluorescence turn-off response to Zn2+ (Figures 1A and 1C).
Development of Red Fluorescent Zn2+ Indicators
Narrow photoemission bandwidth is another important feature of genetically encoded single-FP based indicators because it enables visualization of multiple concurrent biological processes with spectrally compatible indicators. Multiple indicators can be employed in the same live cell to independently and simultaneously detect distinct cellular species, be anchored to different subcellular compartments, and be detected by compatible fluorescence emission. With many well-optimized green hue indicators being available, there has recently been a push toward developing red-shifted indicators with longer (red-shifted) wavelengths to facilitate multiplex imaging of biological events. Compared to a green indicator, a red indicator offers the advantage of increased tissue penetration, reduced photodamage upon imaging, and minimized background autofluorescence. Using the same strategy to create ZnGreen1, we developed a single red FP (RFP)-based GEZI. We first utilized Zap1 to link the N- and C-termini of a circularly permuted mApple (cpmApple) from a red fluorescent Ca2+ indicator, R-GECO127, and re-established the original N- and C-termini of mApple, thereby creating a chimeric protein template, ZnRed0.1. Next, similar linker optimization and screening procedures, as in ZnGreen1 development, were applied to four different libraries (Table S3) from which a fluorescent turn-on sensor with 3.8-fold dynamic range was derived—designated as ZnRed (Figures 1A, 1D, and S2). The creation of ZnRed has not only expanded the palette of GEZIs, but is also suggestive that our approach to indicator design and screening can be transferred to other FP templates to further derive indicators with distinct photophysical properties39,40.
Characterization and Initial Validation of Zn2+ Indicators
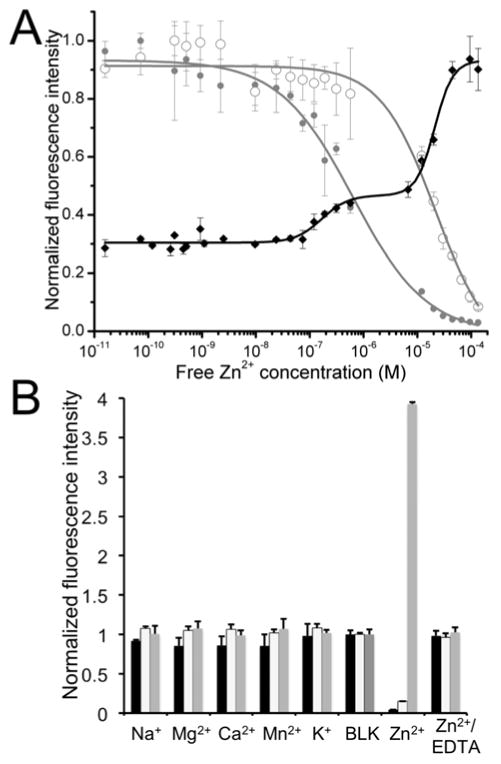
We next characterized the aforementioned indicators in vitro and validated their use in live mammalian cells using fluorescence microscopy. Titrating ZnGreen1 and ZnGreen2 with buffered Zn2+ solutions revealed apparent binding affinities (Kd’s) of 633 ± 161 nM and 20 ± 2.7 μM, respectively, whereas plotting ZnRed fluorescence intensities versus Zn2+ concentrations revealed two different Zn2+-interacting modes showing apparent binding affinities of 166 ± 56 nM and 20 ± 2.0 μM (Figure 2A). The affinities we obtained here are significantly lower than the reported values for the corresponding free Zn2+-binding domains 38,41. To assure our procedure was properly carried out, we also measured the Zn2+ affinity of eCALWY1 and were able to derived a Kd value similar to the reported.42 It is important to note that these apparent Kd values do not directly reflect the absolute Zn2+-binding affinity, but rather only the Zn2+-induced conformational changes in the chimeric proteins that were able to result in fluorescence changes. Such discrepancies may be attributable to protein fusions that could possibly rigidify the domains and affect Zn2+ binding, as was analogously observed in previously reported “camgaroo” type Ca2+ indicators 29, and the FRET-based GEZI employing a canonical Cys2His2 zinc finger from the mammalian transcription factor, Zif26843. It may also be possible that some residues in the engineered linkers of ZnGreen1 and ZnRed provide new coordination sites for Zn2+. Specific interactions between Zn2+ and the linker residues of ZnGreen2 are unlikely, because the side chains of these linker residues (Ser, Leu and Gly) are typically inert to Zn2+ binding.
Figure 2. In vitro characterizations of engineered Zn2+ sensors.
(A) Zn2+ titration curves for ZnGreen1 (gray, filled circle), ZnGreen2 (gray, open circle) and ZnRed (black, filled circle). Curves were fit according to the equations described in Supporting Information, assuming a single-binding mode for ZnGreen1 and ZnGreen2 and a double-binding mode for ZnRFP. (B) Fluorescence responses of ZnGreen1 (black bar), ZnGreen2 (white bar) and ZnRed (gray bar) to various metal ions, showing excellent selectivity toward Zn2+. The final metal concentrations were 20 μM for Zn2+ and 100 μM for all other metals and EDTA. Fluorescence intensities were normalized to blank samples (BLK, no metal ion added).
Upon Zn2+ chelation, the absorbance of ZnGreen indicators decreases at 470 nm and concurrently increases at 400 nm, corresponding to the transformation of deprotonated to protonated chromophores (Figures S3). Other highly abundant cellular metals, including Mg2+, Ca2+, K+, and Na+, triggered little to no fluorescence response of ZnGreen1, ZnGreen2, and ZnRed (Figure 2B), confirming the Zn2+-specific responses of our newly engineered GEZIs. Moreover, addition of EDTA to Zn2+-treated proteins brought their fluorescence back to the original levels, suggesting the reversibility of these GEZIs.
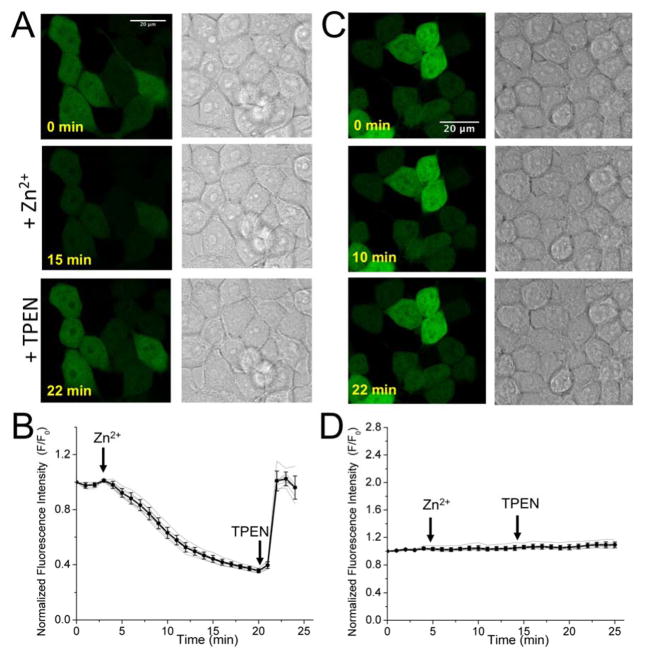
Since ZnGreen1 has a larger dynamic range and higher Zn2+-affinity than ZnGreen2, we utilized ZnGreen1 as the green fluorescent GEZI in all further experiments with live mammalian cells. When expressed in live HEK 293T cells, ZnGreen1 showed dramatic fluorescence decrease upon addition of ZnCl2 and a pyrithione ionophore which facilitates the delivery of Zn2+ into cells. Subsequent addition of a cell-permeable zinc chelator, TPEN (N,N,N′,N′-tetrakis(2-pyridylmethyl)ethane-1,2-diamine), quickly rescued the fluorescence (Figures 3A and 3B). The fluorescence of the cells reached a level close to their initial intensities, suggesting that almost no Zn2+ was associated with ZnGreen1 in the rest state. We also treated our cells first with TPEN, and next, with Zn2+. TPEN caused very little fluorescence change and Zn2+ drastically quenched the fluorescence (Figure S4), supporting that the responses of ZnGreen1 were not affected by the order of addition of TPEN and Zn2+. A large dynamic range of the ZnGreen1, as revealed in the time-lapse fluorescence imaging and single cell fluorescence analysis, is consistent with our in vitro characterization. To further assure that the changes were not caused by pH fluctuations, we treated HEK 293T cells expressing the Zn2+-irresponsive control probe, ZnGreen-N, with ZnCl2/pyrithione and TPEN, and observed no obvious fluorescence response (Figures 3C and 3D). Because ZnGreen-N and ZnGreen1 respond to a similar range of pH changes (Figure S5), the results further support that our observed fluorescence responses of ZnGreen1 were caused by changes in Zn2+ concentrations in HEK 293T cells. Similar validation experiments were carried out with ZnRed in HEK293T, and time-lapse fluorescence imaging further confirmed its Zn2+-dependent fluorescence turn-on response (Figure S6).
Figure 3. Responses of ZnGreen1 and ZnGreen-N to Zn2+ in HEK 293T cells.
(A) Fluorescence and brightfield images of cells expressing ZnGreen1 before and after sequential treatment of Zn2+ and TPEN (scale bar: 20 μm). (B) Representative traces demonstrating intensity changes of ZnGreen1 in HEK 293T. The black line shows the average and standard deviation of seven cells. The gray lines show the traces for individual cells. The arrows indicate the time points for addition of Zn2+/pyrithione or TPEN. (C, D) Data for a control experiment in which ZnGreen-N expressing HEK 293T cells were similarly treated.
Dual-color Imaging of Zn2+ Uptake in Live Mammalian Cells
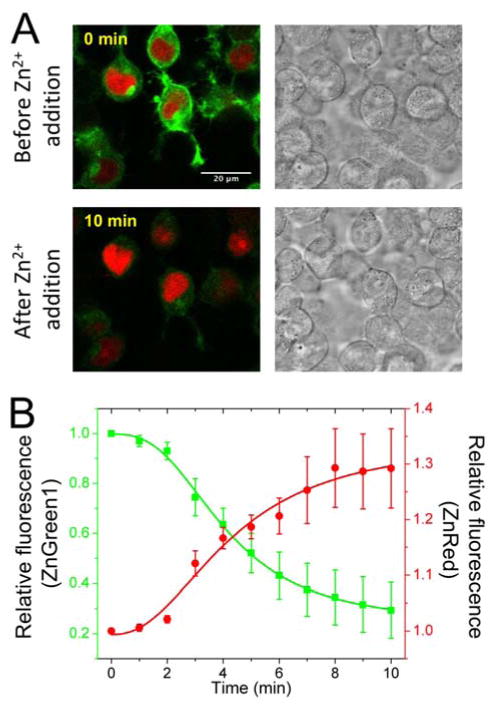
Multicolor imaging has proven to be indispensable in pushing the boundaries of what fluorescence microscopy can ultimately achieve. Optically compatible ZnGreen and ZnRed indicators have enabled us, at great convenience, to image Zn2+ dynamics at distinct subcellular locations within a single cell. Compared to other alternatives, such as dual FRET imaging22–24 and possibly a linear unmixing system of overlapping spectra44, ZnGreen and ZnRed, which utilize less photoemission bandwidth, can greatly simplify experiments. To perform dual-color imaging of Zn2+ dynamics in live mammalian cells, we localized ZnGreen1 to the cell surface with a pDisplay vector (pDisplay-ZnGreen1) and co-transfected HEK293T cells with a nuclear localized ZnRed plasmid, pNuc-ZnRed. Soon after Zn2+ addition, the membrane fluorescence from ZnGreen1 decreased dramatically, followed by an increase of nuclear red fluorescence from ZnRed, suggesting Zn2+ translocation from the extracellular space to nucleus (Figure 4 and Movie S1). Overall, the large dynamic ranges of these GEZIs, especially that of ZnGreen1, make the tracking of Zn2+ dynamics in live cells far less challenging and with high spatiotemporal resolution.
Figure 4. Fluorescence monitoring of Zn2+ diffusion in HEK 293T cells.
(A) Fluorescence and brightfield images of cells expressing ZnGreen1 at cell surface and ZnRed in the nucleus before and after addition of Zn2+/pyrithione (scale bar: 20 μm). (B) Represented traces (n = 6 cells) for intensity changes, showing Zn2+-induced decrease of ZnGreen1 fluorescence (extracellular) and increase of ZnRed fluorescence (nuclear).
Live-Cell Imaging of Glucose-Stimulated Zn2+ Secretion in Pancreatic β-Cells
While high affinity Zn2+ indicators, such as eCALWY and ZapCY118,19, have proven to be instrumental in gauging free Zn2+ concentrations in mammalian cells, their high-affinity Zn2+ binding raises concerns of perturbing intracellular Zn2+ homeostasis when expressed at high concentrations. Additionally, they are not suitable for use in certain biological contexts, such as in insulin-secreting pancreatic β-cells45,46, and synaptic vesicles of certain glutamatergic axon terminals47, which all feature fluctuations of relatively high Zn2+ concentrations. For example, it is estimated that Zn2+ released from vesicles of hippocampal mossy fiber synapses can reach to ~100 μM48. Our single FP-based GEZIs, although not suitable for measuring steady-state free intracellular Zn2+ concentrations due to their relative low affinity and intensiometric nature, would, however, fill an important technical gap to permit monitoring of the dynamics of Zn2+ in the high nanomolar to micromolar range. These new probes complement current FRET-based indicators18, thereby enabling imaging of Zn2+ in a large array of biological conditions. To validate this application, we localized ZnGreen1 to the extracellular membrane of INS-1E β-cells, a rat insulinoma cell line with enhanced sensitivity to glucose49. In pancreatic β-cells, Zn2+ is required for the biosynthesis and secretion of insulin. Typically, Zn2+ forms co-crystals with insulin in vesicular granules to aid their storage. During insulin secretion, Zn2+ and insulin are co-released to the extracellular space. These storage and secretion events are tightly regulated, with deregulation contributing to the pathogenesis of diabetes50. To image Zn2+ secretion, INS-1E cells transfected with pDisplay-ZnGreen1 was transiently stimulated with high concentrations of extracellular glucose. As expected, we observed a drastic fluorescence decrease from the plasma membrane (Figure 5A and Movie S2), indicating activation of Zn2+ release to the extracellular space, which is consistent with previous reports using small molecule probes51,52. As secreted Zn2+ diffuses into the extracellular milieu53, ZnGreen1 fluorescence gradually recovers (Figure 5B). Localizing pDisplay-ZnGreen1 to the extracellular membrane of HEK293T resulted in no fluorescence signal change upon high glucose stimulation (Figure S7), suggesting that ZnGreen1 specifically detects intracellular Zn2+ release from pancreatic β-cells. Unlike most other small molecule probes, such as FluoZin-3 53 and a recently reported ZIMIR-HaloTag52 for imaging of Zn2+ secretion, ZnGreen1 is fully genetically encoded, and therefore, may be used in live cells, tissues, or disease models for long term monitoring of Zn2+ dynamics, which will undoubtedly facilitate mechanistic understanding of paracrine signaling and diabetes.
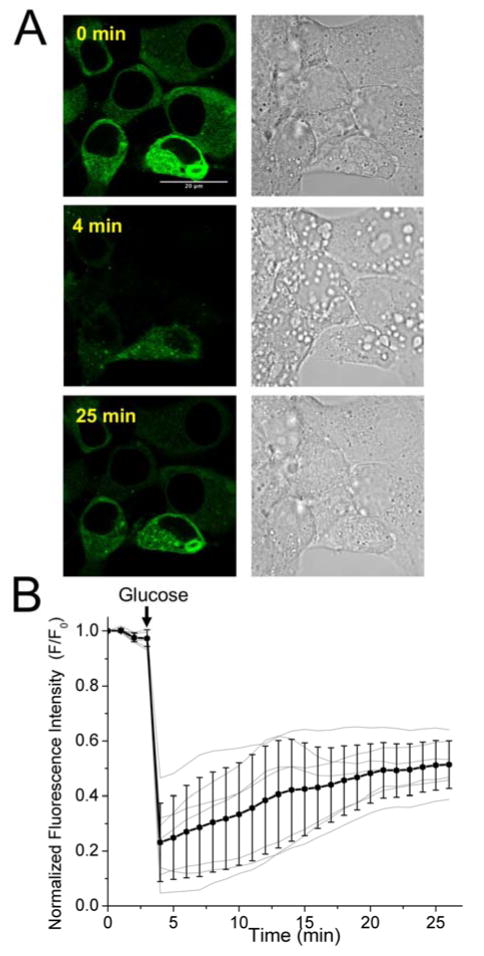
Figure 5. Fluorescence monitoring of glucose-induced Zn2+ release in pancreatic INS-1E β-Cells.
(A) Fluorescence and brightfield DIC images of cells expressing ZnGreen1 at cell surface before and after addition of glucose (scale bar: 20 μm). (B) Representative traces demonstrating intensity changes of ZnGreen1 induced by glucose. The black line shows the average and standard deviation of seven cells. The gray lines show the traces for individual cells. The arrows indicate the time points for addition of glucose.
CONCLUSIONS
We have engineered novel GEZIs by screening and developing green and red GEZIs based on single fluorescent proteins. ZnGreen1 and ZnGreen2 are two green fluorescent turn-off probes with 26.3-fold and 8.7-fold dynamic ranges, respectively, whereas ZnRed is a red fluorescent probe with a 3.8-fold turn-on response. These new GEZIs have many attractive features, including a relatively small molecular size, large dynamic range, narrow emission bandwidth, and spectrally compatible fluorescence. When expressed in live mammalian cells, these GEZIs can monitor transient, cellular Zn2+ dynamics. We have shown that ZnGreen1 and ZnRed could be utilized for dual-color imaging of Zn2+ diffusion with a relatively simple experimental setup. Furthermore, given than the estimated zinc concentrations in biological systems, which span several orders of magnitude, from 1–10 mM in zinc granules to micromolar in synapse, and down to nanomolar and picomolar under rest conditions, GEZIs with various binding affinities are needed for different biological applications (i.e. high affinity probes are suited for use under rest conditions, and low affinity probes are suited for use in zinc vesicles or under stimulated conditions).18 Our new sensors, which have high nanomolar to micromolar affinities to Zn2+, fill this unique technical gap to complement existing eCALWY and ZapCY sensors. We have demonstrated that ZnGreen1 can readily detect glucose-stimulated Zn2+ secretion in insulin-secreting pancreatic β-cells. The availability of these new probes not only creates new opportunities for multicolor, multiplex imaging, but also fills the gap in the current toolbox for Zn2+ imaging in living cells, tissues, and other disease models. Considering the importance of Zn2+ in the brain, these GEZIs may be useful for dissecting neural circuits and understanding neuronal disorders. Moreover, the probes reported here may serve as intriguing templates for further engineering of next-generation GEZIs or other types of FP-based biosensors.
Supplementary Material
Acknowledgments
We are grateful to Dr. Pierre Maechler (University of Geneva, Switzerland) for the kind gift of the INS-1E cell line, and Dr. Robert E. Campbell (University of Alberta, Canada) for plasmids pTorPE-G-GECO1 (Addgene Plasmid # 32466) and pTorPE-R-GECO1 (Addgene Plasmid # 32465). We thank the University of California-Riverside and the National Institutes of Health (R03EB020211 and R01GM118675) for financial support.
Footnotes
Supporting Information. Supplemental Information includes the sequences of oligonucleotides used in this work, sequence alignments of sensor proteins, amino acid sequences of the linkers of libraries, additional characterization and images of sensors, additional experimental procedures, and videos showing time-lapse fluorescence responses of biosensors. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Maret W. Adv Nutr. 2013;4:82–91. doi: 10.3945/an.112.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallee BL, Falchuk KH. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 3.Fukada T, Yamasaki S, Nishida K, Murakami M, Hirano T. J Biol Inorg Chem. 2011;16:1123–1134. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 5.Prasad AS. Nutrition. 2001;17:685–687. doi: 10.1016/s0899-9007(01)00598-6. [DOI] [PubMed] [Google Scholar]
- 6.Jiang DM, Sullivan PG, Sensi SL, Steward O, Weiss JH. J Biol Chem. 2001;276:47524–47529. doi: 10.1074/jbc.M108834200. [DOI] [PubMed] [Google Scholar]
- 7.Wenzlau JM, Juhl K, Yu LP, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC. Proc Natl Acad Sci U S A. 2007;104:17040–17045. doi: 10.1073/pnas.0705894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello LC, Feng P, Milon B, Tan M, Franklin RB. Prostate Cancer P D. 2004;7:111–117. doi: 10.1038/sj.pcan.4500712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szewczyk B. Front Aging Neurosci. 2013:5. doi: 10.3389/fnagi.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colvin RA, Holmes WR, Fontaine CP, Maret W. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 11.Frederickson CJ, Koh JY, Bush AI. Nat Rev Neurosci. 2005;6:449–462. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 12.Walkup GK, Burdette SC, Lippard SJ, Tsien RY. J Am Chem Soc. 2000;122:5644–5645. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T, Kikuchi K, Urano Y, Higuchi T, Nagano T. J Am Chem Soc. 2000;122:12399–12400. [Google Scholar]
- 14.Huang Z, Lippard SJ. Meth Enzymol. 2012;501:445–468. doi: 10.1016/B978-0-12-388448-0.00031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomat E, Lippard SJ. Curr Opin Chem Biol. 2010;14:225–230. doi: 10.1016/j.cbpa.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter KP, Young AM, Palmer AE. Chem Rev. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domaille DW, Que EL, Chang CJ. Nat Chem Biol. 2008;4:168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- 18.Hessels AM, Merkx M. Metallomics. 2015;7:258–266. doi: 10.1039/c4mt00179f. [DOI] [PubMed] [Google Scholar]
- 19.Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. Proc Natl Acad Sci U S A. 2011;108:7351–7356. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean KM, Palmer AE. Nat Chem Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindenburg L, Merkx M. Sensors. 2014;14:11691–11713. doi: 10.3390/s140711691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hessels AM, Chabosseau P, Bakker MH, Engelen W, Rutter GA, Taylor KM, Merkx M. ACS Chem Biol. 2015;10:2126–2134. doi: 10.1021/acschembio.5b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenburg LH, Hessels AM, Ebberink EHTM, Arts R, Merkx M. ACS Chem Biol. 2013;8:2133–2139. doi: 10.1021/cb400427b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miranda JG, Weaver AL, Qin Y, Park JG, Stoddard CI, Lin MZ, Palmer AE. PLoS One. 2012;7:e49371. doi: 10.1371/journal.pone.0049371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai J, Ohkura M, Imoto K. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 26.Nagai T, Sawano A, Park ES, Miyawaki A. Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. J Am Chem Soc. 2002;124:3522–3524. doi: 10.1021/ja0176954. [DOI] [PubMed] [Google Scholar]
- 29.Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen ZJ, Ren W, Wright QE, Ai HW. J Am Chem Soc. 2013;135:14940–14943. doi: 10.1021/ja408011q. [DOI] [PubMed] [Google Scholar]
- 31.Chen ZJ, Ai HW. Biochemistry. 2014;53:5966–5974. doi: 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- 32.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 33.Qiao W, Mooney M, Bird AJ, Winge DR, Eide DJ. Proc Natl Acad Sci U S A. 2006;103:8674–8679. doi: 10.1073/pnas.0600928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ai HW, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akerboom J, Calderon NC, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen TW, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL. Front Mol Neurosci. 2013;6 doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan WB, Hires SA, Looger LL. Nat Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Nat Methods. 2009;6:875–U113. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochanczyk T, Jakimowicz P, Krezel A. Chem Commun. 2013;49:1312–1314. doi: 10.1039/c2cc38174e. [DOI] [PubMed] [Google Scholar]
- 39.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 40.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 41.Bird AJ, McCall K, Kramer M, Blankman E, Winge DR, Eide DJ. EMBO J. 2003;22:5137–5146. doi: 10.1093/emboj/cdg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Nat Methods. 2009;6:737–U710. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. J Biol Chem. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiraoka Y, Shimi T, Haraguchi T. Cell Struct Funct. 2002;27:367–374. doi: 10.1247/csf.27.367. [DOI] [PubMed] [Google Scholar]
- 45.Rorsman P, Braun M. Annu Rev Physiol. 2013;75:155–179. doi: 10.1146/annurev-physiol-030212-183754. [DOI] [PubMed] [Google Scholar]
- 46.Zalewski PD, Millard SH, Forbes IJ, Kapaniris O, Slavotinek A, Betts WH, Ward AD, Lincoln SF, Mahadevan I. J Histochem Cytochem. 1994;42:877–884. doi: 10.1177/42.7.8014471. [DOI] [PubMed] [Google Scholar]
- 47.Cole TB, Wenzel HJ, Kafer KE, Schwartzkroin PA, Palmiter RD. Proc Natl Acad Sci U S A. 1999;96:1716–1721. doi: 10.1073/pnas.96.4.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogt K, Mellor J, Tong G, Nicoll R. Neuron. 2000;26:187–196. doi: 10.1016/s0896-6273(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 49.Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, Maechler P. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- 50.Tamaki M, Fujitani Y, Hara A, Uchida T, Tamura Y, Takeno K, Kawaguchi M, Watanabe T, Ogihara T, Fukunaka A, Shimizu T, Mita T, Kanazawa A, Imaizumi MO, Abe T, Kiyonari H, Hojyo S, Fukada T, Kawauchi T, Nagamatsu S, Hirano T, Kawamori R, Watada H. J Clin Invest. 2013;123:4513–4524. doi: 10.1172/JCI68807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li DL, Chen SW, Bellomo EA, Tarasov AI, Kaut C, Rutter GA, Li WH. Proc Natl Acad Sci U S A. 2011;108:21063–21068. doi: 10.1073/pnas.1109773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li DL, Liu L, Li WH. ACS Chem Biol. 2015;10:1054–1063. doi: 10.1021/cb5007536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gee KR, Zhou ZL, Qian WJ, Kennedy R. J Am Chem Soc. 2002;124:776–778. doi: 10.1021/ja011774y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.