Abstract
GINS complex subunit 2 (GINS2), a member of the GINS complex, is involved in DNA replication. GINS2 is upregulated in a variety of aggressive tumors. However, its role in cervical cancer carcinogenesis remains to be elucidated. We investigated the clinical significance of GINS2 in patients with early-stage cervical cancer and its biological functions in cervical cancer progression. GINS2 expression was analyzed in cervical cancer cell lines and in 8 matched cervical cancer samples at the mRNA and protein levels using real-time PCR and western blotting, respectively. GINS2 protein expression in 155 paraffin-embedded cervical cancer specimens was validated using immunohistochemistry. Statistical analysis was used to evaluate its clinicopathological significance. Short hairpin RNA interference, anchorage-independent growth ability, colony formation assay, wound healing ability, Transwell assays and western blotting were used to determine the effects of GINS2 on the aggressive phenotype of cervical cancer cells. There was obvious upregulation of GINS2 in the cervical cancer cell lines and tumor specimens compared to that in the normal cervical tissues. Significant correlations were identified between GINS2 expression and squamous cell carcinoma antigen (SCC-Ag; P<0.001), deep stromal invasion (P=0.021), vital status (P<0.001), recurrence (P<0.001) and pelvic lymph node metastasis (PLNM; P<0.001). Moreover, patients with higher GINS2 expression had shorter overall survival (OS) compared to patients with low GINS2 expression. Multivariate analysis revealed that GINS2 may serve as an independent risk factor of poor prognosis in early-stage cervical cancer. In addition, GINS2 downregulation markedly suppressed cell proliferation and tumorigenic ability, as well as cell migration and invasion. Our findings suggest that GINS2 is a novel indicator of PLNM and a valuable prognostic biomarker in early-stage cervical cancer, and subsequently is a valuable molecular target for cervical cancer diagnosis and treatment.
Keywords: GINS2, cervical cancer, prognosis, proliferation, tumorigenesis, migration, invasion
Introduction
Cervical cancer, a prevalent gynecological malignancy, is the fourth leading cause of cancer-related death in women worldwide, with an estimated 527,600 new cases and 265,700 deaths worldwide in 2012 (1). The majority of cases occur in developing countries and shows a trend for younger patients (2). Despite the decreasing incidence and mortality rate of cervical cancer in recent years owing to improved diagnosis and treatment, the clinical outcome for patients with advanced-stage disease remains bleak (2). Moreover, lymph node metastasis (LNM) can cause higher mortality and recurrence rates, even in patients with early-stage cervical cancer, and definite information on lymph node status is essential for tailoring adjuvant treatment (3,4). However, no sensitive biomarkers specific for indication of LNM, and the early detection and prognosis of cervical cancer are available to date. Therefore, it is urgent to identify novel molecular markers of cervical cancer to facilitate a more accurate prediction of clinical outcome and to prescribe effective treatment.
GINS complex subunit 2 (GINS2), also known as PSF2, encodes a protein with a molecular weight of ~21 kDa. GINS2 belongs to the GINS complex family that also consists of GINS2, GINS3 and GINS4 (5). The GINS complex has been identified as playing a critical role in the initiation of DNA replication and the cell cycle. Stably interacting with minichromosome maintenance (MCM) 2–7 complex and CDC45, the GINS family functions to correctly establish and maintain DNA replication forks (5). Moreover, GINS components may play a role in cell division, and more accurately, in chromosome segregation (6). GINS2 was reported to be involved in tumorigenesis in several types of cancers. For example, genome-wide gene expression profile analysis revealed that GINS2 is highly expressed in lung carcinoma (7). Zheng et al indicated that GINS2 is correlated with aggressive characteristics of breast cancer, and speculated that it is involved in lung metastasis (8). In addition, enhanced expression of GINS2 was found to promote leukemia cell proliferation and desensitize cells to apoptosis (9). These findings all suggest that GINS2 plays an important role in cancer progression. However, the clinical significance of GINS2 in cervical cancer has not been investigated.
In the present study, we explored the GINS2 expression pattern and its clinical implication and prognostic significance in early-stage cervical cancer. Furthermore, we gained insight into the important functions of this protein in cervical cancer development.
Materials and methods
Cell lines
Six cervical cancer cell lines, SiHa, HeLa, C33A, Caski, MS751 and ME180, were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA), and HCC94 and HeLa299, were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cell lines were all cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT, USA) and 1% antibiotics.
Patients and samples
In the present study, we enrolled 155 patients with cervical cancer who underwent radical hysterectomy and lymphadenectomy at The First Affiliated Hospital, Sun Yat-sen University. All patients had stage IA2-IIA disease and received treatment from January 2007 to December 2009. The clinical staging and clinicopathological classifications were determined according to the International Federation of Obstetrics and Gynecology (FIGO), 2009. The clinicopathological characteristics of the enrolled cases are summarized in Table I. The follow-up duration for all patients was >5 years and the last follow-up date was January 2014. Survival was counted from the date of surgery to the date of death or the last follow-up. Eight paired fresh cervical tumor tissues and the adjacent normal tissues were collected for real-time PCR and western blotting. All paraffin-embedded and fresh tissues used in the present study were obtained with the consent of each patient and with institutional research ethics committee approval.
Table I.
Clinicopathological features and GINS2 expression of patients (n=155) with early-stage cervical cancer.
| Characteristics | No. of cases (%) |
|---|---|
| Age (years) | |
| ≤47 | 78 (50.3) |
| >47 | 77 (49.7) |
| FIGO stage | |
| Ia | 18 (11.6) |
| Ib | 83 (53.5) |
| IIa | 54 (34.9) |
| Histological type | |
| Squamous cell carcinoma | 149 (96.1) |
| Adenocarcinoma | 6 (3.9) |
| Pelvic lymph node metastasis (PLNM) | |
| No | 99 (63.9) |
| Yes | 56 (36.1) |
| Tumor size (cm) | |
| ≤4 | 132 (85.2) |
| >4 | 23 (14.8) |
| SCC-Ag (ng/ml) | |
| ≤1.5 | 91 (58.7) |
| >1.5 | 64 (41.3) |
| Deep stromal invasion | |
| No | 49 (31.6) |
| Yes | 106 (68.4) |
| Lymphovascular space involvement | |
| No | 146 (94.2) |
| Yes | 9 (5.8) |
| Positive surgical margin | |
| No | 149 (96.1) |
| Yes | 6 (3.9) |
| Positive parametrium | |
| No | 149 (96.1) |
| Yes | 6 (3.9) |
| Tumor recurrence | |
| No | 137 (88.4) |
| Yes | 18 (11.6) |
| Vital status (at follow-up) | |
| Alive | 126 (81.3) |
| Dead | 29 (18.7) |
| Expression of GINS2 | |
| Low or no expression | 48 (31.0) |
| High expression | 107 (69.0) |
GINS2, complex subunit 2; SCC-Ag, squamous cell carcinoma antigen; FIGO, International Federation of Obstetrics and Gynecology.
Plasmids
To silence endogenous GINS2 expression, the following 2 short hairpin RNAs (shRNAs) were synthesized and purchased: GINS2 shRNA1, CCGGATCCCGAAGGCA GACGAAATCCTCGAGGATTTCGTCTGCCTTCGGGAT TTTTTG; GINS2 shRNA2, CCGGGAATGGATTCAGGAT GTTGTTCTCGAGAACAACATCCTGAATCCATTCTTT TTG. The shRNA sequences were cloned into pSuper-retro-neo plasmids to generate the respective pSuper-retro-GINS2-RNAi(s). After 48 h infection, the SiHa and HeLa cell lines stably expressing the GINS2 shRNAs were selected with puromycin (0.5 µg/ml).
Real-time PCR
Total RNA from cervical cancer cells and fresh tumor tissues was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The isolated RNA was pretreated with RNase-free DNase, and 2 µg RNA/sample was used for complementary DNA (cDNA) synthesis. For the PCR amplication of GINS2 cDNA, an initial amplification step using CINS2-specific primers was performed with denaturation at 95̊C for 10 min, followed by 28 denaturation cycles at 95̊C for 60 sec, primer annealing at 58̊C for 30 sec, and a primer extension phase at 72̊C for 30 sec. Upon completion of the cycling steps, a final extension step at 72̊C for 5 min was performed before the reaction mixture was stored at 4̊C. Quantitative PCR (qPCR) was then conducted to determine the increase in GINS2 mRNA in each of the primary cervical tumors relative to the paired normal cervical tissue from the same patient and in the 8 cervical cancer cell lines relative to that in normal cervical tissue. Expression data were normalized to the geometric mean of the expression level of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers were designed using Primer Express v 2.0 software (Applied Biosystems, Foster City, CA, USA). The GINS2 forward and reverse primer sequences were: 5′-GCTGGCGATTAACCTGAAAC-3′ and 5′-TTCCTTTCGTTCATGATCCC-3′, respectively. The GAPDH forward and reverse primers were: 5′-TTGAGGTCAATGAAGGGGTC-3′ and 5′-GAAGGTGAAGGTCGGAGTCA-3′, respectively.
Western blotting
Cells at 80–90% confluence were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed on ice in radioimmunoprecipitation assay buffer (RIPA; Cell Signaling Technology, Danvers, MA, USA) containing complete protease inhibitor cocktail (Roche Applied Sciences, Mannheim, Germany). Fresh tissue samples were ground to powder in liquid nitrogen and lysed with SDS-PAGE sample buffer. Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal protein samples (30 µg) extracted from the cervical cancer cell lines and tissues were electrophoretically separated on 10.5% sodium dodecyl sulfate (SDS)/polyacrylamide gels, and transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, MA, USA). The membranes were blocked with 5% fat-free milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature. The membranes were then incubated with anti-GINS2 rabbit monoclonal antibody (1:2,000; HPA057285; Sigma, St. Louis, MO, USA) overnight at 4̊C. After washing 3 times with TBST, the membranes were probed with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:2,000; SC-2004; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and protein expression was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer's suggested protocols. An anti-α-tubulin mouse monoclonal or an anti-GAPDH mouse monoclonal antibody (1:2,000; Sigma) was used as a loading control.
Immunohistochemistry (IHC)
IHC was used to examine GINS2 expression in 155 human cervical cancer specimens. Briefly, the paraffin-embedded specimens were cut into 4-µm sections and baked at 60̊C for 1 h, deparaffinized with xylenes and rehydrated, submerged in EDTA antigen retrieval buffer, and microwaved for antigen retrieval. The samples were treated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity, followed by incubation with 1% bovine serum albumin to block non-specific binding. The sections were then incubated with anti-GINS2 rabbit monoclonal antibody (1:600; HPA057285; Sigma) overnight at 4̊C. Normal goat serum was used as the negative control. After washing, the sections were incubated with a biotinylated anti-rabbit secondary antibody, followed by incubation with streptavidin-horseradish peroxidase complex (both from Abcam, Cambridge, MA, USA). The sections were immersed in 3-amino-9-ethylcarbazole and counterstained with 10% Mayer's hematoxylin, dehydrated and mounted in crystal mount.
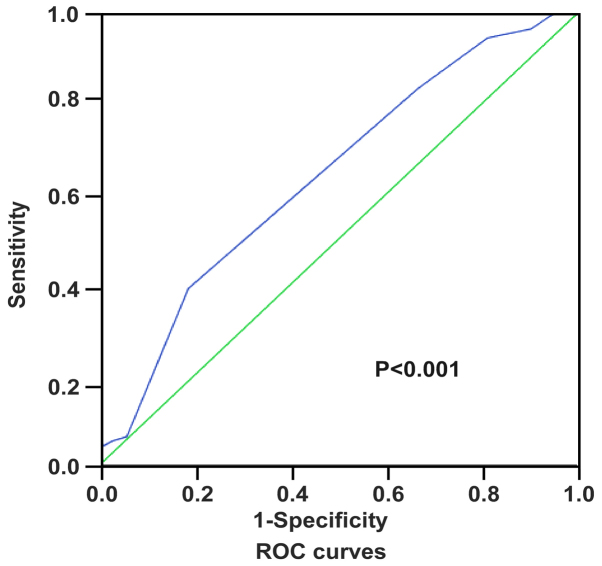
Two independent observers blinded to the histopathological features and patient data of the samples evaluated and scored the degree of immunostaining. The scores were based on the proportion of positively stained tumor cells [graded as: 1 (<10% positive), 2 (10–50% positive), 3 (51–75% positive) or 4 (>75% positive)] and staining intensity [categorized as 1 (no staining), 2 (weak staining, light yellow), 3 (moderate staining, yellow brown) or 4 (strong staining, brown)]. The staining index was generated by multiplying the scores for staining intensity and for the proportion of positive cells (scored as 1, 2, 3, 4, 6, 8, 9, 12 and 16). Staining index score ≥8 indicated tumors with high GINS2 expression; a score of <8 defined low GINS2 expression. Receiver operating characteristics (ROC) curve analysis was conducted using variables including GINS2 expression and patient outcomes to determine the optimum cut-off values of the scores (Fig. 1).
Figure 1.
Receiver operating characteristics (ROC) curve analysis to determine the optimal cut-off values for IHC staining index scores and 7 was defined as the cut-off point. Accordingly, scores ≥8 were judged as high GINS2 expression and scores <8 were categorized as low GINS2 expression.
Colony formation and anchorage-independent growth ability assays
Cells (5×102/well) were plated in 6-well plates and cultured for 2 weeks. The colonies were washed with PBS 3 times and fixed with 4% formaldehyde for 10 min. Then, the colonies were stained with 1% crystal violet for 10 min. After washing, the colonies were counted. For the anchorage-independent growth ability assay, 500 cells were trypsinized and suspended in 2 ml complete medium plus 0.3% agar (Sigma). The agar-cell mixture was plated on 1% agar complete medium mixture. For ~10 days, viable colonies that were >0.1 mm in diameter were counted. The experiment was carried out in triplicate for each cell line.
Wound healing and Transwell migration and invasion assays
In the wound healing assay, cells (2×106/well) were seeded in 6-well plates. When the cells were 90% confluent, they were serum-starved for 24 h. A linear wound was created in the confluent monolayer using a 10-µl pipette tip. The wounds were observed and photographed immediately (time 0) and thereafter at 24 and 48 h (magnification, ×200). Each experiment was repeated at least 3 times. For the Transwell migration and invasion assays, 2×104 cells were seeded in 8-µm pore inserts coated with (for invasion) or without (for migration) 50 µl Matrigel in triplicate wells. After 24 h incubation at 37̊C, cells that had passed through the filter into the bottom chamber were fixed with 1% paraformaldehyde, stained with hematoxylin, and counted under a magnification of ×200 (10 random fields/well).
Statistical analysis
All statistical analyses were conducted using the SPSS software package (standard version 16.0; IBM). The Chi-square and Fisher's exact tests were used to evaluate the relationship between GINS2 expression and the clinicopathological features. Spearman's rank correlation coefficients were performed to calculate bivariate correlations between the study variables. Survival curves were estimated using the Kaplan-Meier method and the log-rank test. The independent prognostic indicator in all of the clinical parameters was determined using univariate and multivariate analyses. P<0.05 was considered statistically significant in all cases.
Results
GINS2 is overexpressed in cervical cancer
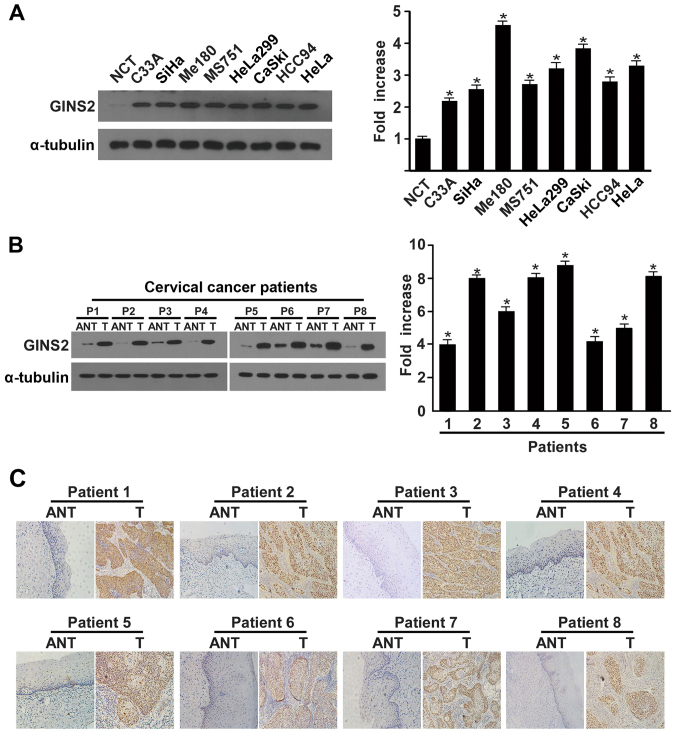
We used real-time PCR and western blotting to determine the GINS2 expression pattern in cervical carcinoma cell lines and samples. There were higher levels of both GINS2 transcription and translation in the cervical carcinoma cell lines in comparison to normal cervical tissue (Fig. 2A). Consistent with the findings in the cervical cancer cell lines, GINS2 mRNA and protein levels were clearly differentially increased in the 8 cervical cancer tissue samples than these levels in the matched adjacent non-cancerous tissues (Fig. 2B). Furthermore, IHC confirmed GINS2 overexpression in the cervical cancer clinical samples (Fig. 2C).
Figure 2.
Western blotting, qPCR, and IHC determination of GINS2 mRNA and protein expression. (A) Western blotting and qPCR of GINS2 expression in normal cervical tissues and cervical cancer cell lines. (B) Western blotting of GINS2 expression in 8 pairs of matched cervical cancer (T) and adjacent non-cancerous cervical tissues (ANT) and average T/ANT ratios of GINS2 mRNA expression quantified by qPCR in 8 pairs of matched cervical cancer tissues. Expression levels were normalized to α-tubulin expression. Error bars represent standard deviation (SD) calculated from 3 parallel experiments. *P<0.05. (C) IHC assay of GINS2 protein expression in 8 pairs of matched cervical cancer tissues.
GINS2 expression correlates with clinical characteristics in early-stage cervical cancer
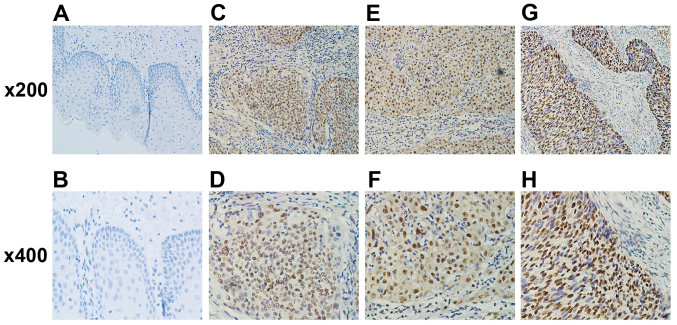
To investigate the clinical relevance of GINS2 expression and cervical cancer progression, IHC was performed on 155 paraffin-embedded, archived clinical cervical cancer samples, which included 18, 83 and 54 cases of stage Ia2, Ib and IIa disease, respectively. We detected strong positive expression of GINS2 in 48 (31%) cervical cancer specimens, whereas there was no or marginally detectable staining in the remaining 107 (69%; Table I) clinical tumor samples. GINS2 primarily localized in the tumor cell nuclei and was absent from the adjacent normal cervical tissues (Fig. 3).
Figure 3.
IHC assay of GINS2 expression in cervical cancer tissues. Original magnification ×200 or ×400. Positive GINS2 staining was observed mainly in cervical cancer cell nuclei. (A and B) GINS2 was not detected in normal cervical cancer tissues. (C and D) Representative images of weak GINS2 staining in cervical cancer tissues. (E and F) Representative images of moderate GINS2 staining in cervical cancer tissues. (G and H) Representative images of strong GINS2 staining in cervical cancer tissues.
Notably, GINS2 expression correlated with several clinical features of cervical cancer, including SCC-Ag (P<0.001), pelvic LNM (PLNM; P<0.001), deep stromal invasion (P=0.021), vital status (P<0.001) and recurrence (P<0.001). In contrast, GINS2 and other clinical characteristics, including FIGO stage, surgical margin, lymphovascular invasion, parauterine organ infiltration and age, were not obviously related (Table II). Spearman correlation analysis (Table III) further confirmed the strong association of GINS2 expression and significant prognostic risk factors, including SCC-Ag (R=0.430; P<0.001), deep stromal invasion (R=0.185; P=0.021), vital status (R=0.287; P<0.001), recurrence (R=0.323; P<0.001), and in particular, PLNM (R=0.455; P<0.001).
Table II.
Correlation of clinicopathological characteristics and GINS2 expression in the early-stage cervical cancer patients.
| GINS2 | |||||
|---|---|---|---|---|---|
| Characteristics | Total (n=155) | Low expression | High expression | Chi-square test P-value | Fishers exact test P-value |
| Age (years) | 0.688 | 0.730 | |||
| ≤47 | 78 | 55 (35.5) | 23 (14.8) | ||
| >47 | 77 | 52 (33.5) | 25 (16.2) | ||
| FIGO stage | 0.571 | ||||
| Ia | 18 | 14 (9.0) | 4 (2.6) | ||
| Ib | 83 | 58 (37.4) | 25 (16.1) | ||
| IIa | 54 | 35 (22.6) | 19 (12.3) | ||
| Pelvic lymph node metastasis (PLNM) | <0.001 | <0.001 | |||
| No | 99 | 84 (54.2) | 15 (9.7) | ||
| Yes | 56 | 23 (14.8) | 33 (21.3) | ||
| Histological types | 0.440 | 0.667 | |||
| Squamous cell carcinoma | 149 | 102 (65.8) | 47 (30.3) | ||
| Adenocarcinoma | 6 | 5 (3.2) | 1 (0.7) | ||
| SCC-Ag (ng/ml) | <0.001 | <0.001 | |||
| ≤1.5 | 91 | 78 (50.3) | 13 (8.4) | ||
| >1.5 | 64 | 29 (18.7) | 35 (22.6) | ||
| Tumor size (cm) | 0.058 | 0.085 | |||
| ≤4 | 132 | 95 (61.3) | 37 (23.9) | ||
| >4 | 23 | 12 (7.7) | 11 (7.1) | ||
| Positive surgical margin | 0.304 | 0.374 | |||
| No | 149 | 104 (67.1) | 45 (29.0) | ||
| Yes | 6 | 3 (1.9) | 3 (1.9) | ||
| Deep stromal invasion | 0.021 | 0.025 | |||
| No | 49 | 40 (25.8) | 9 (5.8) | ||
| Yes | 106 | 67 (43.2) | 39 (25.2) | ||
| Positive parametrium | 0.898 | 1.000 | |||
| No | 149 | 103 (66.5) | 46 (29.7) | ||
| Yes | 6 | 4 (2.6) | 2 (1.3) | ||
| Lymphovascular space involvement | 0.874 | 1.000 | |||
| No | 146 | 101 (65.2) | 45 (29.0) | ||
| Yes | 9 | 6 (3.9) | 3 (1.9) | ||
| Vital status (at follow-up) | <0.001 | 0.001 | |||
| Alive | 126 | 95 (61.3) | 31 (20.0) | ||
| Dead | 29 | 12 (7.7) | 17 (11.0) | ||
| Recurrence | <0.001 | <0.001 | |||
| No | 137 | 102 (65.8) | 35 (22.6) | ||
| Yes | 18 | 5 (3.2) | 13 (8.4) | ||
GINS2, complex subunit 2; SCC-Ag, squamous cell carcinoma antigen; FIGO, International Federation of Obstetrics and Gynecology.
Table III.
Spearman analysis of the correlation of GINS2 expression and clinicopathological features.
| GINS2 expression level | ||
|---|---|---|
| Variables | Spearman correlation | P-value |
| PLNM | 0.455 | <0.001 |
| SCC-Ag (ng/ml) | 0.430 | <0.001 |
| Vital status (at follow-up) | 0.287 | <0.001 |
| Deep stromal invasion | 0.185 | 0.021 |
| Recurrence | 0.323 | <0.001 |
GINS complex subunit 2; PLNM, pelvic lymph node metastasis; SCC-Ag, squamous cell carcinoma antigen.
GINS2 expression is associated with poor survival outcomes in early-stage cervical cancer
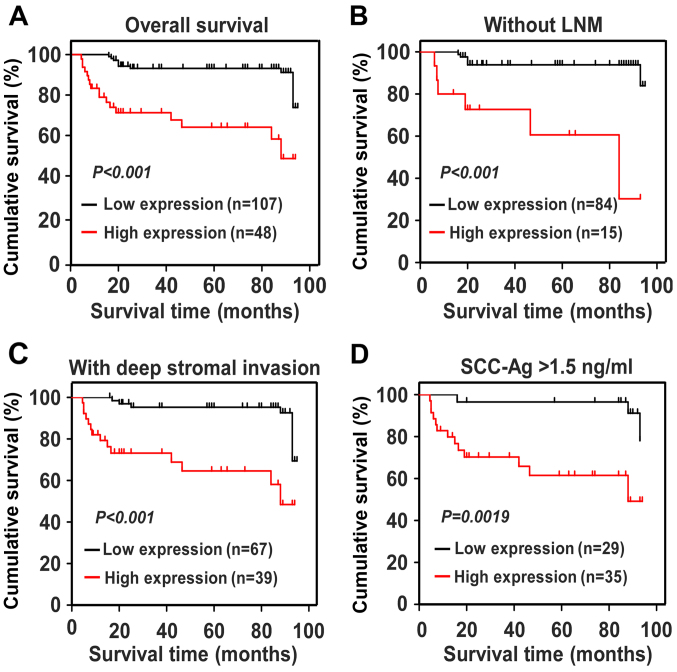
We used Kaplan-Meier survival curves and the log-rank test to evaluate the impact of GINS2 on predicting the prognosis of patients with early-stage cervical cancer. Fig. 4A shows that patients with higher levels of GINS2 had shorter OS (P<0.001), whereas those with lower GINS2 expression survived longer. For the patients with low GINS2 expression, the cumulative OS rates were 89.36% [95% confidence interval (CI), 85.68–93.05%], but were drastically decreased to 65.19% (95% CI, 53.89–76.49%) in the high GINS2 group. In addition, we calculated the prognostic value of GINS2 expression in specific patient subgroups, which were stratified based on age, FIGO stage, PLNM, SCC-Ag, tumor size, differentiation grade, deep stromal invasion, properties of the surgical margin, parauterine organ infiltration and lymphovascular space involvement. Fig. 4B-D shows that there was a markedly negative association between GINS2 expression and OS in the patients without LNM (log-rank test; P<0.001), with SCC-Ag >1.5 ng/ml (log-rank test; P=0.0019), and with deep stromal invasion (log-rank test; P<0.001). The Cox regression model indicated that GINS2 expression and recurrence were independent prognostic risk factors of cervical cancer (Table IV). Taken together, our results indicate that GINS2 plays an important role in cervical cancer progression and may serve as a valuable prognostic predictor for patients with early-stage cervical cancer.
Figure 4.
Kaplan-Meier curves of univariate analysis data (log-rank test). (A) OS curves of patients with high vs. low GINS2 expression. (B) OS curves of patients without LNM with high vs. low GINS2 expression. (C) OS curves of patients with deep stromal invasion with high vs. low GINS2 expression. (D) OS curves of patients with SCC-Ag >1.5 ng/ml with high vs. low GINS2 expression.
Table IV.
Univariate and multivariate analysis of the prognostic parameters in early-stage cervical cancer using Cox-regression model.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| P-value | Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | |
| GINS2 expression | <0.001 | 5.160 | 0.011 | 3.643 |
| Low | (2.438–10.925) | (1.347–9.850) | ||
| High | ||||
| Pelvic lymph node metastasis (PLNM) | 0.008 | 2.716 | 0.127 | 1.827 |
| No | (1.299–5.680) | (0.843–3.962) | ||
| Yes | ||||
| SCC-Ag (ng/ml | 0.099 | 1.854 | 0.862 | 0.922 |
| ≤1.5 | (0.891–3.855) | (0.369–2.303) | ||
| >1.5 | ||||
| Recurrence | <0.001 | 10.201 | <0.001 | 8.523 |
| No | (4.883–21.352) | (3.944–18.416) | ||
| Yes | ||||
| Deep stromal invasion | 0.543 | 1.304 | 0.669 | 0.823 |
| No | (0.555–3.063) | (0.337–2.008) | ||
| Yes | ||||
| Positive parametrium | 0.840 | 1.228 | 0.510 | 2.076 |
| No | (0.166–9.092) | (0.236–18.222) | ||
| Yes | ||||
| Lymphovascular space involvement | 0.735 | 1.282 | 0.971 | 1.032 |
| No | (0.304–5.400) | (0.190–5.623) | ||
| Yes | ||||
| Tumor size (cm) | 0.523 | 1.371 | 0.481 | 0.675 |
| ≤4 | (0.521–3.609) | (0.227–2.013) | ||
| >4 | ||||
CI, confidential interval; GINS2, complex subunit 2; SCC-Ag, squamous cell carcinoma antigen.
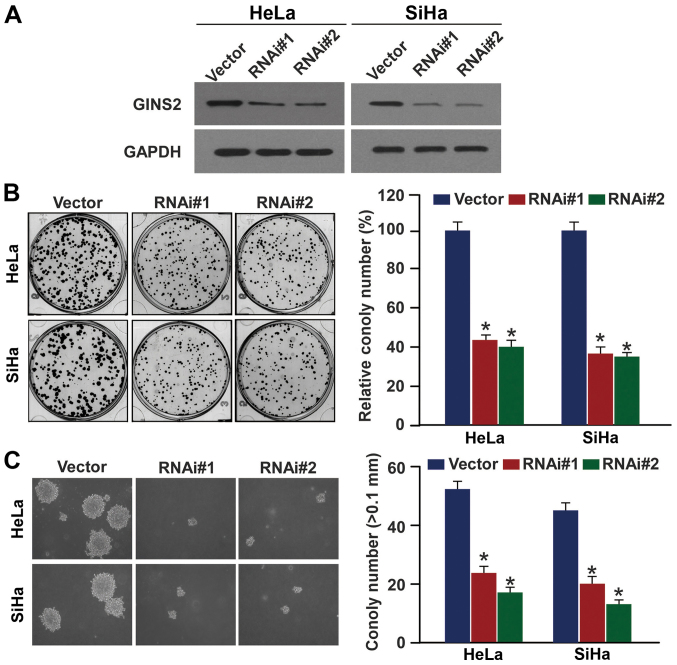
GINS2 downregulation inhibits cervical cancer cell proliferation and tumorigenic ability
Given the involvement of GINS2 in cervical cancer progression and its potential as a biomarker for identifying patients with a more aggressive cervical cancer phenotype, we explored the effects of GINS2 on the proliferation and tumorigenicity of cervical cancer cell lines. Two shRNAs against GINS2 were transduced into the SiHa and HeLa cervical cancer cell lines to stably suppress endogenous GINS2 expression. The downregulation efficiency was verified using western blotting (Fig. 5A). As expected, the colony formation assay revealed that GINS2 depletion induced the formation of much smaller and fewer colonies as compared to the vector control cells (Fig. 5B). Furthermore, silencing of GINS2 markedly decreased the anchorage-independent growth ability of the two cell lines in soft agar (Fig. 5C). These results suggest that GINS2 promotes proliferation and tumorigenesis of cervical cancer cells.
Figure 5.
GINS2 promotes cervical cancer cell proliferation and tumorigenicity. (A) Western blot analysis of GINS2 expression in GINS2-silenced SiHa and HeLa cells. GAPDH was used as the loading control. (B) Colony formation assay showing that silencing of endogenous GINS2 inhibited proliferation. (C) Anchorage-independent growth ability assay showing that GINS2 knockdown suppressed cervical cancer cell tumorigenicity. The number of colonies >0.1 mm in diameter was quantified after 10 days of culture. Original magnification, ×200. Bars represent the mean ± SD of 3 independent experiments; *P<0.05.
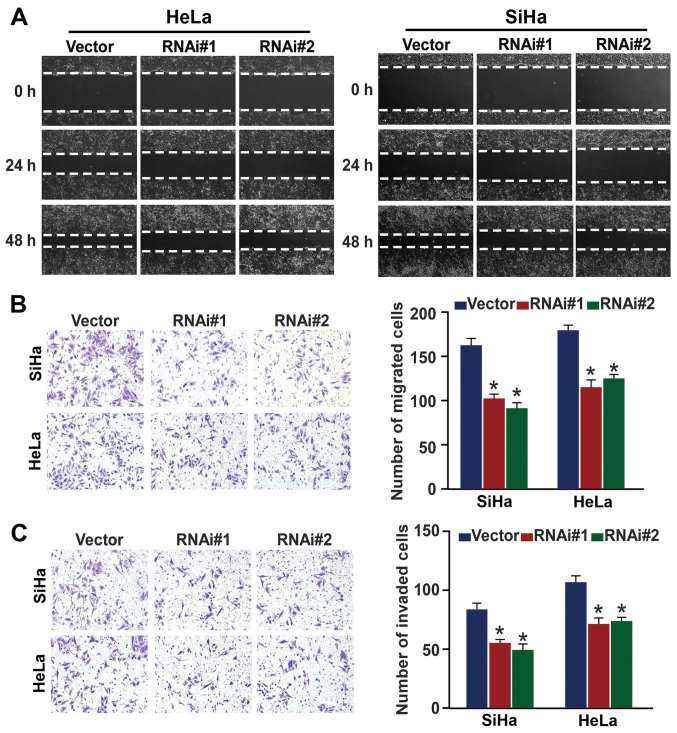
GINS2 inhibition reduces cervical cancer cell migration and invasion
After demonstrating that GINS2 is closely connected with PLNM, we then investigated whether GINS2 regulates cervical cancer cell motility and invasiveness. Fig. 6A shows that in the wound healing assay, GINS2 ablation slowed the speed with which the SiHa and HeLa cells filled the gap in comparison to the control in an obvious manner. The Transwell migration and matrix invasion assays yielded similar results (Fig. 6B and C), as indicated by the smaller number of migrated and invaded cells that passed through the filter in the GINS2 downregulation group as compared to the negative control group. Collectively, our findings confirmed that GINS2 enhances the migratory and invasive properties of cervical cancer cells.
Figure 6.
GINS2 is essential for cervical cancer cell migration and invasion. (A) Cell mobility was measured by examining the rate of wound closure at 0, 24 and 48 h. Original magnification, ×200. (B) Transwell migration assay investigation of the mobility properties induced by fetal bovine serum. Original magnification, ×200; *P<0.05. (C) Transwell invasion assay of the invasive properties induced by fetal bovine serum. Original magnification, ×200; *P<0.05. Bars represent the mean ± SD of 3 independent experiments
Discussion
In the present study, we demonstrated for the first time that GINS2 upregulation correlated with poor prognosis and reduced survival in early-stage cervical cancer. In addition, GINS2 downregulation hampered the cellular capabilities of proliferation, tumorigenesis, migration and invasion in an obvious manner. These findings are convincing evidence that GINS2 plays a significant role in the progression of cervical cancer and has the potential to be a neoteric prognostic biomarker of early-stage cervical cancer.
Clinical evidence has confirmed the role of GINS2 as an oncogene in various malignancies. GINS2 is elevated in intrahepatic cholangiocarcinoma, lung adenocarcinoma and breast cancer, and induces intrahepatic cholangiocarcinoma cell proliferation (7,8,10). Moreover, GINS2 overexpression contributes to maintenance of the cancer stem cell population and has been correlated with progression and poor prognosis in breast cancer (8). The above mentioned studies indicated that GINS2 plays a critical role in carcinogenesis and cancer progression. Herein, we investigated its role in cervical cancer. Consistent with the above studies, GINS2 mRNA and protein expression was higher in the 8 cervical cancer cell lines and clinical samples. IHC analysis indicated that GINS2 expression was associated with well-known prognostic parameters: SCC-Ag (P<0.001), deep stromal invasion (P=0.021), vital status (P<0.001) and recurrence (P<0.001), particularly PLNM (P<0.001), providing strong evidence that GINS2 plays an oncogenic role in cervical cancer development and that it may act as a biomarker for assessing patients with a more aggressive form of the disease. More importantly, the present study demonstrated that GINS2 overexpression correlates with poor clinical outcome and is potentially an independent prognostic factor of poor OS in cervical cancer. Taken together, our results implicate GINS2 as a crucial contributing factor in tumor progression.
Clearly, the fundamental characteristic of tumor cells is their malignant uncontrolled growth, the key process of which is DNA replication (11,12). Correct DNA replication requires the proper combination of replication-associated proteins and prereplication complexes. Defects in the proper function of these proteins are responsible for chromosome instability and the subsequent generation of neoplasms (13–16). Indeed, aberrant expression of several DNA replication-related proteins has been demonstrated in certain cancers. Gan et al proposed MCM genes, which represent primary helicase for unwinding DNA during replication, as diagnostic or prognostic markers in cervical carcinoma (17). Tane et al showed that elevated levels of PSF3, a GINS family member, predicted poor prognosis of non-small cell lung cancer (18). In addition, silencing of PSF3 inhibited cell proliferation by inducing G1-S arrest, suggesting that PSF3 may be required for the development of lung cancer. In accordance with these previous studies, we established the clinical significance of GINS2 in cervical cancer and demonstrated that GINS2 knockdown markedly impaired the proliferative and tumorigenic properties of cervical cancer cells. GINS2 was previouly found to alter the percentages of the apoptosis-relevant genes BAX and BCL2, and the levels of the cell cycle regulators ataxia-telangiectasia mutated (ATM), checkpoint kinase 2 (CHK2), and P53 to stimulate G2/M transition (9). Accordingly, we similarly hypothesized that GINS2 accelerated G2/M transitional entry by circumventing cell cycle checkpoints, consequently exerting its oncogenic potential in cervical cancer. However, further investigations are needed to verify the hypothesis and to clarify the carcinogenesis mechanisms in greater detail.
Currently, radical hysterectomy plus pelvic lymph node dissection or chemoradiation is the criterion standard of treatment for patients with early-stage cervical cancer (19,20). Lymph node status is critical for determining the appropriate treatment strategies. Patients with LNM require chemoradiation, which may render the initial surgical intervention unnecessary in retrospect (19,21). Meanwhile, unnecessary lymphadenectomy in patients with negative LNM can cause unfavorable complications such as infection, lymphocysts, and lymphedema (22). However, imaging methods and sentinel lymph node biopsy both fail to accurately detect LNM (21,23,24). Worse, there is no existing preoperative marker for sensitive and efficient LNM diagnosis. Surprisingly, our data showed that GINS2 was closely associated with LNM; moreover, inhibition of GINS2 significantly suppressed the aggressive cervical cancer cell phenotype. Therefore, we speculated that GINS2 is a valuable marker for predicting PLNM. However, a larger cohort of patients with LNM should be enrolled in future studies, and the definite regulatory mechanisms involved warrant further investigation.
SCC-Ag is currently most widely applied in clinical diagnosis, evaluation of therapeutic effects, and for predicting the clinical outcomes of patients with cervical cancer (25,26). SCC-Ag levels are associated with the extent of cervical carcinoma (27–33). Sustained high or continuous elevation of SCC-Ag represents persistent disease or recurrence (34,35). However, the clinical practical value of serum SCC-Ag is still controversial. The numerical changes of SCC-Ag do not specifically reflect cervical cancer; increased SCC-Ag levels have also been observed in patients with lung cancer, esophageal SCC and other benign diseases (36–38). More unfortunately, the autofit cut-off level of SCC-Ag is still a matter of debate partially due to the different disease stages of patient groups (39). Therefore, it appears that SCC-Ag is not an ideal indicator for predicting outcomes in cervical cancer.
Notably, we reported that GINS2 plays an important role in the development of cervical cancer and may serve as a valuable prognostic marker for assessing survival in cervical cancer. Therefore, determining the expression of GINS2 in cervical tissues via biopsy can provide significant guidance for deciding preferred treatment modality for patients with early-stage cervical cancer.
To date, persistent human papillomavirus (HPV) infection is recognized as the main cause of cervical cancer, and HPV 16 and HPV 18 are considered the most carcinogenic HPVs. The HPV oncoproteins E6 and E7 target the tumor-suppressor gene P53 and retinoblastoma protein (Rb), respectively, simultaneously with the activation of certain oncogenes and pathways, thereby coordinately contributing to the initiation and progression of cervical cancer (40–42). Clinical evidence has demonstrated that GINS2 targets P53, and silencing of GINS2 upregulated P53 expression (9). We hypothesized that GINS2 cooperates with the HPV oncoprotein E6 jointly to promote tumor transformation and development. However, we did not investigate their interaction in the present study, and it warrants further investigation.
While the prognostic value of GINS2 in early-stage cervical cancer was validated in our research. However, there still existed limitations, one of which was the small sample size. Furthermore, as all the patients involved were all in the early-stage of cervical cancer, their prognosis was relatively favorable and few developed pathological risk features including LNM, positive parametrium, positive surgical margin, large tumor size and recurrence. Thus, to validate our results, it is urgent to incorporate a larger cohort of patients with aggressive clinical factors in our subsequent study.
In summary, the present study, confirms the aberrant expression and clinical significance of GINS2 in early-stage cervical cancer. Additionally, we found that GINS2 knockdown suppressed cervical cancer cell proliferation, tumorigenicity, migration and invasion. GINS2 may represent a novel indicator for identifying patients at high-risk, and potentially serve as a clinically relevant biomarker for predicting the patient outcome of early-stage cervical cancer, which could aid gynecologists in determining more appropriate therapeutic strategies.
Acknowledgements
The present study was funded by the Natural Science Foundation of Guangdong Province (S2013010015552), the National Natural Science Foundation of China (nos. 81602723, 81302550 and 81671805), the Postdoctoral Science Foundation of China (2016A020215214), and the Guangdong Medical Research Foundation (A2015130).
Glossary
Abbreviations
- GINS2
GINS complex subunit 2
- OS
overall survival
- PLNM
pelvic lymph node metastasis
- LNM
lymph node metastasis
- MCM
minichromosome maintenance
- shRNAs
short hairpin RNAs
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PBS
phosphate-buffered saline
- PVDF
polyvinylidene difluoride
- IHC
immunohistochemistry
- SCC-Ag
squamous cell carcinoma antigen
- HPV
human papilloma virus
- ATM
ataxia-telangiectasia mutated
- CHK2
checkpoint kinase 2
- Rb
retinoblastoma protein
- FIGO
International Federation of Obstetrics and Gynecology
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Noordhuis MG, Fehrmann RS, Wisman GB, Nijhuis ER, van Zanden JJ, Moerland PD, Ver Loren van Themaat E, Volders HH, Kok M, ten Hoor KA, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res. 2011;17:1317–1330. doi: 10.1158/1078-0432.CCR-10-2320. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Jia X, Li N, Chen C, Liu Y, Wang H. Comprehensive clinic-pathological characteristics of cervical cancer in southwestern China and the clinical significance of histological type and lymph node metastases in young patients. PLoS One. 2013;8:e75849. doi: 10.1371/journal.pone.0075849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 6.Rantala JK, Edgren H, Lehtinen L, Wolf M, Kleivi K, Vollan HK, Aaltola AR, Laasola P, Kilpinen S, Saviranta P, et al. Integrative functional genomics analysis of sustained polyploidy phenotypes in breast cancer cells identifies an oncogenic profile for GINS2. Neoplasia. 2010;12:877–888. doi: 10.1593/neo.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M, Pan H, Zhang F, Zhang Y, Zhang Y, Xia H, Zhu J, Fu W, Zhang X. Identification of TNM stage-specific genes in lung adenocarcinoma by genome-wide expression profiling. Oncol Lett. 2013;6:763–768. doi: 10.3892/ol.2013.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Zhou Y, Yang X, Tang J, Wei D, Zhang Y, Jiang JL, Chen ZN, Zhu P. High GINS2 transcript level predicts poor prognosis and correlates with high histological grade and endocrine therapy resistance through mammary cancer stem cells in breast cancer patients. Breast Cancer Res Treat. 2014;148:423–436. doi: 10.1007/s10549-014-3172-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Zhong L, Liu BZ, Gao YJ, Gao YM, Hu XX. Effect of GINS2 on proliferation and apoptosis in leukemic cell line. Int J Med Sci. 2013;10:1795–1804. doi: 10.7150/ijms.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obama K, Ura K, Satoh S, Nakamura Y, Furukawa Y. Up-regulation of PSF2, a member of the GINS multiprotein complex, in intrahepatic cholangiocarcinoma. Oncol Rep. 2005;14:701–706. [PubMed] [Google Scholar]
- 11.Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825. doi: 10.1158/2159-8290.CD-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Dutta A. Chaotic license for genetic instability and cancer. Nat Genet. 2007;39:10–11. doi: 10.1038/ng0107-10. [DOI] [PubMed] [Google Scholar]
- 16.Hermand D, Nurse P. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol Cell. 2007;26:553–563. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Gan N, Du Y, Zhang W, Zhou J. Increase of Mcm3 and Mcm4 expression in cervical squamous cell carcinomas. Eur J Gynaecol Oncol. 2010;31:291–294. [PubMed] [Google Scholar]
- 18.Tane S, Sakai Y, Hokka D, Okuma H, Ogawa H, Tanaka Y, Uchino K, Nishio W, Yoshimura M, Maniwa Y. Significant role of Psf3 expression in non-small-cell lung cancer. Cancer Sci. 2015;106:1625–1634. doi: 10.1111/cas.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benedet JL, Bender H, Jones H, III, Ngan HY, Pecorelli S. FIGO Committee on Gynecologic Oncology: FIGO staging classifications and clinical practice guidelines in the management of gynecologic cancers. Int J Gynaecol Obstet. 2000;70:209–262. doi: 10.1016/S0020-7292(00)90001-8. [DOI] [PubMed] [Google Scholar]
- 20.Koh WJ, Greer BE, Abu-Rustum NR, Apte SM, Campos SM, Chan J, Cho KR, Cohn D, Crispens MA, DuPont N, et al. National Comprehensive Cancer Network: Cervical cancer. J Natl Compr Canc Netw. 2013;11:320–343. doi: 10.6004/jnccn.2013.0043. [DOI] [PubMed] [Google Scholar]
- 21.Selman TJ, Mann C, Zamora J, Appleyard TL, Khan K. Diagnostic accuracy of tests for lymph node status in primary cervical cancer: A systematic review and meta-analysis. CMAJ. 2008;178:855–862. doi: 10.1503/cmaj.071124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura Y, Kawagoe T, Toki N, Tanaka M, Kashimura M. Long-standing complications after treatment for cancer of the uterine cervix - clinical significance of medical examination at 5 years after treatment. Int J Gynecol Cancer. 2006;6:294–297. doi: 10.1111/j.1525-1438.2006.00354.x. [DOI] [PubMed] [Google Scholar]
- 23.Altgassen C, Hertel H, Brandstädt A, Köhler C, Dürst M, Schneider A. AGO Study Group: Multicenter validation study of the sentinel lymph node concept in cervical cancer: AGO Study Group. J Clin Oncol. 2008;26:2943–2951. doi: 10.1200/JCO.2007.13.8933. [DOI] [PubMed] [Google Scholar]
- 24.Slama J, Dundr P, Dusek L, Cibula D. High false negative rate of frozen section examination of sentinel lymph nodes in patients with cervical cancer. Gynecol Oncol. 2013;129:384–388. doi: 10.1016/j.ygyno.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Bonfrer JM, Gaarenstroom KN, Korse CM, Van Bunningen BN, Kenemans P. Cyfra 21–1 in monitoring cervical cancer: A comparison with tissue polypeptide antigen and squamous cell carcinoma antigen. Anticancer Res. 1997;17:2329–2334. [PubMed] [Google Scholar]
- 26.Esajas MD, Duk JM, de Bruijn HW, Aalders JG, Willemse PH, Sluiter W, Pras B, ten Hoor K, Hollema H, van der Zee AG. Clinical value of routine serum squamous cell carcinoma antigen in follow-up of patients with early-stage cervical cancer. J Clin Oncol. 2001;19:3960–3966. doi: 10.1200/JCO.2001.19.19.3960. [DOI] [PubMed] [Google Scholar]
- 27.Avall-Lundqvist EH, Sjovall K, Nilsson BR, Eneroth PH. Prognostic significance of pretreatment serum levels of squamous cell carcinoma antigen and CA 125 in cervical carcinoma. Eur J Cancer. 1992;28A:1695–1702. doi: 10.1016/0959-8049(92)90071-9. [DOI] [PubMed] [Google Scholar]
- 28.Duk JM, Groenier KH, de Bruijn HW, Hollema H, ten Hoor KA, van der Zee AG, Aalders JG. Pretreatment serum squamous cell carcinoma antigen: A newly identified prognostic factor in early-stage cervical carcinoma. J Clin Oncol. 1996;14:111–118. doi: 10.1200/JCO.1996.14.1.111. [DOI] [PubMed] [Google Scholar]
- 29.Yuan CC, Wang PH, Ng HT, Tsai LC, Juang CM, Chiu LM. Both TPA and SCC-Ag levels are prognostic even in high-risk stage Ib-IIa cervical carcinoma as determined by a stratification analysis. Eur J Gynaecol Oncol. 2002;23:17–20. [PubMed] [Google Scholar]
- 30.Strauss HG, Laban C, Lautenschläger C, Buchmann J, Schneider I, Koelbl H. SCC antigen in the serum as an independent prognostic factor in operable squamous cell carcinoma of the cervix. Eur J Cancer. 2002;38:1987–1991. doi: 10.1016/S0959-8049(02)00159-4. [DOI] [PubMed] [Google Scholar]
- 31.Molina R, Filella X, Lejarcegui JA, Pahisa J, Torné A, Rovirosa A, Mellado B, Ordi J, Puig-Tintore LM, Alicarte J, et al. Prospective evaluation of squamous cell carcinoma and carcinoembryonic antigen as prognostic factors in patients with cervical cancer. Tumour Biol. 2003;24:156–164. doi: 10.1159/000073846. [DOI] [PubMed] [Google Scholar]
- 32.Reesink-Peters N, van der Velden J, Hoor Ten KA, Boezen HM, de Vries EG, Schilthuis MS, Mourits MJ, Nijman HW, Aalders JG, Hollema H, et al. Preoperative serum squamous cell carcinoma antigen levels in clinical decision making for patients with early-stage cervical cancer. J Clin Oncol. 2005;23:1455–1462. doi: 10.1200/JCO.2005.02.123. [DOI] [PubMed] [Google Scholar]
- 33.Gaarenstroom KN, Kenter GG, Bonfrer JM, Korse CM, Van de Vijver MJ, Fleuren GJ, Trimbos JB. Can initial serum cyfra 21–1, SCC antigen, and TPA levels in squamous cell cervical cancer predict lymph node metastases or prognosis? Gynecol Oncol. 2000;77:164–170. doi: 10.1006/gyno.2000.5732. [DOI] [PubMed] [Google Scholar]
- 34.Montag TW. Tumor markers in gynecologic oncology. Obstet Gynecol Surv. 1990;45:94–105. doi: 10.1097/00006254-199002000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Duk JM, de Bruijn HW, Groenier KH, Hollema H, ten Hoor KA, Krans M, Aalders JG. Cancer of the uterine cervix: Sensitivity and specificity of serum squamous cell carcinoma antigen determinations. Gynecol Oncol. 1990;39:186–194. doi: 10.1016/0090-8258(90)90430-S. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Chen P, Mao CM, Tang XP, Zhu LR. Evaluation of diagnostic value of four tumor markers in bronchoalveolar lavage fluid of peripheral lung cancer. Asia Pac J Clin Oncol. 2014;10:141–148. doi: 10.1111/ajco.12066. [DOI] [PubMed] [Google Scholar]
- 37.Cao X, Zhang L, Feng GR, Yang J, Wang RY, Li J, Zheng XM, Han YJ. Preoperative Cyfra21-1 and SCC-Ag serum titers predict survival in patients with stage II esophageal squamous cell carcinoma. J Transl Med. 2012;10:197. doi: 10.1186/1479-5876-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torre GC. SCC antigen in malignant and nonmalignant squamous lesions. Tumour Biol. 1998;19:517–526. doi: 10.1159/000030045. [DOI] [PubMed] [Google Scholar]
- 39.Ryu HK, Baek JS, Kang WD, Kim SM. The prognostic value of squamous cell carcinoma antigen for predicting tumor recurrence in cervical squamous cell carcinoma patients. Obstet Gynecol Sci. 2015;58:368–376. doi: 10.5468/ogs.2015.58.5.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moody CA, Laimins LA. Human papillomavirus oncoproteins: Pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 41.Ferenczy A, Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11–16. doi: 10.1016/S1470-2045(01)00617-9. [DOI] [PubMed] [Google Scholar]
- 42.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]