Abstract
The aim of the present study was to examine the regulatory mechanism underlying the depression in Ski-related novel protein N (SnoN) in diabetic nephrology (DN). NRK-52E cells, a rat primary renal tubular epithelial cell line, were cultured to clarify the effect of small mothers against decapentaplegic (Smad) ubiquitination regulatory factor 2 (smurf2) on SnoN in a low glucose environment in vitro. NRK-52E cells and DM rats were injected with adenoviruses AD-smurf2 and AD-shsmurf2, respectively, and the protein expression profiles of SnoN, smurf2 and phosphorylated (p)-Smad2 were then detected. In addition, the protein levels of smurf2, p-Smad2 and SnoN were analyzed following treatment with transforming growth factor (TGF)-β1 or TGF-β1 inhibitor to validate the effect of the TGF-β1/Smad signaling pathway. The effect of smurf2 on the degradation of SnoN by ubiquitination was found to be a key factor in DN, which was mediated by the TGF-β1/Smad signaling pathway.
Keywords: diabetic nephropathy, Ski-related novel protein N, Smad ubiquitination regulatory factor 2, transforming growth factor-β1/small mothers against decapentaplegic signaling pathway, NRK-52E, ubiquitination
Introduction
Diabetic nephropathy (DN) is the most severe and most common long-term microvascular complication in diabetes mellitus (DM), and 20–40% of diabetic patients are at risk of developing DN (1). In addition with changing lifestyle and dietetic habits, the incidence of diabetes has increased annually worldwide (2). Chronic renal failure resulting from DN is the leading cause for dialysis and renal transplantation in Europe, America and developed areas of China (3). However, the pathogenesis of DN remains to be fully elucidated. Renal tubular lesions appear in the early stage of DN and tubulointerstitial fibrosis, which is closely associated with renal failure, has a substantial effect on renal function in DN (4,5).
The activity of the transforming growth factor (TGF)-β1 signaling pathway can be negatively regulated by Ski-related novel protein N (SnoN), which is a member of the Ski family of protooncoproteins, which are involved in binding to small mothers against decapentaplegic (Smad) complexes (6). SnoN has been found to be expressed at high levels and localized exclusively to the nucleus in normal renal tissues (7), which strictly limits the activity of the TGF-β1 signaling pathway. In this respect, the elevated expression of SnoN is considered a critical gatekeeper and may be responsible for the restriction of Smad signaling in normal conditions. However, the expression level of SnoN is progressively reduced in a time-dependent manner in the fibrotic kidney (8), which renders the restriction mechanism ineffective and can lead to renal fibrosis resulting from the enhanced TGF-β1 signaling pathway. The TGF-β1/Smad signaling pathway has been recognized as a pathway closely associated with renal hypertrophy and tubulointerstitial fibrosis (9). SnoN is an important negative regulator of TGF-β1/Smad signaling, and its expression level has a marked effect on the TGF-β1/Smad pathway (9). Several E3 ubiquitin ligases have been shown to control the TGF-β1/Smad signaling pathway with dual-directional regulation, including Smad ubiquitination regulatory factor 2 (smurf2). Upon TGF-β1/Smad stimulation, the phosphorylated (p-) Smad2 interacts with smurf2 and the degradation of SnoN degradation by ubiquitination is activated by binding to smurf, which enhances the activity of TGF-β1/Smad. Smurf2 also functions as a mediator to promote the degradation of TGF-β1 receptor and Smad2, leading to the weakened or depressed biological function of TGF-β1 (10–12). These studies demonstrated that smurf2 is important in balancing regulation of the TGF-β1/Smad signaling pathway.
In our previous study, SnoN depression was found in DN. To clarify the underlying mechanism, DM rats were treated as in vivo models. The results indicated that the protein level of smurf2 was upregulated and that of SnoN was downregulated in the DM rat renal tissues. The expression level of smurf2 was negatively correlated with SnoN, however, the mRNA level of SnoN showed no significant difference. Based on these results, it was hypothesized that the depressed expression of SnoN was due to increased degradation, which was induced by smurf2. To further examine the regulatory mechanism, NRK-52E rat primary renal tubular epithelial cells and DM rats were utilized in the present study to determine the effect of smurf2 on the expression of SnoN, and to elucidate the molecular mechanism underlying SnoN degradation by TGF-β1 stimulation and inhibition assays.
Materials and methods
Reagents
Streptozotocin (STZ) was purchased from Sigma-Aldrich; Merck Millipore (Darmstadt, Germany). DMEM (low glucose), DMEM (high glucose) and fetal bovine serum (FBS) were purchased from HyClone; GE Healthcare Life Sciences (Logan, UT, USA). Primary antibodies against smurf2 (cat. no. 12024), Smad2 (cat. no. 3103) and p-Smad2 (cat. no. 3108) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA), whereas primary antibody against SnoN (cat. no. NBP1-77306PEP) was purchased from Novus Biologicals (Littleton, CO, USA).
Cell culture
The NRK-52E rat primary renal tubular epithelial cells and 293 human embryonic kidney cell line were purchased from Stem Cell Bank (Shanghai, China). The NRK-52E cells were cultured in DMEM high medium (HyClone; GE Healthcare Life Sciences) containing 5% FBS. The 293 cells were cultured in DMEM high medium containing 10% FBS. All cells were grown in a humidified 5% CO2 incubator at 37°C.
Cell treatment
Effects of glucose on cell culture
To examine the reversibility of the effect of glucose, 2×105 cells were cultured in medium with 24.5 mM glucose at 37°C for 24 h and then switched to medium with 5.5 mM glucose for 0–4 h. The cells were harvested at different time points (0, 15 and 30 min for RT-qPCR and 0, 2 and 4 h for western blot) and the expression levels of smurf2, SnoN, TGF-β1 and Smad2 were detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analyses.
Effects of TGF-β1 in cell culture
To investigate the effect of TGF-β1, 2×105 cells were treated with 2 ng/ml TGF-β1 (HumanZyme, Inc., Chicago, IL, USA) at 37°C for 0–4 h. To inhibit the TGF-β1 signaling pathway, 5 µM TGF-β1 inhibitor (SB431542; Selleck Chemicals, Houston, TX, USA) was added 2 h prior to the switch to high glucose. DMSO solvent was used as a control for the TGF-β1 inhibitor. The cells were harvested at different time points (0, 15 and 30 min for RT-qPCR and 0, 2 and 4 h for western blot), and the expression levels of smurf2, SnoN, TGF-β1 and Smad2 were detected using RT-qPCR and western blot analyses.
Infection with adenovirus
To induce the overexpression of smurf2, 2.5×105 NRK-52E cells were infected with the AD-smurf2 and AD-negative control (NC) adenovirus which were purchased from GeneChem Co., Ltd. (Shanghai, China) according to the manufacturer's protocol, and the medium containing the adenovirus was replaced with fresh medium 12 h following infection. The cells were harvested 48 h following infection, and the expression levels of smurf2, SnoN and the TGF-β1 signaling pathway (smad and p-smad) were detected using RT-qPCR and western blot analyses. Similarly, for smurf2 knockdown, the NRK-52E cells were infected with the AD-short hairpin (sh)smurf2 and AD-shNC adenovirus, respectively, and the expression levels of smurf2, SnoN and the TGF-β1 signaling pathway (smad and p-smad) were detected.
Animal model
Male SD rats (n=50; 8 weeks old; 180–200 g; Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were intraperitoneally injected with 60 mg/kg STZ to establish the DN rat model. The rats were fasted 12 h prior to model establishment without water deprivation. The 1% STZ was prepared with citrate buffer (pH 4.5; 0.1 M) on ice. Prior to injection, the urine glucose and caudal vein glucose levels of the rats were confirmed to be normal. Caudal vein glucose was detected 48 and 72 h following injection and rats with glucose concentrations maintained ≥6.7 mmol/l were recognized as diabetic (DM) rats and the standard remained unchanged during the experiment. Overexpression and knockdown models were established through left renal vein (LRV) injection, prior to which the left renal artery and vein of the DM rats required closure prior to injection, according to surgical methods described by Guadalupe et al (13). The experimental rats were randomly divided into six groups: i) smurf2 knockdown group, in which 5×109 pfu/ml AD-shsmurf2 was injected into the LRV; ii) AD-shNC group, in which 4×109 pfu/ml AD-shNC was injected into the LRV; iii) smurf2 overexpression group, in which 5×10 9 pfu/ml AD-smurf2 was injected into the LRV; iv) AD-NC group, in which 4×109 pfu/ml AD-NC was injected into the LRV; v) sham group, in which normal saline was injected into the LRV; vi) control group of untreated DM rats. The rats were sacrificed by cervical dislocation and the protein expression levels of smurf2 and SnoN were detected in the renal tissues of the different groups.
All animal experiments were performed in strict accordance with the recommendations in the guidelines for the Animal Care and Use Committee of Beijing Friendship Hospital (Beijing, China). The rats were housed at 18–26°C on a 12-h light/dark cycle with free access to water and standard rat chow. The animals were allowed to acclimatize for a minimum of 1 week. The environment was maintained at a relative humidity of 30–70%. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Detection of expression levels of smurf2, SnoN, TGF-β1 and Smad2 using RT-qPCR analysis
RNA isolation and RT
The total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The isolated RNA was reverse transcribed into cDNA using a ReverTra Ace qPCR RT kit (Toboyo Co., Ltd., Osaka, Japan). Briefly, a total of 1,000 ng RNA was used for the initial RT reaction.
qPCR
The qPCR analysis to measure the expression of mRNA was performed using a KAPA SYBR FAST qPCR kit (Kapa Biosystems, Inc., Wilmington, MA, USA) according to the manufacturer's protocol. cDNA was diluted 1:19 with ddH2O and 5 µl was added into each well. The qPCR data collection was performed on an ABI 7500 apparatus (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction was performed for 30 sec at 95°C, 40 cycles at 95°C for 30 sec, and 60°C for 30 sec. RNA 18S used as an endogenous control in each sample. The relative quantification of mRNA expression was calculated using the 2−ΔΔCq method (12) relative to the level of 18S. All reactions were replicated three times.
The qPCR primers for mRNA and 18S were as follows: Smurf2, forward 5′-AACAAGACCGGCGTCAGAAT-3′ and reverse 5′-TGTTCATAGCCTTCGGGTAG-3′; SnoN, forward 5′-GACAAATTTCTCCTTGGTTCCG-3′ and reverse 5′-CTCTCCATCCGTTTCCATCAGT-3′; Smad2, forward 5′-GCCGCCCGAAGGGTAGAT-3′ and reverse 5′-TTCTGTTCTCCACCACCTGC-3′; 18S, forward 5′-CAGCCACCCGAGATTGAGCA-3′ and reverse 5′-TAGTAGCGACGGGCGGTGTG-3′.
Statistical analysis
Statistical comparisons between two different groups were determined using Student's t-test with GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The results are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Western blot analysis
Following the various treatments for the indicated intervals, the cells and tissues were lysed in lysis buffer. The protein concentrations were determined using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. The proteins (50 µg) were separated on a 10% SDS-polyacrylamide gel and transferred electrophoretically onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% nonfat milk in Tris-buffered saline/0.1% Tween 20 for 1 h at room temperature, and subsequently incubated with primary antibodies with the dilution of 1:1,000 at 4°C overnight. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit immunoglobulin G; cat. no. BS13278; and goat anti-mouse immunoglobulin G; cat. no. BS12478, respectively; Bioworld Technology, Inc., St Louis Park, MN, USA) with the dilution of 1:5,000 at room temperature for 1 h. The protein bands were visualized using an enhanced chemiluminescence detection system (GE Healthcare Life Sciences, Chalfont, UK).
Histology and immunohistochemistry (IHC)
Alterations in renal morphology were examined in methyl Carnoy-fixed, paraffin-embedded tissue sections (4 µm) stained with hematoxylin and eosin or periodic acid-Schiff stain. Immunostaining was performed with the paraffinized sections using a microwave-based antigen retrieval technique. The antibodies used included smurf2, SnoN, Smad2 and pSmad2 and were all diluted at 1:50. The primary antibodies were incubated at 4°C overnight. An isotype-matched rabbit IgG was used as a negative control throughout the experiment. All slides were counterstained with hematoxylin for visualization of the nuclei.
Results
Protein levels of SnoN are negatively correlated with smurf2 in low glucose
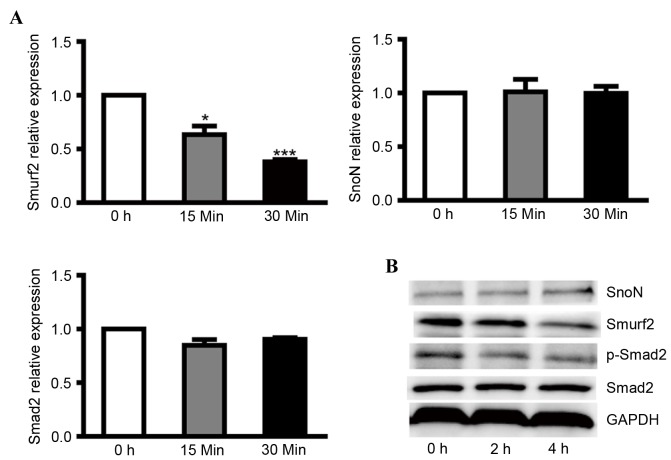
To clarify the regulatory effect of smurf2 on SnoN in DN, NRK-52E cells were cultured overnight with 24.5 mM glucose (normal) and then treated with medium containing 5.5 mM glucose (low). After 30 min, low glucose treatment induced a lower mRNA expression level of smurf2, however, no significant differences were found in the mRNA levels of SnoN and Smad2 (Fig. 1A). At the protein level, low glucose caused higher expression of SnoN and lower expression of smurf2 and p-Smad2 in the NRK-52E cells (Fig. 1B). These results revealed that low glucose inactivated the TGF-β1/Smad signaling pathway and that depressed expression of smurf2 in NRK-52E cells caused the degradation of SnoN by ubiquitination.
Figure 1.
Expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells incubated with low glucose medium. (A) Reverse transcription-quantitative polymerase chain reaction analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2 in NRK-52E cells incubated with low glucose medium at different time points. (B) Western blot analysis was performed to detect the expression levels of smurf2, SnoN, Smad2 and p-Smad2 in NRK-52E cells incubated with low glucose medium at different time points. *P<0.05 and ***P<0.001, vs. control. SnoN, ski-related novel protein N; p-Smad2, phosphorylated small mothers against decapentaplegic; smurf2, Smad ubiquitination regulatory factor 2.
SnoN degradation through ubiquitination is mediated by smurf2 in vitro
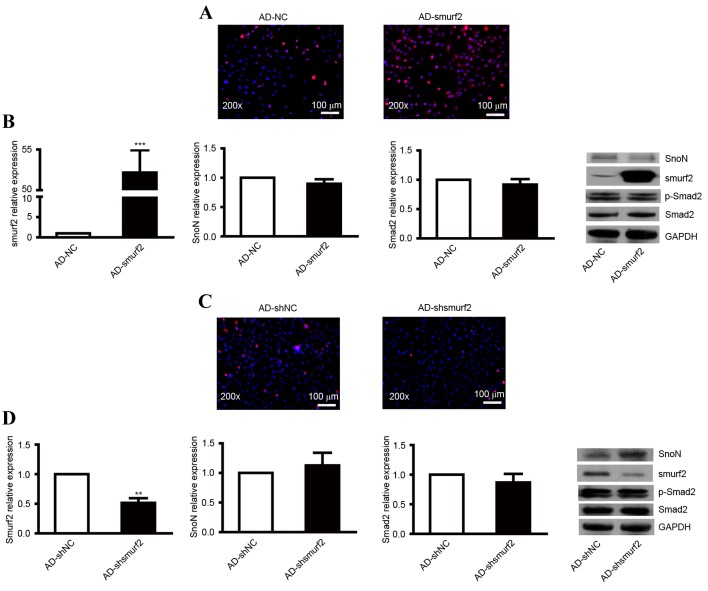
To evaluate whether the degradation of SnoN through ubiquitination was mediated by smurf2, the NRK-52E cells were transfected with smurf2 recombinant adenovirus. An immunofluorescent assay was used to detect the expression of smurf2. Compared with the control, the NRK-52E cells transfected with AD-smurf2 showed enhanced red fluorescence (Fig. 2A). No significant difference in the ectopic overexpression of smurf2 was found in SnoN and Smad2 at the mRNA level (Fig. 2B). The protein level of SnoN was reduced, but no effect on the expression of p-Smad2 was observed (Fig. 2B).
Figure 2.
Expression levels of SnoN and p-Smad2 in NRK-52E cells transfected with recombinant adenovirus. (A) Immunofluorescence analysis of the expression of smurf2 in NRK-52E cells infected with AD-smurf2. (B) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2, and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells infected with AD-smurf2. (C) Immunofluorescence analysis of the expression of smurf2 in NRK-52E cells treated with AD-shsmurf2. (D) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2, and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells infected with AD-shsmurf2. ***P<0.001, vs. control. SnoN, ski-related novel protein N; p-Smad2, phosphorylated small mothers against decapentaplegic; smurf2, Smad ubiquitination regulatory factor 2; AD, adenovirus; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
The present study also infected NRK-52E cells with recombinant adenovirus AD-shsmurf2 to knockdown the expression of smurf2. An immunofluorescent assay was used to detect the expression of smurf2. Compared with the control, NRK-52E cells infected with AD-shsmurf2 showed weakened red fluorescence (Fig. 2C). RT-qPCR analysis was performed to detect the expression levels of smurf2, SnoN and Smad2 in the NRK-52E cells infected with AD-shsmurf2. The results indicated no significant differences in the expression of SnoN or Smad2; however, the expression of smurf2 was decreased, compared with that in NRK-52E cells infected with AD-shNC (Fig. 2D). By performing a western blot assay, it was found that the downregulated expression of smurf2 increased the protein level of SnoN, however, there was no obvious alteration in the expression of p-Smad2 (Fig. 2D). Taken together, these results indicated that smurf2 led to the degradation of SnoN by ubiquitination when the TGF-β1/Smad signaling pathway was activated continuously.
Smurf2 induces SnoN degradation in vivo
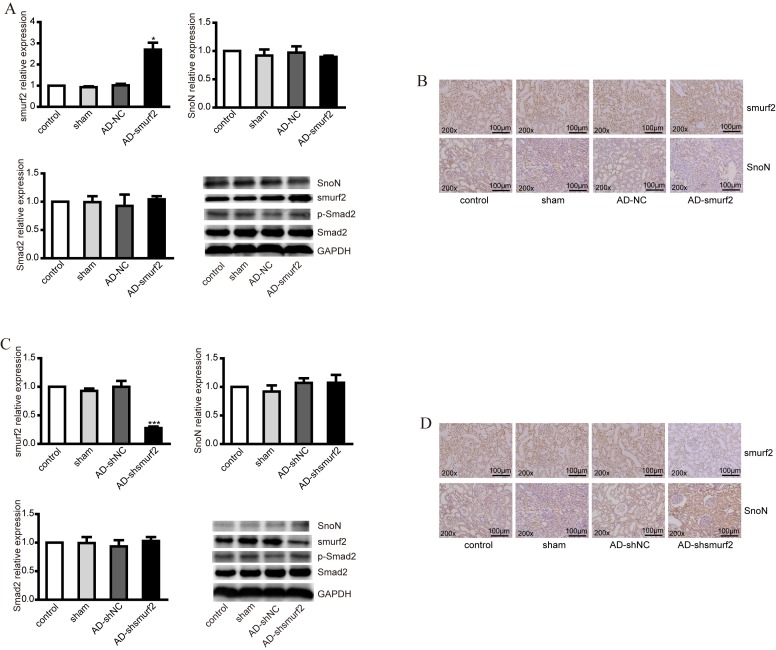
To further validate the effect of smurf2 on the degradation of SnoN in vivo, smurf2 overexpression and knockdown DM rat models were established. DM rats were injected with adenovirus AD-smurf2 and AD-shsmurf2, respectively. The smurf2 expression profile was upregulated in tissues collected from the AD-smurf2 group, compared with those from the control, however, no significant difference in the expression levels of SnoN or Smad2 were observed at the mRNA level (Fig. 3A). At protein level, the overexpression of smurf2 decreased the expression of SnoN, but had no effect on the expression of p-Smad2 (Fig. 3A). This difference was further confirmed using immunohistochemistry (IHC), which revealed the expression of smurf2 and SnoN in renal tubules. Smurf2 was localized to the nucleus and plasma membrane; however, SnoN was predominantly expressed in the plasma membrane. The results of the IHC staining showed that the overexpression of smurf2 decreased the expression of SnoN (Fig. 3B), which was consistent with the results of the western blot analysis.
Figure 3.
Expression levels of smurf2, SnoN and p-Smad2 in DM rats injected with recombinant adenovirus. (A) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2, and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in animal models established via injection of recombinant adenovirus AD-smurf2 into DM rats. (B) Immunohistochemical staining showed the expression of smurf2 and SnoN in renal tubules of DM rats in different groups. (C) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2 and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in the DM rats injected with recombinant adenovirus AD-shsmurf. (D) Immunohistochemical staining showed the expression of smurf2 and SnoN in renal tubules of the DM rats in different groups. *P<0.05 and ***P<0.001, vs. control. DM, diabetes mellitus; SnoN, ski-related novel protein N; p-Smad2, phosphorylated small mothers against decapentaplegic; smurf2, Smad ubiquitination regulatory factor 2; AD, adenovirus; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
In addition, the DM rats were injected with adenovirus AD-shsmurf2 to knockdown the expression of smurf2 in vivo. However, no significant differences were observed in SnoN or Smad2 at mRNA level following RT-qPCR analysis (Fig. 3C). From the western blot assays, it was found that the decreased expression of smurf2 increased the protein level of SnoN; however, no alterations were observed in the expression of p-Smad2 (Fig. 3C). The IHC staining showed a higher expression level of SnoN, compared with control group (Fig. 3D).
TGF-β1/Smad signaling is required for the degradation of SnoN
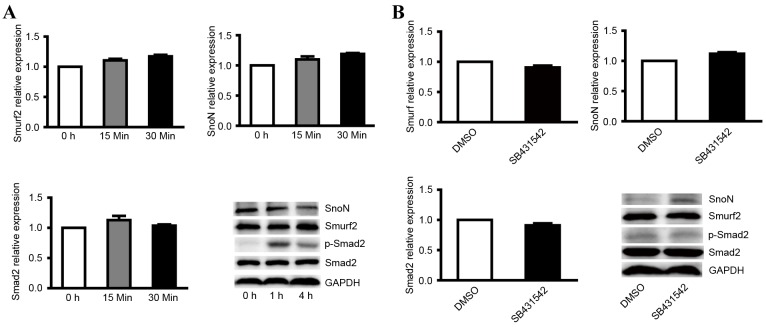
Although it was confirmed that the degradation of SnoN is important in the development of DN and that smurf2 was the key factor in SnoN degradation by ubiquitination, the mechanism of SnoN degradation remains to be fully elucidated. The TGF-β1/Smad signaling pathway is considered to be pathway associated with renal hypertrophy and tubulointerstitial fibrosis, whereas SnoN is the key negative regulator of TGF-β1/Smad and it has been widely recognized that SnoN has the ability to inhibit the signaling pathway (7–9). To verify the role of the TGF-β1/Smad signaling pathway in the degradation of SnoN, the present study treated NRK-52E with 2 ng/ml TGF-β1. The results indicated that TGF-β1 did not affect the mRNA levels of smurf2, SnoN or Smad2 (Fig. 4A), however, the ectopic expression of TGF-β1 resulted in decreased expression of SnoN and increased expression of p-Smad2 following treatment with TGF-β1 for 4 h (Fig. 4A).
Figure 4.
Expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells treated with TGF-β1 and its inhibitor. (A) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2, and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells treated with TGF-β1 at different time points. (B) RT-qPCR analysis was performed to detect the mRNA levels of smurf2, SnoN and Smad2, and western blot analysis was performed to detect the expression levels of smurf2, SnoN and p-Smad2 in NRK-52E cells treated with TGF-β1 inhibitor. SnoN, ski-related novel protein N; p-Smad2, phosphorylated small mothers against decapentaplegic; smurf2, Smad ubiquitination regulatory factor 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TGF-β1, transforming growth factor-β1.
In order to verify the expression level of active genes following inhibition of the TGF-β1/Smad signaling pathway, NRK-52E cells were also treated with the TGF-β1 kinase inhibitor, SB431542, which specifically targets the TGF-β1 receptor. The results indicated that the TGF-β1 kinase inhibitor did not affect the mRNA levels of smurf2, SnoN or Smad2 in cells treated with 5 µM SB431542 (Fig. 4B). Western blot analysis showed that inhibiting the TGF-β1/Smad signaling pathway enhanced the expression of SnoN and decreased the expression of p-Smad2 (Fig. 4B). These results suggested that the TGF-β1/Smad signaling is indispensable in the degradation of SnoN by ubiquitination mediated by smurf2.
Discussion
DN is one of the most important microvascular complication of diabetes and is the leading cause of end-stage kidney failure worldwide, contributing to morbidity and mortality rates (14). Due to the complexity of metabolic disorders, the treatment of DN is more difficult, compared with other kidney diseases. As the pathogenesis of DN remains to be fully elucidated and there is no effective drugs, it is important to investigate the pathogenesis for the prevention and further treatment of DN. Fukasawa et al (15) suggested that the reduction in the protein expression of SnoN is critical in the progression of obstructive nephropathy and results in dysregulation of the TGF-β1/Smad signaling pathway, which is important in the pathogenesis of renal fibrosis. However, ureteral obstruction is the initiating factor of obstructive nephropathy accompanied with rapid onset and apparent inflammatory reaction. Hyperglycemia is the initiating factor of DN, in addition to glucose metabolic disturbance, slow onset and marginal inflammatory reaction, and DN has a more complex pathogenesis and higher morbidity rates, compared with obstructive nephropathy. There have been no reports on whether the progression of DN is associated with the protein expression of SnoN. In a DM rat model, it was found that the protein expression of SnoN was significantly downregulated and showed a negative correlation with smurf2 in the rat renal tissue (16,17). However, there was no apparent difference in the mRNA expression of SnoN between DM rat and normal rat, indicating that downregulation of the protein expression of SnoN was possibly caused by degradation mediated by smurf2. To verify this, the present study infected NRK-52E or rat renal tissues with adenovirus to knock down or overexpress smurf2, and then detected the protein expression of SnoN, the level of which was decreased following infection.
In general, the activated TGF-β1 signaling pathway causes degradation of SnoN protein and leads to the activation of TGF-β1 downstream targets. The protein expression of SnoN is then increased and affects the transcriptional activity of the Smad complex (18). It has been reported that the activation of TGF-β1 reduces the half-life of SnoN from 4 h to 45 min through degradation by ubiquitination, in which E3 ubiquitin ligase was involved. There are different ubiquitination pathways between cells and diseases (9). TGF-β1 is important in the process of DN. SnoN effectively inhibits TGF-β1/Smad signaling activity through several mechanisms (19,20). Several reports have indicated that SnoN directly interacts with intranuclear Smad and inhibits the gene transcriptional activity induced by Smad; secondly, SnoN recruits other nuclear transcription co-repressors, including nuclear hormone receptor co-repressor; finally, SnoN prevents Smad from binding to the uclear transcription co-activator, p300/CBP. As a significant negative regulator of the TGF-β1/Smad signaling pathway, SnoN inhibits the target gene of TGF-β1 pre-transcriptionally and decreases the potency and duration of TGF-β1/Smad signaling. Although it is generally recognized that SnoN specifically inhibits the TGF-β1/Smad signaling pathway and decreased SnoN protein is important in the process of DN, the mechanism remains to be fully elucidated. The present study treated NRK-52E with TGF-β1 or TGF-β1 inhibitor and detected the gene expression of the TGF-β1/Smad signaling pathway. The results indicated that the TGF-β1/Smad negatively regulated the protein expression of SnoN, demonstrating that the degradation of SnoN by smurf2 was mediated by the TGF-β1/Smad signaling pathway during the process of DN.
In the present study, NRK-52E cells and DM rat renal tissues were also infected with adenovirus to knock down or overexpress smurf2, to demonstrate that the degradation of SnoN protein by ubiquitination was mediated by smurf2. The NRK-52E cells were also infected with TGF-β1 or its inhibitor, and it was confirmed that the reduction in nuclear transcription of the co-repressor SnoN protein was regulated by the TGF-β1/Smad signaling pathway during the process of DN. These results provide a theoretical basis for the treatment of DN.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81300607).
References
- 1.Standards of medical care in diabetes-2012. Diabetes Care. 2012;35:S11–S63. doi: 10.2337/dc12-s011. (Suppl 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.InterAct Consortium: Dietary fibre and incidence of type 2 diabetes in eight European countries: The EPIC-InterAct Study and a meta-analysis of prospective studies. Diabetologia. 2015;58:1394–1408. doi: 10.1007/s00125-015-3585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan J, Li P, Liu DW, Chen Y, Mo HZ, Liu BG, Chen WJ, Lu XQ, Guo J, Zhang Q, et al. GSK-3β inhibitor attenuates urinary albumin excretion in type 2 diabetic db/db mice, and delays epithelial-to-mesenchymal transition in mouse kidneys and podocytes. Mol Med Rep. 2016;14:1771–1784. doi: 10.3892/mmr.2016.5441. [DOI] [PubMed] [Google Scholar]
- 4.Vallon V, Thomson SC. Renal function in diabetic disease models: The tubular system in the pathophysiology of the diabetic kidney. Annu Rev Physiol. 2012;74:351–375. doi: 10.1146/annurev-physiol-020911-153333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang SC, Leung JC, Lai KN. Diabetic tubulopathy: An emerging entity. Contrib Nephrol. 2011;170:124–134. doi: 10.1159/000325647. [DOI] [PubMed] [Google Scholar]
- 6.Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.ASN.0000033611.79556.AE. [DOI] [PubMed] [Google Scholar]
- 7.Krakowski AR, Laboureau J, Mauviel A, Bissell MJ, Luo K. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci USA. 2005;102:12437–12442. doi: 10.1073/pnas.0504107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Zhang X, Li Y, Liu Y. Downregulation of Smad transcriptional corepressors SnoN and Ski in the fibrotic kidney: An amplification mechanism for TGF-beta1 signaling. J Am Soc Nephrol. 2003;14:3167–3177. doi: 10.1097/01.ASN.0000099373.33259.B2. [DOI] [PubMed] [Google Scholar]
- 9.Deheuninck J, Luo K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009;19:47–57. doi: 10.1038/cr.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y, Zhou CH, Fu D, Shen XZ. Overexpression of Smad ubiquitin regulatory factor 2 suppresses transforming growth factor-β mediated liver fibrosis. J Dig Dis. 2012;13:327–334. doi: 10.1111/j.1751-2980.2012.00592.x. [DOI] [PubMed] [Google Scholar]
- 11.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99:2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng XM, Chung AC, Lan HY. Role of the TGF-β/BMP-7/Smad pathways in renal diseases. Clin Sci (Lond) 2013;124:243–254. doi: 10.1042/CS20120252. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz-Muñoz G, Mallavia B, Lopez-Franco O, Hernandez-Vargas P, Egido J, Gomez-Guerrero C. Renal delivery of adenovirus and antisense oligonucleotides in rats by retrograde renal vein injection. Methods Mol Biol. 2012;886:321–329. doi: 10.1007/978-1-61779-851-1_29. [DOI] [PubMed] [Google Scholar]
- 14.Marchant V, Droguett A, Valderrama G, Burgos ME, Carpio D, Kerr B, Ruiz-Ortega M, Egido J, Mezzano S. Tubular overexpression of Gremlin in transgenic mice aggravates renal damage in diabetic nephropathy. Am J Physiol Renal Physiol. 2015;309:F559–F568. doi: 10.1152/ajprenal.00023.2015. [DOI] [PubMed] [Google Scholar]
- 15.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, et al. Ubiquitin-dependent degradation of SnoN and Ski is increased in renal fibrosis induced by obstructive injury. Kidney Int. 2006;69:1733–1740. doi: 10.1038/sj.ki.5000261. [DOI] [PubMed] [Google Scholar]
- 16.Rui-Xia L, Bing G, Ying X, Ming-jun S, Yuan-yuan W. Expression of Smurf 2 in kidney of diabetic rat and its relationship with downregulation of SnoN. Chinese Journal of Pathophysiology. 2010;26:1743–1748. [Google Scholar]
- 17.Liu R, Wang Y, Xiao Y, Shi M, Zhang G, Guo B. SnoN as a key regulator of the high glucose-induced epithelial-mesenchymal transition in cells of the proximal tubule. Kidney Blood Press Res. 2012;35:517–528. doi: 10.1159/000339172. [DOI] [PubMed] [Google Scholar]
- 18.Tecalco-Cruz AC, Sosa-Garrocho M, Vázquez-Victorio G, Ortiz-García L, Domínguez-Hüttinger E, Macías-Silva M. Transforming growth factor-β/SMAD Target gene SKIL is negatively regulated by the transcriptional cofactor complex SNON-SMAD4. J Biol Chem. 2012;287:26764–26776. doi: 10.1074/jbc.M112.386599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Dai C, Liu Y. A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J Am Soc Nephrol. 2005;16:68–78. doi: 10.1681/ASN.2003090795. [DOI] [PubMed] [Google Scholar]
- 20.Javelaud D, van Kempen L, Alexaki VI, Le Scolan E, Luo K, Mauviel A. Efficient TGF-β/SMAD signaling in human melanoma cells associated with high c-SKI/SnoN expression. Mol Cancer. 2011;10:2. doi: 10.1186/1476-4598-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]