Abstract
To remedy the drug resistance of natural tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and enhance its antitumor effects, we prepared a type of TRAIL mutant membrane penetrating peptide alike (TMPPA)-TRAIL mutant R6 (MuR6-TR) by mutating the N-terminal of the soluble TRAIL gene sequence. The expressed MuR6-TR protein was purified to treat pancreatic carcinoma cell lines BxPC-3 and PANC-1. The inhibitory effects on the proliferation of BxPC-3 and PANC-1 cells was assessed with CCK-8 assay and compared with natural TRAIL. The antitumor effect of MuR6-TR was assessed on implant tumors derived from PANC-1 cells in nude mice and compared with gemcitabine. Finally, the soluble MuR6-TR gene was successfully mutated with 4 amino acids in the N-terminal of TRAIL and had a molecular size of 513 bp. The mutant MuR6-TR was connected to pET32a and verified by enzymatic digestion and sequencing. The recombinant MuR6-TR was transformed and expressed in Escherichia coli. The CCK-8 assay results indicated that MuR6-TR inhibited the growth of BxPC-3 and PANC-1 cells in a dose-dependent manner, with IC50 values of 4.63 and 7.84 ng/ml, respectively, which were much lower than that of natural TRAIL. MuR6-TR demonstrated a higher inhibitory effect on tumor growth (24.2%) than natural TRAIL (14.4%) and an effect similar to that of gemcitabine at an early period. Thus, the mutant MuR6-TR exhibited a stronger antitumor effect than that of natural TRAIL both in vivo and in vitro and may have potential therapeutic value for pancreatic carcinoma, which requires further validation.
Keywords: tumor necrosis factor-related apoptosis-inducing ligand, cell-penetrating peptides, antitumor effect, pancreatic carcinoma, apoptosis
Introduction
Pancreatic carcinoma is a neoplasm formed by abnormal proliferation of pancreatic cells due to the dysregulation of cellular growth under the effect of multiple tumorigenic factors. Due to its biological complexity and serious threat to patients, it is necessary to develop new therapeutic strategies for pancreatic carcinoma since current approaches have limited efficacy. For example, gemcitabine is associated with serious side effects and resistance is observed in various cases (1). Among tumorigenic factors, a deficiency in cellular apoptosis, the programmed death of cells, plays a critical role in the onset and development of tumors (2). Caspase activation is a key step in apoptosis and can be activated by both intrinsic and extrinsic pathways to induce a catalytic reaction and mediate cellular apoptosis (3). It has been proposed that an increase in the apoptotic threshold through alteration of molecules contributes to the therapeutic resistance of pancreatic carcinoma including apoptosis inducers or antitumor medications (4). Therefore, enhancing the sensitivity of tumor cells to apoptosis inducers is a potential strategy for the development of novel therapeutic strategies.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), also termed as Apo2 ligand (Apo2L), is a member of the tumor necrosis factor (TNF) superfamily and the immune regulator of congenital and acquired immunity. TRAIL can initiate apoptotic signaling by binding to the death receptor (DR) to induce apoptosis targeting multiple tumor cells but without obvious killing of normal cells (5). However, treating different types of tumors simply dependent on TRAIL/Apo2L has limited efficacy. Studies have indicated that normal cells and more than half of passaged tumor cells (even >60%) demonstrate tolerance to TRAIL (4,6). The reason for the tolerance is due to the existence of a deficiency and mutations in the apoptotic signaling pathways of tumor cells, which include pro-apoptotic factors or anti-apoptotic factors to increase the apoptotic threshold of drug-resistant tumor cells to escape apoptotic scavenging. Increasing the sensitivity of DR to TRAIL in tumor cells and the activity of pro-apoptotic factors, or removing the inhibition of anti-apoptotic factors can promote the efficacy of TRAIL to induce the apoptosis of tumor cells and reverse the tolerance of tumor cells to TRAIL.
Cell-penetrating peptides (CPPs) exhibit high transportation efficacy, low toxicity and no permanent damage to the cellular membrane, and therefore show potential value in reprogramming and gene editing (7). As one member of the CPPs, the basic amino acid polyarginine (pAr) carries cations under physiological pH and can bind to the negative glycosaminoglycan or lipids on the cellular membrane. Because of the small structure and higher penetrating efficacy, pAr can transport molecules into the cytoplasm to bind to downstream signals and to display its biological effects. The features of CPPs demonstrate that the targeting protein can be transported to the cytoplasm through CPPs to decrease the clearance rate of drugs in the blood and to ensure that the drugs directly react with the targeting signal in cells, which simultaneously activate extracellular and intracellular signaling to play a role in the effects of drugs. Therefore, pAr has been used to design antitumor peptides (8,9).
In the present study, we modified the N-terminal of TRAIL to R6 by mutating 4 loci at domain 114-281aa which is enriched in arginine to form a CPP-like amino acid sequence [TRAIL mutant R6 (MuR6-TR)] and investigated the in vivo and in vitro antitumor effects of MuR6-TR in pancreatic carcinoma. This study provides evidence for the modification of TRAIL in order to enhance the sensitivity of pancreatic carcinoma cells to apoptosis inducers.
Materials and methods
Design and synthesis of the primers
According to the literature (10) and GeneBank (http://www.ncbi.nlm.nih.gov/nuccore/), the soluble TRAIL sequence 114-281 at the N-terminal was selected. In reference to the prefered adjusting coding sequence of Escherichia coli (E. coli) synonym codon, the TRAIL sequence was inserted with the initiation codon ATG (M) and termination codon TAA to harvest E. coli preference codon with 513 bp. The amino acid sequence VRERGP located at 114-119 was mutated into RRRRRR, i.e., the N-terminal of natural TRAIL was mutated into R6 with 4 mutation loci to obtain the TRAIL mutant R6 (MuR6-TR). The amino acid sequence of MuR6-TR at 114-281 was: MRRRRRRQRVAAHITGTRGRSNTLSSPNSKNEKALGR KINSWESSRSGHSFLSNLHLRNGELVIHEKGFYYIYSQ TYFRFQEEIKENTKNDKQMVQYIYKYTSYPDPILLMK SARNSCWSKDAEYGLYSIYQGGIFELKENDRIFVSVTN EHLIDMDHEASFFGAFLVG.
The existing TRAIL sequence was used as a template for PCR to achieve local mutation with upstream NdeI and downstream EcoRI as restriction enzyme cleavage sites. The primers were upstream MuR6-TR-NdeI (48 bp), GGTCATATGCGT CGTCGTCGTCGTCGTCAGCGTGTGGCTGCTCACATC and downstream TR-Eco-R (41 bp) GTTGAATTCTTATTAACCAACAAGGAAAGCACCGAAGAAAG.
Amplification of the MuR6-TR segment with PCR
The MuR6-TR was amplified with PCR using 50 µl of the total reaction system containing plasmid DNA (Pmd19/TRAIL) 1 µl, 10× PCR buffer for KOD-Plus-Neo 5 µl, dNTPs (2 mM) 5 µl, 25 mM MgSO4 3 µl, KOD-Plus-Neo 1 µl, MuR6-TR-NdeI/TR-Eco-R (10 pmol/µl) 1 µl, MuR6-TR-NdeI/TR-Eco-R (10 pmol/µl) 0.5 µl of each and RNase-free water 33 µl. The reaction condition included an initial denaturation at 94°C for 2 min followed by 25 cycles of denaturation at 94°C for 15 sec and annealing/extension at 68°C for 30 sec, with a final extension at 68°C for 5 min.
Transformation and identification of MuR6-TR
The vector and target gene were digested with NdeI and EcoRI, harvested with OMEGA recovery kit, eluted with 30 µl deionized water, electrophoresed and images were captured, respectively. The target segment and vector were linked together and 10 µl of the linked product was added to 100 µl Top10 competent cells for transformation. Then the transformed competent cells were smeared on LB solid medium containing ampicillin (Amp) at 37°C overnight. The bacterial colonies were selected and digested with enzyme for identification. These positive colonies were stored for sequencing.
Expression of MuR6-TR in bacterial colony
The E. coli BL21(DE3) (in 1,000 µl) treated with pET3a-MuR6-TR at 37°C overnight was added to 50 ml LB-Amp+ medium and incubated on a shaking plate (250 rpm) at 37°C for 3 h and then the temperature was decreased to 24°C. Then, IPTG (0.1 M) was added at a 1% ratio for culture induction overnight. Samples of 0.5 and 0.15 ml collected before and after induction were centrifuged and the sediments after removal of the supernatant were re-suspended with 50 µl H2O, followed by addition of 50 µl 2× loading buffer for electrophoresis. The remaining bacterial solution was centrifuged at 12,000 rpm for 5 min to obtain the bacterial collection which was re-suspended with 8 ml Na2HPO4 (50 mM) and lysed with ultrasound. The ultrasound lysis condition was: Φ6 probe, sonication with 200 W pulses for 2 sec with an interval of 2 sec, and repetition for 10 min. The 1 ml bacterial lysis was centrifuged at 12,000 rpm for 10 min to harvest the supernatant and sediment. Then, the supernatant and sediment re-suspended with 1 ml H2O (20 µl for each) were added to 30 µl H2O and 50 µl 2× loading buffer for electrophoresis. The samples for electrophoresis were heated in boiled water for 10 min and centrifuged at 12,000 rpm for 10 min. Finally, 10 µl of the supernatants was used for SDS-PAGE electrophoresis.
Purification of the targeting protein
The protein was firstly purified with cation SP Sepharose FF XK16 column (GE Healthcare, Piscataway, NJ, USA), according to the manual, followed by elution with an anion XK26/20 column filled with Sephadex G-25 medium and anion exchanging buffer (GE Healthcare). Then the protein was purified using a Q Sepharose FF XK16 column (GE healthcare), according to the manufacturer's manual. The penetrating and eluting components were collected, respectively, and 50 µl 2× loading buffer was added in a 1:1 ratio for electrophoresis. The same volume (50 µl) of original solution served as the control. The purified MuR6-TR and TRAIL proteins were subjected to western blotting for identification.
Activity of MuR6-TR protein in inhibiting proliferation of tumor cells
TRAIL-insensitive pancreatic carcinoma cell lines (BxPC-3 and PANC-1) were cultured in a 5% CO2 incubator at 37°C with different media and density in a 96-well plate as shown in Table I. The medium was changed every 2–3 days, and the cells were passaged with medium containing 0.25% trypsin 0.02% EDTA at a 1:1 ratio. Cells in a logarithmic growth stage were used for experiments.
Table I.
Conditions for cell culture.
| Cell line | Medium | Density |
|---|---|---|
| BxPC-3 | RPMI-1640 + 10% FBS + 1.0 mM sodium pyruvate | 4×103/well |
| PANC-1 | Low-glucose DMEM + 10% FBS | 5×103/well |
The MuR6-TR and TRAIL proteins were diluted with sterile PBS buffer to a final concentration of 5 mg/ml and sterilized with a filter. The cells were then treated with the different proteins with initial concentrations of 1 µg/ml and 200 µg/ml (the concentration of natural TRAIL protein was adjusted according to preliminary data) followed by a 3-fold dilution (total 10 dilution concentration). The experiment was repeated triple times.
The inhibitory effects on the proliferation of cells were measured with CCK-8 (cat. no. CK04-13; Dojindo), according to the manufacturer's instructions, analyzed with the equation y = A2 + (A1−A2)/[1 + (x/x0)p] using OriginPro 9.0 software and fitted with growth/digmoidal-logistics for the inhibition rate curve to calculate IC50. The in vitro therapeutic effect of the drugs was defined as sensitive killing with IC50 <10 µg/ml, dose-dependent cytotoxicity and maximal inhibition ratio >80%.
Inhibition of the growth of the implanted tumors in PANC-1- loaded nude mice by MuR6-TR
All animal procedures were approved by the Animal Care and Scientific Committee of Sichuan University. Balb/c nude female mice (SPF, 6–8 weeks, 18–22 g) were provided by Shanghai Sippr-BK Laboratory Animal Ltd. and bred at 23±2°C, with a humidity of 40–70% and a 12/12 light/dark cycle with free access to food and water.
The PANC-1 cells were cultured in DMEM containing 10% FBS in a 5% CO2 incubator at 37°C. The cells at logarithm growth stage were digested with 0.25% trypsin, rinsed with PBS and re-suspended with serum-free medium to adjusted the cell density to 2.5×107 cells/ml (1:1 Matrigel).
Under sterile conditions, each nude mouse was subcutaneously implanted with a 0.2 ml cell suspension (5×106 cells/mouse) in the right axillary. When tumors grew to a size of 150–250 mm3, 32 mice with a healthy appearance and with a similar tumor size (single, global tumor without irregular shape or cluster tumors) were divided into 4 groups: i) saline; ii) gemcitabine; iii) natural TRAIL; and iv) MuR6-TR (n=8 for each group). The mice in the different groups were injected via tail vein with the different agents according to Table II consecutively for 5 days at fixed times.
Table II.
Treatments of mice in the different groups.
| Group | Concentration (mg/ml) |
Volume (ml/kg) |
Strategy |
|---|---|---|---|
| Vehicle (saline) | – | 10 | i.v., q.d. × 5 days |
| Gemcitabine | 5 | 10 | i.v., q.o.d. × 3 times |
| TRAIL | 6 | 10 | i.v., q.d. × 5 days |
| MuR6-TR | 6 | 10 | q.d. × 5 days |
TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
The growth of tumors was measured twice weekly with the long (Y) and short (X) diameters to calculate the tumor growth inhibition (TGI, %), and simultaneously the body weight of mice was determined consecutively for 4 weeks. The TGI (%) was calculated according to the formula:
where volumetreatment is the tumor volume in the treatment groups and tumor volumevehicle is the tumor volume in the vehicle group.
After the last injection of the agents, the animals were sacrificed with an over-dosage of CO2 and the tumors were removed to measure the weight. If the animals appeared moribund or the tumor size was >3,000 mm3 during the experiment, the animal was sacrificed with an over-dosage of CO2 to examine the pathological change in organs.
Statistical analysis
All data are expressed as mean ± SEM only when denoted differently. Comparisons were performed with the Student's t-test and P<0.05 was considered to indicate a statistically significant difference. The mortality rate was expressed as a percentage and compared with the Chi-square test.
Results
Amplification and transformation of the MuR6-TR targeting gene
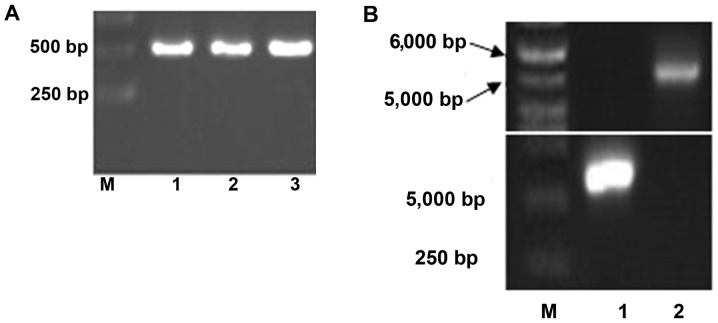
As shown in Fig. 1A, the resulting MuR6-TR segment by one-round PCR with primers MuR6-TR-NdeI/TR-Eco-R was ~510 bp which was in agreement with the size of the designed gene product (513 bp).
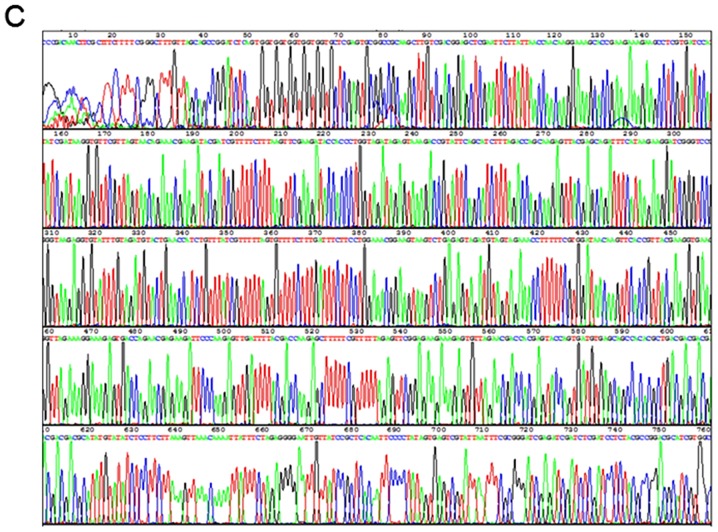
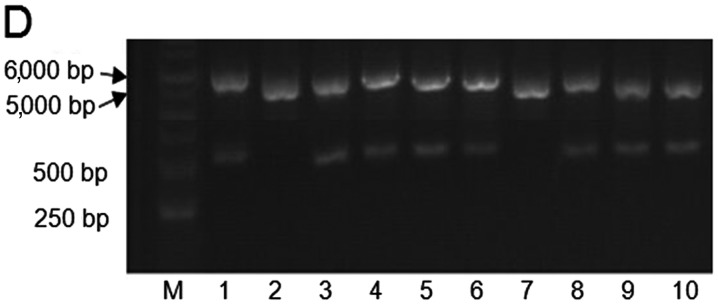
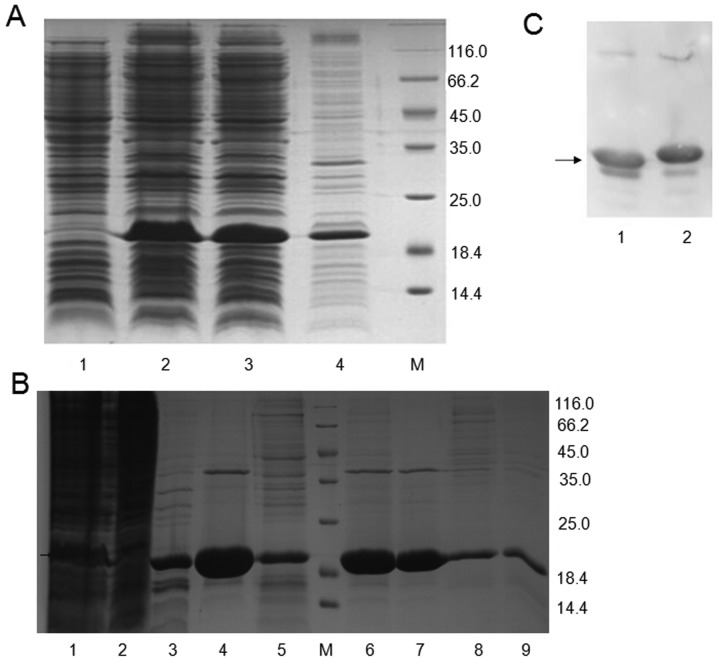
Figure 1.
PCR production of (A) TRAIL mutant R6 (MuR6-TR) digested segment by NdeI/EcoRI, (B) MuR6-TR/pET32a, (C) sequencing and (D) transformation of bacteria. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand.
The segments from MuR6-TR and pET32a digested by NdeI and EcoRI were 550 bp and 5.4 kb, respectively, as shown in Fig. 1B. The sequence of the target gene MuR6-TR was confirmed by Beijing Genomics Institute (Shenzhen, China) and was in agreement with the designed sequence as shown in Fig. 1C.
After transformation by connection of MuR6-TR/pET32a, the bacteria grew well with normal density. The transformed plasmid carrying pET32a/MuR6-TR in the bacteria was digested with XbaI and EcoRI, resulting in an ~5.4 kb vector segment and ~550 bp target segment. As shown in Fig. 1D, the target segment of MuR6-TR was positive in 8 of 10 samples.
Expression and purification of pET32a/MuR6-TR in bacteria
The expression of pET32a/MuR6-TR in bacteria was high. The supernatant of lysate contained 80% target protein and the sediment contained 20% target protein (Fig. 2A).
Figure 2.
Identification of MuR6-TR targeting protein. (A) Electrophoresis of pET32a/MuR6-TR protein from transformed bacteria: lane 1, before induction; lane 2, after induction; lane 3, supernatant; lane 4, sediment. (B) Proteins in different eluting solution: lane 1, original MuR6 solution in SP column; lane 2, penetrating solution in SP column; lane 3, 0.8 M NaCl eluting solution in SP column; lane 4, 1.5 M NaCl eluting solution in SP column; lane 5, NaOH eluting solution in SP column; lane 6, original anion-exchange solution; lane 7, anion-exchange penetrating solution; lane 8, anion-exchange eluting solution; lane 9, 0.5 M NaOH eluting solution; M, marker. (C) Expression of TRAIL (lane 1) and MuR6-TR (lane 2) determined by western blotting. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
The concentration of MuR6-TR protein in the cation-exchange eluting solution, desalination eluting solution and anion-exchange penetrating solution was measured (Table III). According to Table III, the recovery rate from cation-exchange to desalination, from desalination to anion-exchange and from cation-exchange to anion-exchange was 91.33, 87.68 and 80.08%, respectively, which was within the acceptable range. The solution during exchange was clear and the purified protein was subjected to electrophoresis as shown in Fig. 2B. There was a visible protein band in the cation-exchange eluting solution; in the final anion-exchange penetrating solution, there was a clear protein band and few artificial bands.
Table III.
Measurement of protein in the different chromatography solutions.
| Samples | Volume (ml) | Concentration (mg/ml) |
|---|---|---|
| Cation-exchange eluting solution | 26 | 7.7267 |
| Desalination eluting solution | 44 | 4.1701 |
| Anion-exchange penetrating solution | 69 | 2.3315 |
The expression of TRAIL and MuR6-TR proteins was confirmed by western blotting (Fig. 2C).
Inhibition of cell growth in culture by MuR6-TR protein
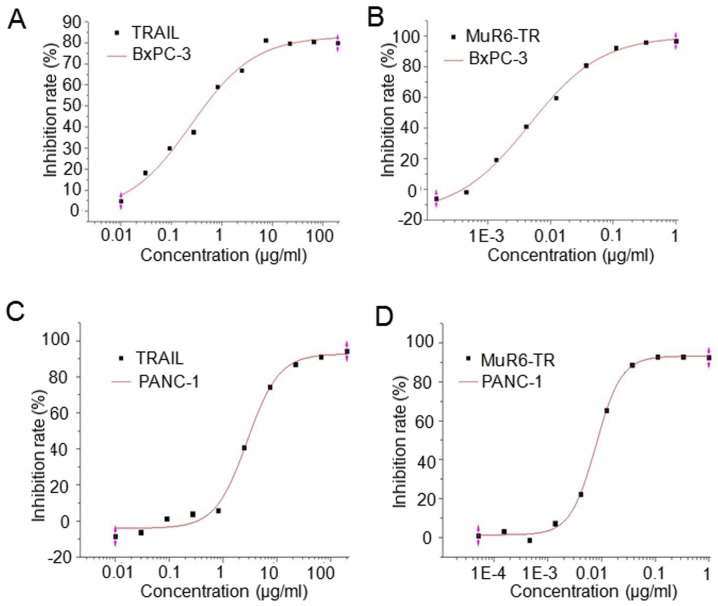
The antitumor activity of MuR6-TR protein was measured in pancreatic carcinoma cells BxPc-3 and PANC-1. The results indicated that the inhibition rate of both cell lines by MuR6-TR was significantly higher when compared to that by natural TRAIL when MuR6-TR was at a similar range of concentrations (P<0.05; Table IV). The fitting curve showed that the IC50 of MuR6-TR for both BxPC-3 and PANC-1 cell lines was significantly lower than that of natural TRAIL (P<0.05; Table V, Fig. 3).
Table IV.
Proliferation inhibition of BxPC-3 and PANC-1 cells by natural TRAIL and MuR6-TR at different concentrations.
| Natural TRAIL
|
MuR6-TR
|
||||
|---|---|---|---|---|---|
| Concentration (µg/ml) | Inhibition rate (%)
|
Concentration (µg/ml) | Inhibition rate (%)
|
||
| BxPc-3 | PANC-1 | BxPc-3 | PANC-1 | ||
| 200 | 79.890 | 94.191 | 1 | 96.659 | 92.408 |
| 66.667 | 80.561 | 91.061 | 0.333333333 | 95.747 | 92.832 |
| 22.222 | 79.657 | 86.793 | 0.111111111 | 92.254 | 92.872 |
| 7.407 | 81.259 | 74.223 | 0.037037037 | 80.832 | 88.602 |
| 2.469 | 66.873 | 40.823 | 0.012345679 | 59.624 | 65.288 |
| 0.823 | 59.125 | 5.747 | 0.004115226 | 41.020 | 22.361 |
| 0.274 | 37.56 | 3.834 | 0.001371742 | 19.372 | 7.291 |
| 0.091 | 29.993 | 1.147 | 0.000457247 | −1.694 | −1.418 |
| 0.030 | 18.294 | −6.277 | 0.000152416 | −5.981 | 3.178 |
| 0.010 | 4.838 | −8.491 | 0.0000508052 | – | 1.037 |
TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
Table V.
IC50 of natural TRAIL and MuR6-TR in the BxPC-3 and PANC-1 cell lines.
| Natural TRAIL
|
MuR6-TRAIL
|
|||
|---|---|---|---|---|
| BxPC-3 | PANC-1 | BxPC-3 | PANC-1 | |
| IC50 (µg/ml) | 0.284 | 2.817 | 4.63×10−3a | 7.84×10−3b |
P<0.05 vs. BxPC-3 in natural TRAIL;
P<0.05 vs. PANC-1 in natural TRAIL. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
Figure 3.
The fitting curves of (A and C) natural TRAIL and (B and D) MuR6-TR on the proliferation inhibition of (A and B) BxPC-3 cells and (C and D) PANC-1 cells. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
Inhibition of tumor growth in nude mice by MuR6-TR
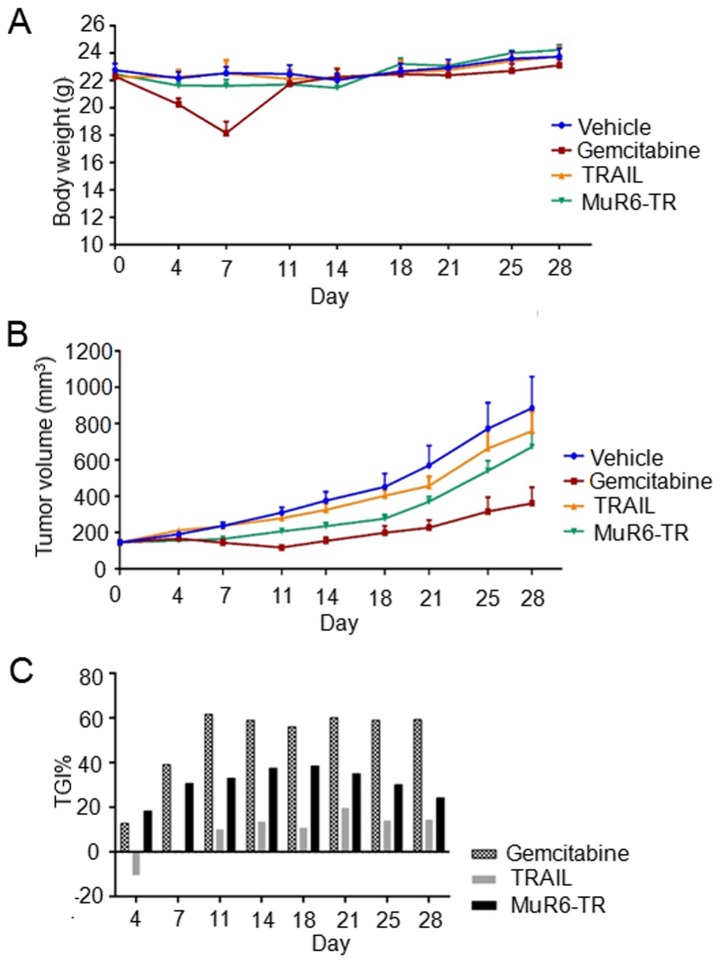
The effects of MuR6-TR on the body weight, tumor growth, tumor inhibition rate and mortality rate of mice are shown in Fig. 4 and Table VI. As shown in Fig. 4A, the body weight of the mice in the 4 groups was not significantly increased during the observation time and there was no significant different among the different groups (P>0.05); but with exception in the gemcitabine group at day 7 when there was a significant decrease noted in body weight (P<0.05).
Figure 4.
Effects of different treatments on (A) body weight, (B) tumor volume and (C) tumor growth inhibition (TGI). TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; MuR6-TR, TRAIL mutant R6.
Table VI.
Mortality rate of mice in different groups.
| Group | Mortality rate (%)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Day
| |||||||||
| 0 | 4 | 7 | 11 | 14 | 18 | 21 | 25 | 28 | |
| Vehicle | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gemcitabine | 0 | 0 | 12.5 | 25 | 25 | 25 | 25 | 25 | 25 |
| Natural TRAIL | 0 | 12.5 | 37.5 | 37.5 | 37.5 | 37.5 | 37.5 | 37.5 | 37.5 |
| MuR6-TR | 0 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25a |
P<0.05 vs. TRAIL. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand. MuR6-TR, TRAIL mutant R6.
The results of tumor growth indicated that the tumor volumes at the different time-points and in the different groups were variable. At day 7 after treatment, the tumor volume in the gemcitabine group was significant smaller than that in the other groups (P<0.05). Furthermore, the tumor volume in the MuR6-TR group was significant smaller than that in the vehicle group and TRAIL group (P<0.05; Fig. 4B). Although the inhibition rate of tumor growth by MuR6-TR was significantly lower than gemcitabine, the inhibition rate of tumor growth by MuR6-TR was significantly higher than that by TRAIL at all observation time-points (P<0.05; Fig. 4C).
For the mortality rate, the results indicated that the mortality rate in the MuR6-TR group was significantly lower than that in the TRAIL group at days 7–28 and similar to that in the gemcitabine group at days 11–28.
Discussion
In the present study, we mutated the natural TRAIL protein at 114-119 residues with CPP-like change to form TRAIL mutant MuR6-TR which was verified by western blotting and demonstrated the antigen determinant of TRAIL. The MuR6-TR protein also displayed antitumor effects in pancreatic carcinoma cell lines and in implanted pancreatic carcinoma tumors in a nude mouse model. These results indicated that natural TRAIL mutant MuR6-TR enhanced the sensitivity of tumor cells to apoptosis inducers and could be a potential targeting therapy for pancreatic cancer.
Human natural TRAIL is a type II transmembrane glycoprotein composed of 281 amino acids. The structural domain of TRAIL C-terminal at residues of 114-281 has several β-motifs to form hollow tubular structure. Three TRAIL monomers can form homogenous trimer through the β-motif to display biological activity (11). The C-terminal of natural TRAIL can be hydrolyzed by metalloprotease into soluble functional segments (sTRAIL) while the trimer of either entire TRAIL or sTRAIL can induce the apoptosis of tumor cells. The defect of N-terminal amino acid of TRAIL has no effect on the protein function, which provides the possibility for the directed reformation of TRAIL.
In a primary study, we modified sTRAIL by forming CPPs in the N-terminal and found that different CPPs in classification and molecular weight demonstrated large variation in the stability, soluble expression and biological activities of TRAIL (data not shown). After studying the structure of TRAIL, we found that the arginine (R) dominant VRERGPQR domain in 114-121 residues of C-terminal has a similar structure with the non-amphipathic pAr of CPPs and is not involved in the specific spatial conformation of TRAIL. Therefore, we proposed to directly reform the N-terminal of sTRAIL with CPPs in 114-121 residues of the C-terminal.
In the study of pAr cell-penetration, R9 is commonly used because both the linear and branched structures have cell-penetrating function (12). A previous study indicated that the cell-penetrating efficacy of the peptide having <5 or >15 R is obviously decreased (13). With the success in mutating VRERGPQR with 5 amino acid into RRRRRRRRR8 (R8) in 114-121 residues of TRAIL (data not shown), we further reduced the mutation by mutating 4 residues of sTRAIL N-terminal (R6) in the present study and this mutant was confirmed by PCR. This soluble R6 mutation increased the stability while minimizing the change of the primary structure of TRAIL to exhibit biological activities.
The expression of exogenous protein by E. coli is affected by many factors, such as IPTG concentration, induction time and stability of exogenous protein in the host. The commonly used concentration of IPTG is 0.1–1.0 mmol/l while a higher concentration of IPTG can increase the cost and inhibit bacterial growth (14). Therefore, minimal IPTG should be used under the prerequisite that there is no effect on the expression of protein. Our primary study indicated that 0.1 mM IPTG could achieve ideal expression of protein while a higher concentration did not increase the expression but decreased the bacterial sum. Therefore, we set the IPTG concentration to 0.1 mM. In addition, the induction time is also critical because earlier induction can inhibit the growth of bacteria while later induction results in aging bacteria and is bad for expression. We found that induction after 3 h is the best time for ideal bacterial production and protein expression. The fermentation of E. coli at a high density requires reduction of the production of acetic acid which can inhibit the growth and expression of bacteria, and the glucose can be replaced by glycerol which produces less acetic acid (15). Under our current condition, the final total expression of MuR6-TR was high (80%) which is helpful for the next purification.
In bacteria, there are nucleic acid, polysaccharose and other proteins which are existent in complex forms with the expressed targeting proteins. Therefore, extraction and purification of the targeting protein is critical. The MuR6-TR protein has an isoelectric point at 9.96 but no disulfide bond, is lowly adhesive to regular purifying gels under low-salt condition and resistant to high-salt (11). While high ion intensity under high-salt condition can deviate the practical pI from theoretical pI, which is helpful to explore ideal condition for ion exchange and eluting to obtain targeting proteins with higher concentration and purity. For the safety of animals in subsequent experiments, it is necessary to remove the pyrogen which is a metabolic endotoxin produced by microbes. The first step of cation-exchange is mainly to elute the targeting protein from the added samples containing many unspecific proteins which are mostly negative-charged. Therefore, it is necessary to perform the second step of anion-exchange for purification to obtain targeting protein with high purity. The western blot result indicated that the purified protein after two-step elution in the present study had the antigenic determinant and was suitable for the next experiment in animals.
TRAIL mainly exhibits its effect by binding to superficial DR4 and DR5 on the cell membrane. In pancreatic tumor tissues, there is universal expression of TRAIL receptor which plays an important role in modulating the apoptosis of pancreatic carcinoma and there is differential expression of the TRAIL receptor. For example, DR4 and DR5 are expressed much higher in pancreatic tumor tissues than pancreatic normal tissues. The expression of DR5 in pancreatic carcinoma tissue is related to the differentiation degree and malignant degree of tumor tissues while the expression of DR4 and decoy receptor 1 and 2 (DcR1, DcR2) in pancreatic carcinoma tissue is not related with the differentiation degree and clinical stage (16,17). According to a review by Di Pietro and Zauli (6), Apo2L/TRAIL was sensitive to 61 of 92 studied primary or passaged tumor cell lines, having a sensitivity rate of 66.3% and a resistance rate of 33.7%. Other studies indicated that approximately 50% tumor cell lines are resistant to TRAIL (18,19). In the present study, both TRAIL-insensitive pancreatic carcinoma cell lines BxPC-3 and PANC-1 showed a higher growth inhibition rate and lower IC50 with MuR6-TR than natural TRAIL, suggesting that the mutant protein with N-terminal CCPs had an in vitro advantage in inhibiting the growth of pancreatic carcinoma.
As a platform to mimic the intrinsic environment of the body, animal experiments can primarily explore the effect and safety of novel drugs in the body. According to previous studies, the TRAIL receptor agonists (including recombinant soluble TRAIL) and the monoclonal antibodies against TRAIL-R1 and TRAIL-R2 which are specific for TRAIL-induced apoptosis have been evaluated in the early stage of clinical experiments in hematologic and solid tumors including pancreatic carcinoma (20,21). However, most pancreatic carcinoma cell lines showed low sensitivity to apoptosis induced by TRAIL although they express basic signaling molecules of the TRAIL system (22). According to the in vitro experiment, we selected the pancreatic carcinoma cell line PANC-1 which is resistant to natural TRAIL but sensitive to MuR6-TR. The present study indicated that MuR6-TR showed an antitumor effect in inhibiting the growth of implanted pancreatic tumors in nude mice. Although lower than gemcitabine, MuR6-TR demonstrated a more effective effect than natural TRAIL protein in inhibiting the growth of tumors. There are several explanations for the lower effect of MuR6-TR than gemcitabine. Firstly, the mutant MuR6-TR had a higher therapeutic effect but a shorter time of bioactivity in the body, which is supported by the finding that MuR6-TR had a similar effect with gemcitabine during the first 4–7 days. We will conduct continuous administration of the drug and to change the frequency in future experiments to observe the antitumor effect. Secondly, the optimal dosage for the mutant protein requires further investigation. Notably, the IC50 of MuR6-TR was much lower than TRAIL in the in vitro experiment and the lower dosage MuR6-TR demonstrated a similar effect with gemcitabine in vivo at 4–7 days. Therefore, we can increase the dosage to enhance the antitumor activity after determining the safe dosage of the drug. Thirdly, the in vitro sensitivity of cell lines to mutant protein may not represent the in vivo sensitivity. It is possible to combine current first-line clinical medicines to investigate the potential antitumor effects.
During the selection of the drug dosage and administration frequency, we considered the tolerance of the mice, the convenience for patients in future clinical experiments and the quantity of MuR6-TR entering cells in order to prolong its half-life period in the body. These considerations approached the scheme of q.d. × 5 days which will be further improved in future experiments. Moreover, 2 of 8 mice (25%) died during the early stage of the experiment (day 1–4 after administration of MuR6-TR); however, the mice did not die immediately after the administration of MuR6-TR. Thus, it was not due to an acute allergic reaction. We tried to ascertain the reasons of the death by dissection of the two dead mice. We identified a large area of white necrotic foci in the livers of the dead mice. Thus, we diagnosed liver toxicity. We will add a toxicity test in future reseach. In addition, a higher requirement for the purification protocol in the future is needed. On the other hand, the mortality rate in the gemcitabine group was the same at 25%, suggesting that the dosage of gemcitabine should be adjusted in future experiments.
In summary, the present study mutated TRAIL with CPPs at the N-terminal and demonstrated that the mutant MuR6-TR had improved antitumor effects both in vitro and in vivo compared to the natural TRAIL. The mechanism was not explored in this study. However, since the structure of mutant MuR6-TR was similar to natural TRAIL, it is likely to also display its antitumor effects by binding to the TRAIL receptors on the tumor cell membrane. There was another TRAIL mutant membrane penetrating peptide alike (TMPPA) which showed significantly stronger affinity to the cancer cell membrane compared with natural TRAIL (data not shown). Thus, we believe that TMPPA enhances the affinity to the cancer cell membrane. Then, it massively aggregates on the cancer cell membrane and increases the signal transduction finally increasing the antitumor effects. Nevertheless, the therapeutic effect of MuR6-TR and the detailed mechanism warrant further research.
Acknowledgments
This study was supported by grants from the National Natural Scientific Foundation of China (nos. 81301962 and 81372444).
References
- 1.Singh D, Upadhyay G, Srivastava RK, Shankar S. Recent advances in pancreatic cancer: Biology, treatment, and prevention. Biochim Biophys Acta. 2015;1856:13–27. doi: 10.1016/j.bbcan.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/S1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 4.Arlt A, Müerköster SS, Schäfer H. Targeting apoptosis pathways in pancreatic cancer. Cancer Lett. 2013;332:346–358. doi: 10.1016/j.canlet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 6.Di Pietro R, Zauli G. Emerging non-apoptotic functions of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/Apo2L. J Cell Physiol. 2004;201:331–340. doi: 10.1002/jcp.20099. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Zeng F, Zhang M, Huang F, Wang J, Guo J, Liu C, Wang H. Emerging landscape of cell penetrating peptide in reprogramming and gene editing. J Control Release. 2016;226:124–137. doi: 10.1016/j.jconrel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Joseph SC, Blackman BA, Kelly ML, Phillips M, Beaury MW, Martinez I, Parronchi CJ, Bitsaktsis C, Blake AD, Sabatino D. Synthesis, characterization, and biological activity of poly(arginine)-derived cancer-targeting peptides in HepG2 liver cancer cells. J Pept Sci. 2014;20:736–745. doi: 10.1002/psc.2665. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Zhang N, Zhao T, Liu M, Zhang T, Li D. Inhibition of tumor growth by polyarginine-fused mutant cytosine deaminase. Appl Biochem Biotechnol. 2015;175:1633–1643. doi: 10.1007/s12010-014-1389-x. [DOI] [PubMed] [Google Scholar]
- 10.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 11.Cha SS, Song YL, Oh BH. Specificity of molecular recognition learned from the crystal structures of TRAIL and the TRAIL:sDR5 complex. Vitam Horm. 2004;67:1–17. doi: 10.1016/S0083-6729(04)67001-4. [DOI] [PubMed] [Google Scholar]
- 12.Alhakamy NA, Dhar P, Berkland CJ. Charge Type, Charge Spacing, and Hydrophobicity of Arginine-Rich Cell-Penetrating Peptides Dictate Gene Transfection. Mol Pharm. 2016;13:1047–1057. doi: 10.1021/acs.molpharmaceut.5b00871. [DOI] [PubMed] [Google Scholar]
- 13.Fujita T, Furuhata M, Hattori Y, Kawakami H, Toma K, Maitani Y. High gene delivery in tumor by intratumoral injection of tetraarginine-PEG lipid-coated protamine/DNA. J Control Release. 2008;129:124–127. doi: 10.1016/j.jconrel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Papaneophytou CP, Kontopidis GA. Optimization of TNF-α overexpression in Escherichia coli using response surface methodology: Purification of the protein and oligomerization studies. Protein Expr Purif. 2012;86:35–44. doi: 10.1016/j.pep.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24:530–536. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Ozawa F, Friess H, Kleeff J, Xu ZW, Zimmermann A, Sheikh MS, Büchler MW. Effects and expression of TRAIL and its apoptosis-promoting receptors in human pancreatic cancer. Cancer Lett. 2001;163:71–81. doi: 10.1016/S0304-3835(00)00660-1. [DOI] [PubMed] [Google Scholar]
- 17.Stadel D, Mohr A, Ref C, MacFarlane M, Zhou S, Humphreys R, Bachem M, Cohen G, Möller P, Zwacka RM, et al. TRAIL-induced apoptosis is preferentially mediated via TRAIL receptor 1 in pancreatic carcinoma cells and profoundly enhanced by XIAP inhibitors. Clin Cancer Res. 2010;16:5734–5749. doi: 10.1158/1078-0432.CCR-10-0985. [DOI] [PubMed] [Google Scholar]
- 18.Stuckey DW, Shah K. TRAIL on trial: Preclinical advances in cancer therapy. Trends Mol Med. 2013;19:685–694. doi: 10.1016/j.molmed.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maksimovic-Ivanic D, Stosic-Grujicic S, Nicoletti F, Mijatovic S. Resistance to TRAIL and how to surmount it. Immunol Res. 2012;52:157–168. doi: 10.1007/s12026-012-8284-8. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys RC, Halpern W. Trail receptors: Targets for cancer therapy. Adv Exp Med Biol. 2008;615:127–158. doi: 10.1007/978-1-4020-6554-5_7. [DOI] [PubMed] [Google Scholar]
- 21.Ashkenazi A, Holland P, Eckhardt SG. Ligand-based targeting of apoptosis in cancer: The potential of recombinant human apoptosis ligand 2/Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL) J Clin Oncol. 2008;26:3621–3630. doi: 10.1200/JCO.2007.15.7198. [DOI] [PubMed] [Google Scholar]
- 22.Vogler M, Dürr K, Jovanovic M, Debatin KM, Fulda S. Regulation of TRAIL-induced apoptosis by XIAP in pancreatic carcinoma cells. Oncogene. 2007;26:248–257. doi: 10.1038/sj.onc.1209776. [DOI] [PubMed] [Google Scholar]