Abstract
The aberrant expression of miR-30d has been reported in several types of human malignancies. However, its biological function in colorectal cancer (CRC) remains largely unknown. In this study, we identified that miR-30d was significantly downregulated in CRC tissues compared to that observed in normal controls as detected by RT-qPCR analysis. Downregulation of miR-30d was significantly associated with aggressive clinicopathological parameters including tumor differentiation, invasive depth, TNM stage, lymph node metastasis, distant metastasis, and poor prognosis. Furthermore, functional analysis revealed that overexpression of miR-30d significantly inhibited cell proliferation, caused cell cycle arrest at the G0/G1 phase, suppressed cell migration and invasion, induced cell apoptosis in vitro, and decreased tumor growth in a xenograft mouse model. Bioinformatic analysis and dual-luciferase reporter assay revealed that liver receptor homologue-1 (LRH-1) is a direct target of miR-30d in CRC cells. Rescue assay showed that LRH-1 overexpression could restore the inhibitory effect of miR-30d on CRC cells. In addition, miR-30d overexpression suppressed the activation of key components of the Wnt/β-catenin signaling pathway, β-catenin, c-Myc and cyclin D1, which contributed to the inhibition of CRC development. Thus, our findings suggest that miR-30d functions as a tumor suppressor against CRC development and miR-30d/LRH-1/Wnt signaling may be novel potential targets for CRC treatment.
Keywords: miR-30d, liver receptor homologue-1, colorectal carcinoma, Wnt signaling, cell proliferation, cell invasion
Introduction
Colorectal cancer (CRC), one of the most common malignancies worldwide, is regarded as the second cause of cancer-related deaths each year (1). The incidence of CRC has rapidly increased in China among both men and women (2). Although advances in treatment strategy and extensive investigation of CRC have been accomplished in the past decade, the mortality rate remains high due to the metastasis-associated poor prognosis (3). Therefore, in order to improve the prognosis and reduce the rate of recurrence of CRC patients, the search for novel potential biomarkers and efficient targets for CRC treatment is a crucial goal.
MicroRNAs (miRNAs or miRs), small non-coding RNA molecules approximately 18–25 nucleotides in length, are highly conserved in most eukaryotic organisms (4). Through binding to the 3′-untranslated regions (3′-UTRs) of their corresponding messenger RNA (mRNA) targets, miRNAs degrade or inhibit the posttranscriptional regulation of their (mRNA) targets (5). miRNAs have been reported to be involved in multiple physiological and pathological processes including human malignancies (6,7). Many studies have confirmed that miRNAs can be identified as oncogenes or tumor suppressors in the initiation and development of cancers by targeting corresponding abnormally expressed proteins (8). Consequently, miRNAs have been proposed to be potential indicators and therapeutic targets in various types of cancers (9,10).
miR-30d is a member of the miR-30 family that is responsible for tumor development and progression. Recently, accumulating evidence has shown that miR-30d plays a role as either a tumor suppressor or an oncogene in the progression of different tumor types (11). Zhang et al found that dysregulation of miR-30d in human anaplastic thyroid carcinoma resulted in the insensitivity to chemotherapeutic drugs by enhancing autophagic survival (12). Xuan et al demonstrated that down-regulation of miR-30d resulted in the pathogenesis of prostate cancer by targeting B-cell specific moloney leukemia virus insertion region homologue-1 (Bmi-1) (13). Ye et al indicated that overexpression of miR-30d blocked transforming growth factor-β (TGF-β1)-induced epithelial-mesenchymal transition (EMT) by targeting Snail in ovarian cancer cells (14). However, the exact role and molecular mechanisms underlying the regulation of the progression of CRC tumorgenesis by miR-30d remains largely unknown.
Liver receptor homologue-1 (LRH-1), also known as NR5A2, is a member of the nuclear receptor (NR) subfamily (15). This NR participates in a variety of biological processes, such as differentiation and development, reverse cholesterol transport, bile-acid homeostasis and steroidogenesis (16,17). Evidence indicates that LRH-1 is responsible for the pathogenesis of multiple types of tumors (18). Recent studies have demonstrated high LRH-1 expression in pancreatic cancer cells and have found that LRH-1 overexpression enhances cell migration and invasion abilities (19). In addition, LRH-1 contributes to intestinal tumor proliferation in gastrointestinal tumors by activating the Wnt/β-catenin pathway (20,21). These findings indicate that LRH-1 can be involved in the development of cancers.
In the present study, we found that miR-30d was down-regulated in CRC tissue samples and a decreased expression level of miR-30d was closely correlated to clinicopathological parameters including poor prognosis, the degree of differentiation, invasive depth, TNM stage, distant metastasis, and lymph node metastasis. LRH-1 was identified as a direct target of miR-30d as detected by the LRH-1 3′-UTR region and luciferase report assay. In addition, ectopic expression of miR-30d inhibited CRC cell proliferation and invasion, as well as the Wnt/β-catenin signaling pathway by suppressing the expression of LRH-1. Therefore, the present study indicates that miR-30d may be a potential prognostic marker for CRC.
Materials and methods
Clinical sample collection
Fresh CRC tissue samples and paired adjacent normal samples were obtained from 80 patients who underwent routine surgery at the Department of General Surgery, Shaanxi Province People's Hospital (Xi'an, China) from March 2005 to July 2010. Patients who underwent radiotherapy or chemotherapy prior to surgery were excluded. All patients involved were divided into two groups according to the median RNA level of miR-30d (high and low). A five-year follow-up was performed to record the difference in prognosis between the low and high miR-30d groups. Informed consent was provided by all patients, and the present study was approved by the Human Ethics Committee of Shaanxi Provincial People's Hospital (Xi'an, China) and the 1964 Helsinki Declaration.
Cell culture
The human CRC cell lines (LoVo, HT-29, RKO, HCT8 and SW480), the normal colorectal mucosa cell line FHC, and the 293T cell line were purchased from the Cell Center of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FBS) (Gibco BRL, Gaithersburg, MD, USA) and 1% penicillin/streptomycin in a humidified incubator containing 5% CO2 at 37°C.
Cell transfection
The miR-30d mimic (miR-30d) and miR-30d mimic NC (miR-NC) were purchased from GenePharma Company (Shanghai, China). LoVo and SW480 cells were selected and seeded into 6-well plates at the density of 3×105 cells/well and transfected with 100 nM oligonucleotides using Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol.
Isolation of total RNA and quantitative RT-PCR
Total RNA or miRNA from the CRC tumor samples and cell lines was extracted using the RNeasy Mini kit or miRNeasy Mini kit (Qiagen, Manchester, UK) according to the manufacturer's instructions. cDNA was synthesized from 10 µg RNA using M-MLV reverse transcriptase (Clontech Laboratories, Mountain View, CA, USA) or miScript reverse transcription kit (Qiagen). Real-time polymerase chain reaction (PCR) was performed with SYBR Premix Ex Taq (Takara Bio, Dalian, China) to examine the mRNA or miRNA level. GAPDH and U6 were used for normalization. The primers for LRH-1 and GAPDH were designed as follows: LRH-1 forward, 5′-CCCAGCATTTTAACACTACCGA-3′ and reverse 5′-GGGCAGGGGAAAGACAACTAA-3′; GAPDH forward, 5′-AAGCGTTGTCCTTACTGTCGT-3′ and reverse 5′-GTCGGTAGTGTTAAAATGCTGGG-3′. ABI 7900 Fast Real-time PCR system (ABI, USA) was used to perform RT-qPCR. The 2−ΔΔCt method was used to analyze the relative gene expression level.
Western blot analysis
CRC tissue samples and cells were washed with ice-cold phosphate-buffered saline (PBS) 3 times and lysed in RIPA buffer supplemented with protease inhibitor. The protein concentration was determined using the bicinchoninic acid method. Equal amounts of proteins (30 µg) were separated by 15% SDS-PAGE and transferred to a PVDF membrane (Immobilon; Millipore Corp., Bedford, MA, USA). After blocking with 5% non-fat milk (diluted with phosphate-buffered saline/Tween-20, TBST) at 37°C for 1 h, the membranes were incubated with primary antibodies: anti-LRH-1 (1:1000, ab41901), anti-matrix metalloproteinase (MMP)2 (1:1,000, ab80737), anti-MMP9 (1:1,000, ab119906), anti-β-catenin (1:500, ab92514), cyclin D1 (1:1,000, ab134175), c-Myc (1:500, ab51156) and GAPDH (1:1,000, ab8245; Abcam, Cambridge, UK) at 4°C overnight. After rinsing with TBST, they were incubated with horseradish peroxidase-conjugated goat anti-mouse or rabbit immunoglobulin G antibodies (1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Protein bands were detected using an enhanced chemiluminescence reagent (Pierce Biotechnology, Inc., Rockford, IL, USA). Band intensity was quantified using Image software (Media Cybernetics, Inc., Rockville, MD, USA).
Cell proliferation assay
3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was used to assess the CRC cell proliferation. Firstly, CRC cells were seeded into a 96-well plate at a density of 2×103 cells/well and transfected with miR-30d or miR-NC for 96 h. Fresh medium with 20 µl MTT (5 mg/ml) was added into each well to replace the old medium and incubation was carried out for another 4 h at 37°C. Then the medium was removed and 150 µl DMSO was added into each well. The optical density was recorded at a wavelength of 490 nm using a microplate reader (Multiskan FC; Thermofisher, Finland).
Cell cycle analysis
Cell cycle distribution was analyzed by flow cytometry as previously reported (22). Briefly, cells trans-fected with miR-30d or miR-NC for 48 h were resuspended and fixed with 70% ice-cold ethanol at -20°C overnight. Afterwards, the cells were washed with PBS and stained with propidium iodide (PI)/RNase fluorescent probe solution (Molecular Probes, Eugene, OR, USA) for 30 min in the dark. DNA contents were assessed with a FACSCalibur flow cytometer (Becton-Dickinson, San Diego, CA, USA).
Apoptosis assay
Apoptosis was measured using the Annexin V-FITC/PI apoptosis kit (Abcam, Cambridge, UK) according to the manufacturer's protocol (22). Cells were collected after transfection with miR-30d or miR-NC for 48 h, washed twice with cold PBS, and resuspended at 1×105 cells/ml and mixed with a binding buffer containing Annexin V-FITC and PI. After incubation for 15 min in the dark, the apoptotic cells were analyzed using a FACSCalibur flow cytometry.
Migration and invasion assays
Cells were pretreated with miR-30d or miR-NC for 48 h. For the migration assay, 1×104 cells in serum-free medium were added to the upper chamber of each transwell (Millipore Corp.) with a non-coated membrane. For the invasion assay, 5×104 cells were added on the upper chamber of each insert coated with Matrigel (Becton-Dickinson). Medium (500 µl) containing 10% FBS as the chemotactic factor was added to the lower chambers. After 24 h of incubation for the migration assay and 48 h of incubation for the invasion assay at 37°C, the cells remaining on the upper membrane of the filter were carefully removed with cotton swabs. Cells that had translocated to the lower chambers were fixed with methanol and stained using 0.1% crystal violet. The migratory or invasive cells were evaluated and imaged by an IX71 inverted microscope (Olympus, Tokyo, Japan).
miRNA target prediction and dual-luciferase reporter assay
The potential targets of miR-30d were predicted using three online prediction programs: microRNA.org (http://www.microrna.org/microrna/home.do), PicTar (http://www.pictar.org/cgi-bin/PicTar_vertebrate.cgi) and TargetScan (http://www.targetscan.org/vert_60/). Based on the prediction results, the 3′-UTR of LRH-1 containing the miR-30d putative binding site and mutant site were designed and produced by GenePharma (Shanghai, China), and subcloned into the pGL3 luciferase vector (Promega Corp., Madison, WI, USA) to construct a 3′-UTR-LRH1-wild-type and mutant 3′-UTR-LRH1. Human 293T cells were seeded into a 24-well plate and co-transfected with miR-30d or miR-NC and LRH-1 WT-3′-UTR or the mutant 3′-UTR vector for 48 h. The cells were then harvested and luciferase activity was assessed using the dual-luciferase reporter assay system (Promega, USA). Renilla luciferase activity was used to normalize the luciferase activity.
Xenograft experiment
Four-week-old BALB/c male nude mice were obtained from the Animal Center of the Fourth Military Medical University (Xi'an, China) and divided into three groups (control, miR-30d and miR-NC, n=10 in each group). SW480 cells with miR-30d or miR-NC transfection were suspended in PBS and subcutaneously injected into each mouse at a total of 5×106 cells. Tumor growth was monitored every week after tumor cell injection. After 7 weeks, all animals were euthanized by cervical dislocation and xenograft tumor volumes were measured using the formula: Volume = 1/2 (length × weight)2. All the animal experimental procedures were approved by the Animal Care and Use Committee of Shaanxi Province People's Hospital (Xi'an, China).
Statistical analysis
All statistical analyses were performed using SPSS 13.0 software. The data are presented as the mean ± standard deviation (SD). Student's t-test was used for comparisons between two groups, whereas more than two groups were compared by one-way analysis of variance followed by Bonferroni test. Pearson correlation was used to identify the correlation between the expression of miR-30d and LRH-1 (R=correlation coefficient). P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least in triplicate.
Results
miR-30d is downregulated and LRH-1 is upregulated in human CRC tissues
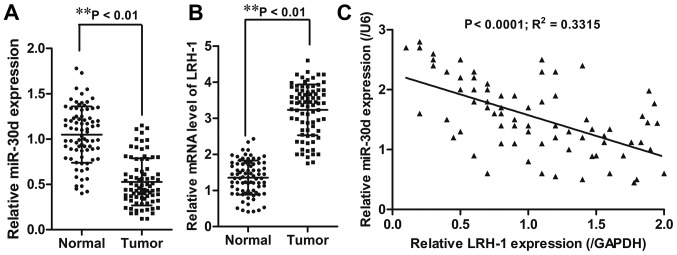
First, the expression level of miR-30d was detected in CRC tissue samples and paired adjacent tissues (n=80) using RT-qPCR. The results showed that CRC tissues had significantly reduced miR-30d mRNA expression (P<0.01) and increased LRH-1 mRNA expression (P<0.01) when compared to the adjacent normal tissues (Fig. 1A and B). Pearson's correlation analyses showed that miR-30d expression was inversely correlated with LRH-1 expression (R2=0.3315; P<0.0001) in the CRC tissue samples (Fig. 1C).
Figure 1.
miR-30d is downregulated and LRH-1 is upregulated in CRC cell tissue. Relative expression levels of (A) miR-30d and (B) LRH-1 in 80 CRC tissue samples and paired adjacent normal tissues were detected by RT-qPCR. (C) Correlation between the expression of miR-30d and LRH-1 in CRC tissues was analyzed by Pearson correlation coefficient test. Each experiment was repeated at least three times. Data are expressed as the mean ± SD. **P<0.01 vs. normal tissues. LRH-1, liver receptor homologue-1; CRC, colorectal cancer.
Downregulation of miR-30d and upregulation of LRH-1 is associated with aggressiveness of CRC progression
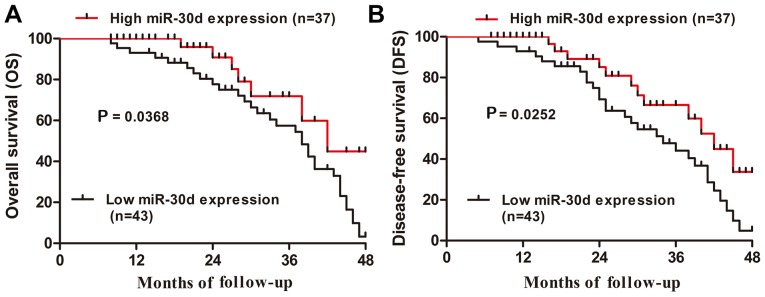
To further verify the significance of aberrant expression of miR-30d and LRH-1 in CRC tissue specimens, clinicopathological analysis was performed accordingly. Based on the median levels (0.527, miR-30d; 3.232, LRH-1) in CRC tissue specimens, all 80 CRC patients were subdivided into a low miR-30d expression group (<0.527, 43 cases), a high miR-30d expression group (>0.527, 37 cases) as well as a high LRH-1 expression group (>3.232, 51 cases) and a low LRH-1 expression group (<3.232, 29 cases). The correlation between miR-30d expression and the clinicopathological factors of the patients was determined. As shown in Table I, the clinicopathological analysis illustrated that downregulation of miR-30d was significantly associated with distant metastasis (P<0.05), differentiation (P<0.05), invasive depth (P<0.05), TNM stage (P<0.05), and lymph node metastasis (P<0.05); whereas, upregulation of LRH-1 was markedly correlated with distant metastasis (P<0.05), TNM stage (P<0.05), invasive depth (P<0.05), and lymph node metastasis (P<0.05), but had no correlation with age, gender or tumor size (P>0.05). Furthermore, Kaplan-Meier analysis revealed that CRC patients with low miR-30d expression had markedly shorter overall survival and disease-free survival rates than those in the high miR-30d expression group (P=0.0368 and P=0.0252, respectively; Fig. 2A and B). Thus, these data indicated that downregulation of miR-30d may be involved in the development of CRC.
Table I.
Correlation between miR-30d expression and clinicopathologic parameters in the patients with CRC.
| Parameters | No. of cases | miR-30d expression
|
P-value | LRH-1 expression
|
P-value | ||
|---|---|---|---|---|---|---|---|
| High (37) | Low (43) | Low (29) | High (51) | ||||
| Age (years) | 0.071 | 0.169 | |||||
| >60 | 44 | 16 | 28 | 19 | 25 | ||
| ≤60 | 36 | 21 | 15 | 10 | 26 | ||
| Gender | 0.262 | 0.486 | |||||
| Male | 41 | 16 | 25 | 13 | 28 | ||
| Female | 39 | 21 | 18 | 16 | 23 | ||
| Distant metastasis | <0.05a | <0.05a | |||||
| M0 | 46 | 30 | 16 | 25 | 21 | ||
| M1 | 34 | 14 | 20 | 10 | 24 | ||
| TNM stage | <0.01b | <0.05a | |||||
| I–II | 34 | 22 | 12 | 7 | 27 | ||
| III–IV | 46 | 15 | 31 | 22 | 24 | ||
| Degree of differentiation | <0.05a | 0.345 | |||||
| Well/moderate | 27 | 17 | 10 | 14 | 13 | ||
| Poor | 53 | 20 | 33 | 21 | 32 | ||
| Invasive depth | <0.05a | <0.05a | |||||
| T1-T2 | 32 | 20 | 12 | 17 | 15 | ||
| T3-T4 | 48 | 17 | 31 | 12 | 36 | ||
| Lymph node metastasis | <0.01b | <0.05a | |||||
| Yes | 36 | 25 | 11 | 18 | 18 | ||
| No | 44 | 12 | 32 | 11 | 33 | ||
| Tumor size (cm) | 0.262 | 0.647 | |||||
| >5 | 41 | 19 | 22 | 16 | 25 | ||
| ≤5 | 39 | 13 | 26 | 13 | 26 | ||
Statistically significant difference;
highly significant difference. CRC, colorectal cancer; LRH-1, liver receptor homologue-1.
Figure 2.
Downregulation of miR-30d is associated with poor prognosis in CRC patients. (A) Overall survival and (B) disease-free survival of CRC patients with low and high miR-30d expression were assessed by Kaplan-Meier method. *P<0.05, vs. the high miR-30d expression group. CRC, colorectal cancer.
Overexpression of miR-30d suppresses CRC cell proliferation and invasion
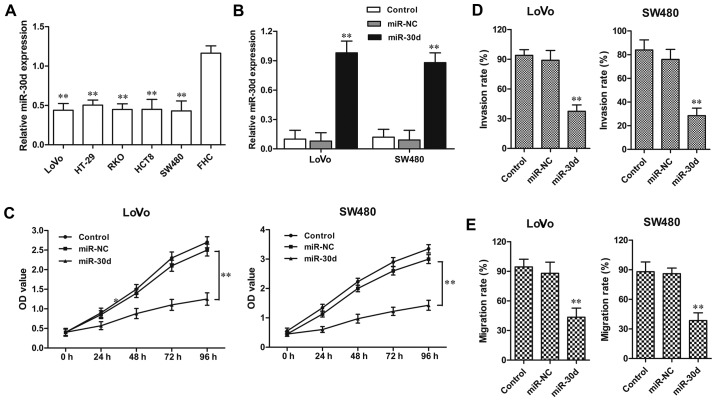
The miR-30d expression level was detected in four CRC cell lines. Consistent with the tissue sample results, the miR-30d expression level was also distinctly downregulated in the CRC HT29, LoVo, RKO, HCT8, SW480 cell lines when compared to the level in the normal colorectal cell line FHC (Fig. 3A). Then LoVo and SW480 CRC cell lines were randomly selected for functional analysis in the following experiments. miR-30d mimic or miR-NC was transfected into the LoVo and SW480 cells for 48 h, respectively. The efficiency of transfection was confirmed by RT-qPCR (Fig. 3B). MTT assay showed that in both the LoVo and SW620 cell lines, the cell proliferation was significantly inhibited by miR-30d overexpression when compared to the NC groups, respectively (Fig. 2C). In addition, the invasion assay demonstrated that the miR-30d mimic markedly suppressed the migratory and invasive abilities of the LoVo and SW480 cells when compared with the NC groups, respectively (Fig. 3D and E). These findings indicate that overexpression of miR-30d suppressed cell proliferation and invasion in CRC cells.
Figure 3.
miR-30d overexpression suppresses CRC cell proliferation and invasion. (A) Relative expression levels of miR-30d in CRC cell lines including LoVo, HT-29, RKO, HCT8 and SW480 and normal colorectal cell line FHC were determined by RT-qPCR. U6 and GAPDH were used for normalization. (B) Relative expression level of miR-30d in LoVo and SW480 cell lines with miR-30d mimics or miR-NC transfection were determined by RT-qPCR. U6 was used for normalization. (C) Cell proliferation of LoVo and SW480 cell lines with miR-30d mimic or miR-NC transfection was detected by MTT assay. (D) Cell invasion and (E) migration abilities of LoVo and SW480 cell lines with miR-30d mimic or miR-NC transfection were detected by invasion and migration assays. The invasion and migration results are expressed as a percentage of total seeded cells. Each experiment was repeated at least three times. Data are the mean ± SD. **P<0.01 vs. the miR-NC group. CRC, colorectal cancer
Upregulation of miR-30d causes cell cycle arrest and induces cell apoptosis in CRC cells
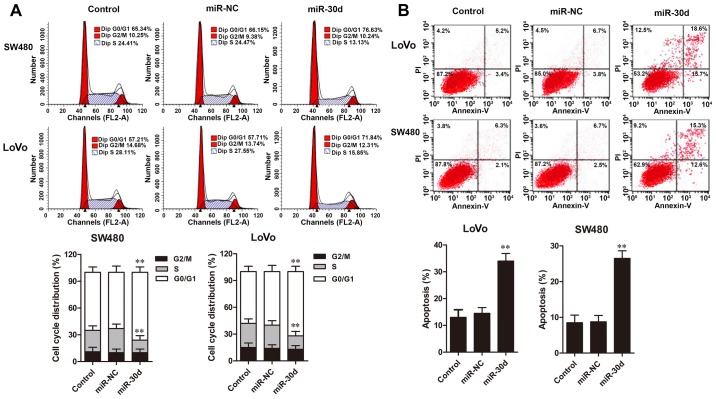
Cell cycle distribution and apoptosis of CRC cells were assessed by flow cytometric analysis. The results showed that overexpression of miR-30d in LoVo and SW480 cells resulted in significant cell accumulation in the G0/G1 phase and a decreased percentage of cells in the S phase when compared to the NC groups (Fig. 4A). Furthermore, flow cytometric analysis revealed that cell apoptosis rates in the LoVo and SW480 cells with miR-30d mimic transfection were significantly promoted compared with the rates noted in the NC groups (Fig. 4B). Taken together, these results demonstrated that upregulation of miR-30d caused cell cycle arrest at the G0/G1 phase and induced cell apoptosis in the CRC cells.
Figure 4.
miR-30d overexpression causes cell cycle arrest at the G0/G1 phase and induces cell apoptosis. (A) Cell cycle distribution and (B) cell apoptosis of LoVo and SW480 cell lines with miR-30d mimics or miR-NC transfection were analyzed by flow cytometry. Each experiment was repeated at least three times. Data are expressed as the mean ± SD. **P<0.01 vs.the miR-NC group.
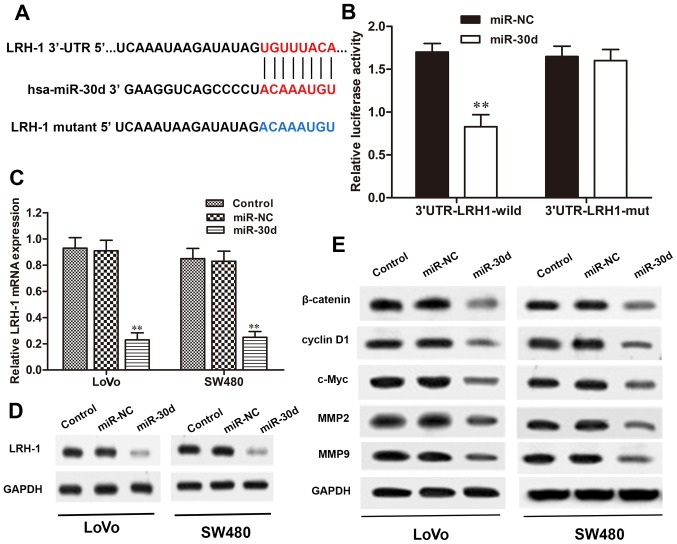
miR-30d directly targets the 3′-UTR of LRH-1 in CRC cells
To verify whether LRH-1 is a direct target of miR-30d, we searched for potential targets of miR-30d using microRNA. org, PicTar, and TargetScan. The binding regions between the miR-30d and LRH-1 3′-UTR were found (Fig. 5A), indicating that LRH-1 may be a potential target of miR-30d. To validate that miR-30d directly targets the 3′-UTR region of LRH-1, a dual-luciferase reporter assay was performed. When 293T cells were transfected with the wild-type LRH-1 3′-UTR vector, the luciferase activity was significantly reduced in the miR-30d mimic co-transfected cells when compared with the NC group (P<0.01). Conversely, the mutant 3′-UTR of LRH-1 caused no significant changes in the luciferase activity (Fig. 5B). We next investigated the interaction between miR-30d and LRH-1 in the LoVo and SW480 cells. As shown in Fig. 5C and D, overexpression of miR-30d significantly reduced LRH-1 expression both at the mRNA and protein level in the LoVo and SW480 cells with miR30d mimic transfection, but not in the miR-NC-transfected cells. These results revealed that miR-30d directly binds to the site of the 3′-UTR region of LRH-1 and regulated LRH-1 expression in the CRC cells.
Figure 5.
LRH-1 is a direct target of miR-30d and regulates Wnt/β-catenin signaling. (A) Bioinformatic analysis of the predicted binding site of miR-30d to the 3′-UTR of LRH-1. (B) Relative luciferase activities of 293T cells co-transfected with miR-30d and wild-type or mutant LRH-1 3′-UTR vector were detected by dual-luciferase reporter assay. The expression levels of LRH-1 in LoVo and SW480 cells with miR-30d transfection were determined by (C) RT-qPCR and (D) western blot analysis. (E) The expression levels of components of Wnt/β-catenin signaling in LoVo and SW480 cells with miR-30d transfection were measured by western blot analysis. GAPDH was used as an internal control. Each experiment was repeated at least three times. Data are expressed as the mean ± SD. **P<0.01 vs. the miR-NC group. LRH-1, liver receptor homologue-1; UTR, untranslated region.
miR-30d regulates the Wnt/β-catenin signaling pathway through LRH-1
It has been reported that miRNAs play crucial roles in regulating tumor cell proliferation, differentiation, and invasion through various signaling pathways, including the Wnt/β-catenin pathway (23). Here, to determine the signaling pathway involved in the regulatory effects of miR-30d on CRC, the Wnt/β-catenin signaling pathway was detected in LoVo and SW480 cells transfected with the miR-30d mimic by western blot analysis. The results showed that the expression levels of Wnt signaling-associated proteins, β-catenin, cyclin D1, c-Myc, MMP2 and MMP9, were markedly reduced in the miR-30d mimic group compared to the levels noted in the NC group (Fig. 5E). Taken together, we conclude that overexpression of miR-30d suppressed activation of the Wnt/β-catenin signaling pathway by targeting LRH-1 to inhibit CRC tumorgenesis.
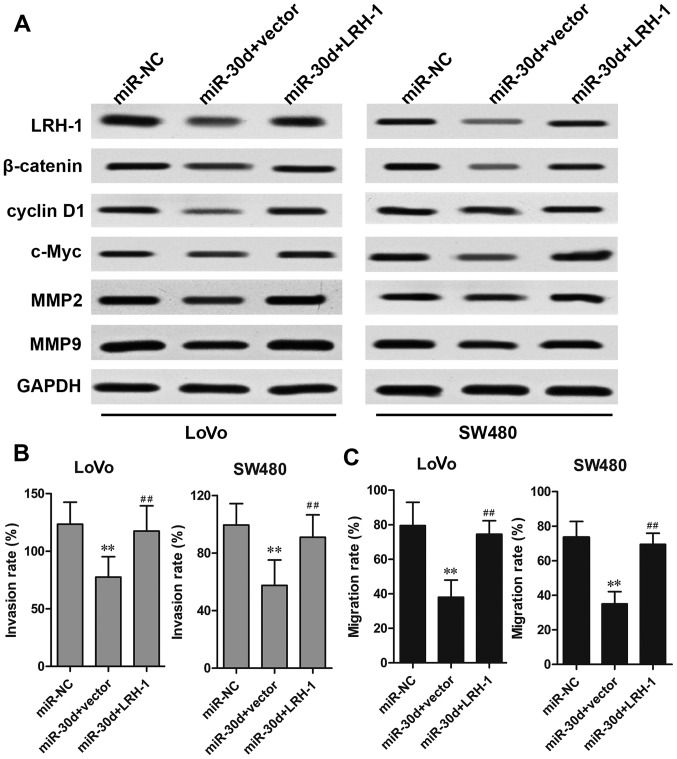
Overexpression of LRH-1 rescues the effects of miR-30d on CRC cells
To investigate the functional relevance of LRH-1 targeting by miR-30d, a rescue assay was performed to determine whether LRH-1 overexpression could rescue the suppressive effects induced by miR-30d. CRC cells were co-transfected with the miR-30d mimics and LRH-1-overexpressing plasmid. The results showed that the reduced protein expression caused by miR-30d mimics was markedly restored by transfection of the LRH-1 overexpressing plasmid in the LoVo and SW480 cells (Fig. 6A). Overexpression of LRH-1 also rescued the inhibitory effect of miR-30d on Wnt signaling-associated proteins, β-catenin, cyclin D1 and c-Myc. In addition, the inhibitory effects of miR-30d on CRC cell metastasis regulatory factors MMP2 and MMP9 were significantly reversed by LRH-1 overexpression, which was further confirmed by invasion and migration assays (Fig. 6B and C). These results indicated that miR-30d suppressed CRC tumorigenesis by inhibiting LRH-1 expression.
Figure 6.
Overexpression of LRH-1 rescues the suppressive effect of miR-30d overexpression. LoVo or SW480 cells were co-transfected with miR-30d mimics and pcDNA-LRH-1 vector or empty vector for 48 h. (A) The protein expression levels of LRH-1, β-catenin, cyclin D1, c-Myc, MMP2, and MMP9 in LoVo and SW480 cells were detected by western blot analysis. The (B) invasion and (C) migration abilities of CRC cells were detected by Transwell assays. The invasion and migration results are expressed as a percentage of total seeded cells. Data are expressed as the mean ± SD. **P<0.01 vs. the miR-NC group. ##P<0.01 vs. miR-30d. LRH-1, liver receptor homologue-1; MMP, matrix metalloproteinase.
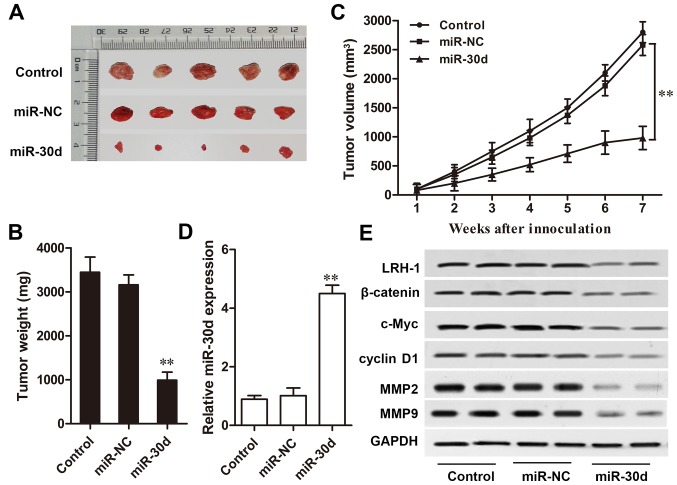
miR-30d inhibits CRC tumor growth in vivo
A CRC mouse model was designed to investigation the effect of miR-30d on CRC tumor growth in vivo. Compared with the NC group, mice injected with miR-30d mimic-transfected cells showed a markedly slow tumor growth rate during the 7-week detection (P<0.01, Fig. 7A–C). Furthermore, RT-qPCR and western blot analysis results showed that the expression level of miR-30d in the xenograft tumors derived from miR-30d mimic-transfected cells was markedly upregulated compared to that in the tumors derived from the miR-NC-transfected cells (P<0.01, Fig. 7D). Western blot analysis results also revealed that the expression levels of key components of the Wnt/β-catenin signaling pathway, β-catenin, c-Myc and cyclin D1; metastasis regulatory factors, MMP2 and MMP9; as well as LRH-1 were significantly reduced in the xenograft tumor tissues with miR-30d overexpression (Fig. 7E). These results indicated that miR-30d suppressed CRC tumor growth in vivo by targeting LRH-1 by regulating the Wnt/β-catenin signaling pathway.
Figure 7.
miR-30d overexpression inhibits CRC tumor growth in vivo. (A) Representative images of xenograft tumors. (B) Tumor weight in the different groups. (C) Growth curves of tumor volume in the xenograft tumors recorded every 7 days. (D) Relative miR-30d expression level in tumor tissue was detected by RT-qPCR. (E) Protein expression levels of LRH-1, β-catenin, cyclin D1, c-Myc, MMP2, and MMP9 were determined by western blot analysis. GAPDH was used as the internal control. Each experiment was repeated at least three times. Data are expressed as the mean ± SD. **P<0.01 vs. the miR-NC group. CRC, colorectal cancer; LRH-1, liver receptor homologue-1; MMP, matrix metalloproteinase.
Discussion
CRC is one of the most prevalent malignancies in the world, with high morbidity and mortality. Patients with advanced stage CRC have a poor survival rate (1). Searching for more efficient prognostic biomarkers for patients with poor prognosis and cancer metastasis is a critical issue for achieving better outcomes (24). Recently, focus on the deregulation of miRNAs during CRC progression has becoming a promising research field, which provides reliable information for the distinct understanding of molecular mechanisms underlying CRC tumorgenesis and development (25). Abnormal miRNA expression is responsible for alterations in CRC cell proliferation, invasion, apoptosis and migration (26). Several oncogenic or tumor-suppressive miRNAs such as miR-21, miR-182, miR-137, and miR-143 have been demonstrated to be associated with the initiation of CRC (27,28), Thus, further investigation of the miRNAs involved in the progression of CRC may contribute to improved diagnostics and more effective therapeutic strategies for CRC patients.
Recently, deregulated miR-30d has been shown to be involved in the development of various types of cancer. Yao et al reported that miR-30d overexpression enhanced tumor invasion and metastasis by targeting galphai 2 in hepatocellular carcinoma (29). Kobayashi et al found that miR-30d was upregulated in prostate cancer cells and appeared to be a potential prognostic marker of prostate progression (30). Moreover, Chen et al recently demonstrated that miR-30d was downregulated in non-small cell lung cancer tissues and serves as a tumor suppressor by inhibiting cell proliferation and motility by directly targeting cyclin E2 (11). Wu et al found that miR-30d induced the apoptosis of renal cell carcinoma through the Akt/FOXO signal transduction pathway (31). In the present study, to avoid the adverse influence of radiotherapy or chemotherapy on the molecular expression and distribution in the tumor tissues, which might indirectly mislead the research results in the study, 80 patients including stage IV patients with distant metastasis who received only routine curative surgery were enrolled. We found that miR-30d expression was significantly downregulated in the CRC tissue specimens and cell lines. Analysis of the clinicopathological parameters showed that the miR-30d expression level was closely associated with poor prognosis, tumor differentiation, invasive depth, lymph node metastasis, distant metastasis, and TNM stage. Functional studies also verified the inhibitory role of miR-30d on cell proliferation, invasion and migration, induction of cell cycle at the G0/G1 phase and increased apoptosis of CRC in vitro. Furthermore, we also found that miR-30d overexpression significantly suppressed tumor growth in vivo, which might suggest the crucial role of miR-30d as a tumor suppressor in CRC development and provide valuable evidence for the development of miR-30d as a novel therapeutic target for CRC treatment.
LRH-1 classified as an orphan NR, is critical to numerous biological processes and is involved in multiple types of cancer, including breast, pancreatic, gastrointestinal and colon cancer. Jiang et al demonstrated that in non-small cell lung cancer, LRH-1 was a direct target of miR-376c to suppress cell proliferation and invasion through the Wnt signaling pathway (32). Liang et al demonstrated that miR-381 promoted colon cell proliferation and growth via upregulation of its target gene LRH-1 (33). A recent investigation also illustrated that LRH-1 functions as a direct target gene of miR-381 in suppressing cell growth and metastasis in hepatocellular carcinoma (34). However, the relationship between miR-30d and LRH-1 has not been elucidated in CRC. Here, we demonstrated that the expression of LRH-1 was negatively correlated with miR-30d expression in CRC tumor tissues, and was verified as a direct target of miR-30d using luciferase reporter assay. Furthermore, restoring the expression of LRH-1 significantly rescued the inhibitory effect of miR-30d overexpression on CRC tumorigenesis. These results imply that miR-30d regulates the progression of CRC by directly binding to LRH-1.
Aberrant activation of the Wnt/β-catenin signaling pathway contributes to the development and progression of various cancers including CRC, and is thus regarded as a potential target in cancer treatment (35,36). The stabilization of β-catenin is the crucial component of the Wnt signaling pathway. During activation of Wnt signaling, the accumulation of β-catenin occurs in the cytoplasm, and then translocates to the nucleus to activate the transcription of Wnt target genes, such as cyclin D1 and c-Myc (37,38). It has been reported that LRH-1 functions as a coactivator of the Wnt signaling pathway during cancer progressions, while c-Myc is required for cell proliferation, which is also shown to be controlled by LRH-1 (23). For example, by upregulating cyclin D1/E1 and c-Myc genes, LRH-1 promoted cancer cell proliferation and tumor growth in pancreatic cancer (19). Furthermore, overexpression of LRH-1 also significantly enhanced cancer cell migration and invasion by upregulating its downstream target genes MMP2 and MMP9 in pancreatic cancer cell lines (39). In the present study, we found that overexpression of miR-30d significantly suppressed the expression of β-catenin, cyclin D1, c-Myc, as well as MMP2 and MMP9 in CRC cell lines and xenograft tumors. These results revealed that upregulation of miR-30d inhibited CRC proliferation and invasion by regulating LRH-1 and its downstream Wnt/β-catenin signaling pathway.
Admittedly, there are limitations concerning the tumor specimen sample size in our study. Thus, in further investigations, a larger sample size involving more patients with various statuses in regards to distant metastasis, TNM stage, and invasive depth are needed to avoid any potential bias effect. In conclusion, our findings demonstrated that the expression level of miR-30d was downregulated in CRC tissues and cell lines, and its expression level was associated with poor prognosis, degree of tumor differentiation, invasive depth, TNM stage, distant metastasis and lymph node metastasis. We also discovered that miR-30d overexpression inhibited cell proliferation, migration and invasion, induced cell cycle arrest at the G0/G1 phase and apoptosis, as well as suppressed tumor growth in vivo. In addition, we identified LRH-1 as a direct target gene of miR-30d and found that miR-30d regulates Wnt/β-catenin signaling in CRC progression. These findings demonstrate that miR-30d plays a tumor-suppressor role in CRC development and inhibits tumorigenesis by regulating LRH-1 and its downstream Wnt/β-catenin signaling pathways. Thus, miR-30d is a potential novel therapeutic target for the treatment of CRC in the clinic.
Acknowledgments
The authors would like to thank Dr L.K. Yan for his valuable suggestions.
References
- 1.Fedewa SA, Ma J, Sauer AG, Siegel RL, Smith RA, Wender RC, Doroshenk MK, Brawley OW, Ward EM, Jemal A. How many individuals will need to be screened to increase colorectal cancer screening prevalence to 80% by 2018? Cancer. 2015;121:4258–4265. doi: 10.1002/cncr.29659. [DOI] [PubMed] [Google Scholar]
- 2.Zheng ZX, Zheng RS, Zhang SW, Chen WQ. Colorectal cancer incidence and mortality in China, 2010. Asian Pac J Cancer Prev. 2014;15:8455–8460. doi: 10.7314/APJCP.2014.15.19.8455. [DOI] [PubMed] [Google Scholar]
- 3.Gellad ZF, Provenzale D. Colorectal cancer: National and international perspective on the burden of disease and public health impact. Gastroenterology. 2010;138:2177–2190. doi: 10.1053/j.gastro.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Chen X, Xiu YL, Sun KX, Zhao Y. MicroRNA-490 3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362:122–130. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Šulc M, Marín RM, Robins HS, Vaníček J. PACCMIT/PACCMIT-CDS: Identifying microRNA targets in 3′ UTRs and coding sequences. Nucleic Acids Res. 2015;43:W474–9. doi: 10.1093/nar/gkv457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423 5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. doi: 10.1186/s13046-015-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikawa R, Goto Y, Kurozumi A, Matsushita R, Enokida H, Kojima S, Naya Y, Nakagawa M, Ichikawa T, Seki N. MicroRNA-205 inhibits cancer cell migration and invasion via modulation of centromere protein F regulating pathways in prostate cancer. Int J Urol. 2015;22:867–877. doi: 10.1111/iju.12829. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Fan XM, Mao L, Zhang JY, Li J, Wu JZ, Tang JH. MicroRNA-224: As a potential target for miR-based therapy of cancer. Tumour Biol. 2015;36:6645–6652. doi: 10.1007/s13277-015-3883-3. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi N, Arora S, Deshmukh SK, Singh S, Marimuthu S, Singh AP. Exploiting nanotechnology for the development of microRNA-based cancer therapeutics. J Biomed Nanotechnol. 2016;12:28–42. doi: 10.1166/jbn.2016.2172. [DOI] [PubMed] [Google Scholar]
- 10.Barger JF, Nana-Sinkam SP. MicroRNA as tools and therapeutics in lung cancer. Respir Med. 2015;109:803–812. doi: 10.1016/j.rmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang L, Liu L, Huang S, Zhao Y, He X. MicroRNA-30d 5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362:208–217. doi: 10.1016/j.canlet.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Yang WQ, Zhu H, Qian YY, Zhou L, Ren YJ, Ren XC, Zhang L, Liu XP, Liu CG, et al. Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem Pharmacol. 2014;87:562–570. doi: 10.1016/j.bcp.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xuan H, Xue W, Pan J, Sha J, Dong B, Huang Y. Downregulation of miR-221, -30d, and -15a contributes to pathogenesis of prostate cancer by targeting Bmi-1. Biochemistry (Mosc) 2015;80:276–283. doi: 10.1134/S0006297915030037. [DOI] [PubMed] [Google Scholar]
- 14.Ye Z, Zhao L, Li J, Chen W, Li X. miR-30d blocked transforming growth factor β1-induced epithelial-mesenchymal transition by targeting snail in ovarian cancer cells. Int J Gynecol Cancer. 2015;25:1574–1581. doi: 10.1097/IGC.0000000000000546. [DOI] [PubMed] [Google Scholar]
- 15.Sablin EP, Blind RD, Uthayaruban R, Chiu HJ, Deacon AM, Das D, Ingraham HA, Fletterick RJ. Structure of Liver Receptor Homolog-1 (NR5A2) with IP3 hormone bound in the ligand binding pocket. J Struct Biol. 2015;192:342–348. doi: 10.1016/j.jsb.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fayard E, Auwerx J, Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Stein S, Schoonjans K. Molecular basis for the regulation of the nuclear receptor LRH-1. Curr Opin Cell Biol. 2015;33:26–34. doi: 10.1016/j.ceb.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Schoonjans K, Dubuquoy L, Mebis J, Fayard E, Wendling O, Haby C, Geboes K, Auwerx J. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci USA. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benod C, Vinogradova MV, Jouravel N, Kim GE, Fletterick RJ, Sablin EP. Nuclear receptor liver receptor homologue-1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc Natl Acad Sci USA. 2011;108:16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayrer JR, Mukkamala S, Sablin EP, Webb P, Fletterick RJ. Silencing LRH-1 in colon cancer cell lines impairs proliferation and alters gene expression programs. Proc Natl Acad Sci USA. 2015;112:2467–2472. doi: 10.1073/pnas.1500978112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SL, Zheng DZ, Lan FH, Deng XJ, Zeng J, Li CJ, Wang R, Zhu ZY. Increased expression of hLRH-1 in human gastric cancer and its implication in tumorigenesis. Mol Cell Biochem. 2008;308:93–100. doi: 10.1007/s11010-007-9616-1. [DOI] [PubMed] [Google Scholar]
- 22.Ray RS, Rana B, Swami B, Venu V, Chatterjee M. Vanadium mediated apoptosis and cell cycle arrest in MCF7 cell line. Chem Biol Interact. 2006;163:239–247. doi: 10.1016/j.cbi.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Gupta S, Sussman DA, Doubeni CA, Anderson DS, Day L, Deshpande AR, Elmunzer BJ, Laiyemo AO, Mendez J, Somsouk M. Challenges and possible solutions to colorectal cancer screening for the underserved. J Natl Cancer Inst. 2014;106:dju032. doi: 10.1093/jnci/dju032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng W, Feng J, Qin H, Ma Y, Goel A. An update on miRNAs as biological and clinical determinants in colorectal cancer: A bench-to-bedside approach. Future Oncol. 2015;11:1791–1808. doi: 10.2217/fon.15.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ress AL, Perakis S, Pichler M. microRNAs and colorectal cancer. Adv Exp Med Biol. 2015;889:89–103. doi: 10.1007/978-3-319-23730-5_6. [DOI] [PubMed] [Google Scholar]
- 27.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015;21:4234–4242. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada A, Cox MA, Gaffney KA, Moreland A, Boland CR, Goel A. Technical factors involved in the measurement of circulating microRNA biomarkers for the detection of colorectal neoplasia. PLoS One. 2014;9:e112481. doi: 10.1371/journal.pone.0112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I, et al. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3:1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Jin B, Chen L, Zhuo D, Zhang Z, Gong K, Mao Z. MiR-30d induces apoptosis and is regulated by the Akt/FOXO pathway in renal cell carcinoma. Cell Signal. 2013;25:1212–1221. doi: 10.1016/j.cellsig.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Tian Y, Jiang S, Liu S, Zhao X, Tian D. MicroRNA-376c suppresses non-small-cell lung cancer cell growth and invasion by targeting LRH-1-mediated Wnt signaling pathway. Biochem Biophys Res Commun. 2016;473:980–986. doi: 10.1016/j.bbrc.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Liang Y, Zhao Q, Fan L, Zhang Z, Tan B, Liu Y, Li Y. Down-regulation of microRNA-381 promotes cell proliferation and invasion in colon cancer through upregulation of LRH-1. Biomed Pharmacother. 2015;75:137–141. doi: 10.1016/j.biopha.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, Zhao S, Pang X, Chi B. MicroRNA-381 suppresses cell growth and invasion by targeting the liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep. 2016;35:1831–1840. doi: 10.3892/or.2015.4491. [DOI] [PubMed] [Google Scholar]
- 35.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 36.Luu HH, Zhang R, Haydon RC, Rayburn E, Kang Q, Si W, Park JK, Wang H, Peng Y, Jiang W, He TC. Wnt/beta-catenin signaling pathway as a novel cancer drug target. Curr Cancer Drug Targets. 2004;4:653–671. doi: 10.2174/1568009043332709. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y, Cao B, Zhang M, Han W, Herman JG, Fuks F, Zhao Y, Guo M. Epigenetic silencing of NKD2, a major component of Wnt signaling, promotes breast cancer growth. Oncotarget. 2015;6:22126–22138. doi: 10.18632/oncotarget.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozaki S, Ikeda S, Ishizaki Y, Kurihara T, Tokumoto N, Iseki M, Arihiro K, Kataoka T, Okajima M, Asahara T. Alterations and correlations of the components in the Wnt signaling pathway and its target genes in breast cancer. Oncol Rep. 2005;14:1437–1443. doi: 10.3892/or.14.6.1437. [DOI] [PubMed] [Google Scholar]
- 39.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]