Abstract
Long non-coding RNAs (lncRNAs) are widely accepted as key players in various biological processes. However, the roles of lncRNA in peripheral nerve regeneration remain completely unknown. Thus, in this study, we performed microarray analysis to measure lncRNA expression in the distal segment of the sciatic nerve at 0, 3, 7 and 14 days following injury. We identified 5,354 lncRNAs that were differentially expressed: 3,788 lncRNAs were differentially expressed between days 0 and 3; 3,314 lncRNAs were differentially expressed between days 0 and 7; and 2,400 lncRNAs were differentially expressed between days 0 and 14. The results of RT-qPCR of two dysregulated lncRNAs were consistent with those of microarray analysis. Bioinformatics approaches, including lncRNA classification, gene ontology (GO) analysis and target prediction, were utilized to investigate the functions of these dysregulated lncRNAs in peripheral nerve damage. Importantly, we predicted that several lncRNA-mRNA pairs may participate in biological processes related to peripheral nerve injury. RT-qPCR was performed for the preliminary verification of three lncRNA-mRNA pairs. The overexpression of NONMMUG014387 promoted the proliferation of mouse Schwann cells. Thus, the findings of our study may enhance our knowledge of the role of lncRNAs in nerve injury.
Keywords: long non-coding RNA, mRNA, microarray, peripheral nerve injury, regeneration
Introduction
Peripheral nerve damage is common worldwide, and patients suffering from this type of injury may partially or completely lose motor, sensory and autonomic function (1). Nevertheless, the treatment of peripheral nerve injury (PNI) remains a majory medical concern due to the lack of satisfactory treatments (2,3).
Following PNI in mammals, the nerve distal to the site of injury may undergo a process known as Wallerian degeneration, during which Schwann cells lose contact with axons and dedifferentiate into stem-like cells or repair cells that play a vital role in repairing PNI (4,5) by forming a bundle that provides a permissive microenvironment for axon regeneration. Along with Schwann cells, macrophages gather at the site of PNI to clear myelin debris. It has also been widely reported that Schwann cells may secrete trophic support molecules, such as neurotrophin-3 (NT-3), fibroblast growth factor (FGF) and nerve growth factor (NGF) (6). Once Schwann cells reach the regrowing axons, they begin to remyelinate the axon apprxomately 7 days following nerve injury (7,8). The molecular mechanism of the regenerative process has not yet been fully elucidated. A better understanding of the mechanisms of Wallerian degeneration would help us to improve the repair process after PNI.
Long non-coding RNAs (lncRNAs) are non-protein-coding RNA molecules ranging from 200 nt to approximately 100 kb in length (9,10). lncRNAs have been proven to regulate gene expression at almost every level of transcription and translation, including genomic imprinting, chromatin modification and cytoplasmic mRNA translation (11,12). Data from an increasing number of studies have indicated that lncRNAs are associated with important regulatory functions during many biological processes. In neurobiology, lncRNAs are well known to be related to neurodevelopmental disorders, neurodegeneration and brain cancers (13). For example, the knockdown of linc-Brn1b has been shown to result in the reduction of intermediate progenitor cells in the brain, indicating that this lncRNA may play an important role in cortical development (14). In another previous study, lncRNAs that were differentially expressed following sciatic nerve resection in rats were found to be involved in regenerating dorsal root ganglion (DRG) neurons, and the downregulation of lncRNA BC089918 was found to promote neurite regeneration in DRG neurons (15). Although lncRNA studies are now common in various fields, the lncRNA expression signature and the possible roles of lncRNAs following Wallerian degeneration in the distal end of the peripheral nerve have not yet been reported, at least to the best of our knowledge.
In this study, the expression of lncRNAs and mRNAs in the distal segment of the sciatic nerve following PNI was profiled using microarray analysis. We identified a group of lncRNAs that were significantly dysregulated at different time points during Wallerian degeneration. The classification of the lncRNAs, validation by RT-qPCR, target prediction, gene ontology (GO) analysis and target prediction were performed. In particular, we predicted several pairs of lncRNAs and their related mRNAs. RT-qPCR analysis was used for the preliminary verification of the lncRNA-mRNA pairs. The overexpression of NONMMUG014387 was also found to promote the proliferation of mouse Schwann cells (MSCs). This study may provide a basis for the further investigation of the function of lncRNAs in peripheral nerve regeneration following injury.
Materials and methods
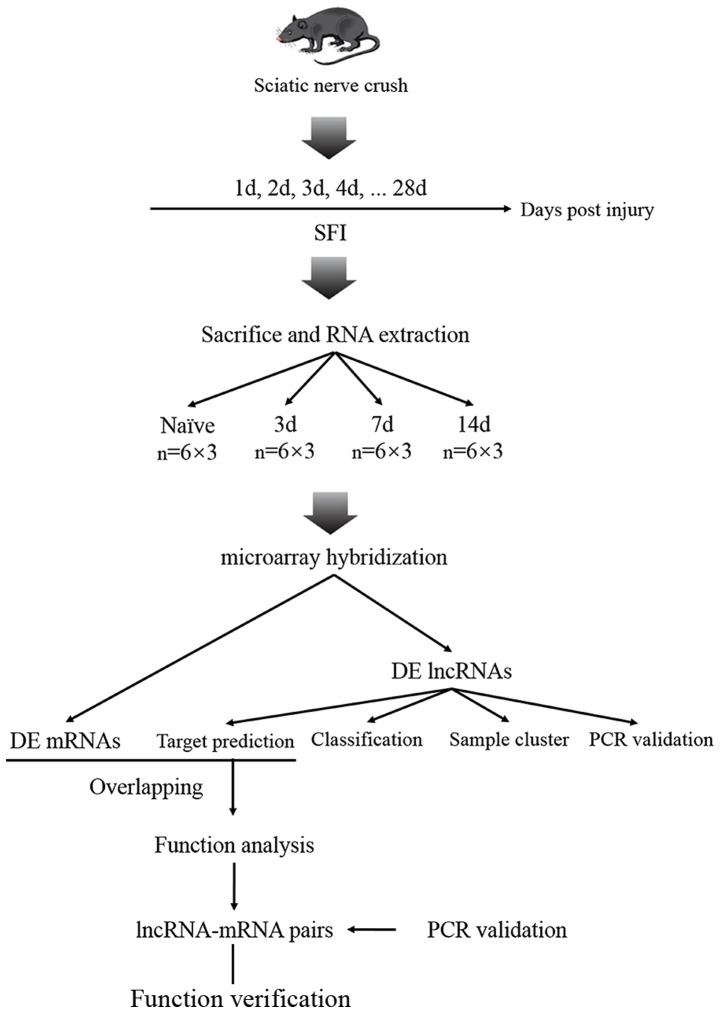
The complete research process used in the present study is summarized and presented in Fig. 1.
Figure 1.
The entire research process is presented. The right sciatic nerves of 15 C57Bl6 mice were crushed using an acutenaculum. To screen key time points for Wallerian degeneration, sciatic functional index (SFI) was performed on the mice. A microarray analysis was performed on 72 mice to detect the expression of long non-coding RNAs (lncRNAs) in the distal segment of the sciatic nerve at 0, 3, 7 and 14 days following injury. Bioinformatic approaches including sample clustering, gene ontology (GO) analysis, and target prediction were performed to investigate the functions of these dysregulated lncRNAs. RT-qPCR was performed on another 12 mice to verify the predicted lncRNA-mRNA pairs. d, day; DE, differentially expressed.
Experimental animals
A total of 99 mice were used in this study; 15 mice were used for the functional assessment of sciatic functional index (SFI), 72 mice were used for microarray analysis and 12 mice were used for further PCR verification. All the mice were obtained from the Laboratory Animal Research Center, Academy of Military Medical Sciences, of the Chinese People's Liberation Army, Beijing, China.
Functional assessment of SFI
The right sciatic nerves of 15 C57Bl6 mice were crushed using an ultra-fine, smooth, straight hemostat (tip width, 0.6 mm, Fine Science Tools) for 20 sec, as previously descrbied (16). After this procedure, the SFI was measured in each mouse daily for 28 days. The hind paws of the mice were immersed in non-toxic ink. The mice moved without assistance along a walking track and generated footprints were recorded. As previously described (17), several parameters were calculated for the SFI: print length (PL), toe spread (TS) and intermediate toe spread (ITS). All parameters were measured for normal (N) and experimental (E) animals. The SFI was determined according to the formula described by Inserra et al (17) as follows: SFI, 118.8 (ETS − NTS/NTS) − 51.2 (EPL − NPL/NPL) − 7.5, where NTS is normal toe spread, ETS is experimental toe spread, EPL is experimental print length, and NPL is normal print length.
Animal preparation and nerve lesion experiments
A total of 72 C57Bl6 mice, approximately 2 months of age were selected and randomly classified into 4 groups according to the time points of 0, 3, 7 and 14 days post-surgery. The 0 day group was used as a control, while all the other groups were the experimental groups. Due to the insufficient volume of a single sciatic nerve, 6 mice were pooled into one sample, and 3 samples for each time point. After the mice were anaesthetized via an intraperitoneal injection of chloral hydrate, an incision was created on the right lateral thigh to expose and lift the sciatic nerve. As described in a previous study (18), right sciatic nerve crush was performed at the upper thigh level using an acutenaculum. The nerve was compressed for 30 sec to ensure that the axon was disconnected while the epineurium remained intact. The crush site was then ligated to generate a marker for the damage site. The incision was then closed. To relieve painful mechanical stimulation and discomfort, the mice were housed in clean cages with sawdust bedding. We provided free access to food and water. These experiments were performed according to the NIH Guidelines for the Care and Use of Laboratory Animals (http://oacu.od.nih.gov/regs/index.htm). All procedures were approved by the Ethics Committee of Tianjin Medical University, Tianjin, China.
Microarray experiment
The microarray experiment was conducted using an Agilent-074622 Mouse lncRNA micro-array (Agilent Technologies, Santa Clara, CA, USA). The mice were sacrificed by cervical dislocation. Tissue perfusion was performed to remove the blood in the sciatic nerve. Distal segments of crushed sciatic nerves (0.5 cm) were isolated and harvested from the mice of each group 0, 3, 7 and 14 days post-surgery. Total RNA was extracted using a Takara RNAiso Plus kit (#9109) according to the manufacturer's instructions. The quality of the total RNA was assessed by the RNA integrity number (RIN) on an Agilent Bioanalyzer 2100 (Agilent Technologies). The total RNA was purified using an RNase-Free DNase kit and an RNeasy micro kit (both Qiagen GmBH, Hilden, Germany). The total RNA samples had a RIN of ≥7.0 and a 28S/18S ratio of ≥0.7.
The preparation of One-Color Spike Mix was performed using an Agilent One-Color RNA Spike-In kit. We amplified and labeled total RNA using a Low Input Quick Amp WT Labeling kit (Agilent Technologies) and purified cDNA using an RNeasy mini kit (Qiagen GmBH).
A total of 1.65 µg of Cy3-labeled cDNA was hybridized onto each microarray using a Gene Expression Hybridization kit in a hybridization oven (both from Agilent Technologies). Microarray slides were washed after a 17-h hybridization using a Gene Expression Wash Buffer kit (Agilent Technologies). All aforementioned procedures were performed in accordance with the manufacturer's instructions.
After scanning the microarrays with an Agilent Microarray Scanner we used Feature Extraction software v10.7 (both from Agilent Technologies) to extract the data. Raw data were normalized by a quantile algorithm in GeneSpring software v11.0 (Agilent Technologies).
Bioinformatics analysis
The microarray data generated in our study were deposited into the NCBI Gene Expression Omnibus (GEO) under accession no. GSE74087 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74087).
Raw data were extracted using Feature Extraction 10.7 software, and the quantiles were normalized using GeneSpring GX 11.0 software (Agilent Technologies). The global distribution characteristics of the sample data were normalized and visualized using a box plot. To comprehensively and clearly depict the associations and differences among the samples, clustering into expressed genes or differentially expressed genes was performed. Samples with similar characteristics may present in the same cluster after processing, and genes presenting in the same cluster may have similar biological functions. Following the normalization of the raw data, the fold change was determined, and multiple hypothesis testing was performed to identify differentially expressed genes. Genes with a fold change (linear) ≤0.5 or ≥2 and a P-value <0.01 in a t-test were selected. Differentially expressed genes that were identified were visually presented using scatter plots and heat maps.
To visualize the time dependence of the differential lncRNA expression, we performed a time series analysis in which expression levels were summed to obtain relative expression patterns over time. lncRNAs were clustered into several distinct profiles using k-means clustering with a distance matrix based on Pearson's correlation.
lncRNAs evidently influence the expression of protein-coding genes; thus, we predicted the target genes of differentially expressed lncRNAs to further investigate their functions in biological processes. The possible target genes for cis- or trans-regulating lncRNAs were predicted by applying two independent algorithms.
For cis-regulating prediction, the genomic positional associations between the lncRNAs and their potential paired target genes were determined from the RefSeq and UCSC Known Genes databases (19). Based on the distance between each lncRNA gene and its neighboring known protein-coding gene, algorithms, including an ORF-Predictor and BLASTP pipeline were used to identify cis-regulating lncRNA-mRNA potential pairs. We selected 10 kb as the cut-off of the distance between lncRNAs and mRNAs based on a previous study (19).
For trans-regulating prediction, the RNAplex method (http://www.tbi.univie.ac.at/RNA/RNAplex.1.html) was applied to identify possible target genes for lncRNAs based on a previous study (20). RNAplex, which is specifically designed to rapidly identify possible hybridization sites for an RNA query in large RNA databases, adopts a slightly different energy model that reduces the computational time significantly.
To predict the function of differentially expressed lncRNAs, we selected predicted target genes that overlapped with the differentially expressed mRNAs for further GO analysis. GO analysis was performed to analyze the primary function of the target coding genes according to the GO database, which defines the terms 'cellular components', 'molecular functions', and 'biological process' for each gene. These analyses were used to determine the functional enrichment of the target mRNA to analyze the functions of the lncRNAs involved in the response to nerve injury.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
To validate the microarray results, the expression level of three randomly selected lncRNAs, ENSMUSG00000087366, NONMMUG018386 and NONMMUG018381, were profiled at different time points after injury. For the preliminary validation of the lncRNA-mRNA pairs, the NONMMUG014387-Cthrc1, NONMMUG042364-Ntm and ENSMUSG00000097535-Icam1 pairs were detected. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control. In total, 12 mice were used for PCR verification. Total RNA was extracted from the sciatic nerve as described above. Reverse-transcribed cDNAs were synthesized using a PrimeScript RT Reagent kit according to the manufacturer's instructions (Shanghai Biotechnology Corp., Shanghai, China). PCR reactions were performed using SYBR Premix Ex Taq according to the manufacturer's instructions in a Rotor-Gene 6000 instrument. The PCR reaction used the following cycling conditions: 10 min at 95°C; 95°C (10 sec), 60°C (60 sec) and 95°C (10 sec) for a total of 40 cycles; and a final temperature increase from 60 to 99°C. The relative quantity of RNA was calculated and analyzed using the 2ΔΔCq method. All experiments were performed in triplicate. The primer sequences are presented in Table I.
Table I.
PCR primers used for expression analyses.
| Gene name and primer sequences (5′→3′) |
|---|
| NONMMUG014387 |
| Forward: AAAGGGATTACAGGCACACG |
| Reverse: CCAGGCCATTTACTCAGCAT |
| Cthrc1 |
| Forward: CGAAATAAAGCCTCTGACGA |
| Reverse: TTAACTTTGCTTTTTCATTCAGC |
| NONMMUG018381 |
| Forward: GCTCTTCTAAAGGTCATGGGTTCA |
| Reverse: CTTGGCTCCCCTGGAACTG |
| NONMMUG018386 |
| Forward: GGTCACATTTCCACATCAGC |
| Reverse: GGTCACTCGGGAATCTTGAA |
| ENSMUSG00000087366 |
| Forward: ACATTTATGGGACCCCCTCT |
| Reverse: AACCACCAACACCACTACCAA |
| Ntm |
| Forward: AGTGCCCCACCATGAAACA |
| Reverse: TTCTTGCTCTGTGCTTGTGTCTATG |
| NONMMUG042364 |
| Forward: TCCTGAAGAGAAGCTGCAAA |
| Reverse: TTCTTCTACCCCAGCTTCCA |
| ENSMUSG00000097535 |
| Forward: TAAGACTCGGGGAATGTGGA |
| Reverse: GGCTTGTCAACACTGAATGC |
| Icam1 |
| Forward: GTTCTCTGCTCCTCCACATCCT |
| Reverse: GGCTGACATTGGGAACAAAAGTAG |
| Gapdh |
| Forward: CGTGTTCCTACCCCCAATGT |
| Reverse: TGTCATCATACTTGGCAGGTTTCT |
Cell culture
Mouse Schwann cells (MSCs) were puchased from Shanghai Cellbio Co. (Shanghai, China). The cells were inoculated in a new culture flask, prepared medium was added, 90% HyClone Dulbecco's modified Eagle's medium (DMEM) [10% fetal bovine serum (FBS); Gibco, Grand Island, NY, USA] 1–1.5% penicillin-streptomycin. The cells were then placed in an incubator chamber at 37°C over a period of 24 h and the density was observed under a microscope. The cells were passaged when they reached 80% confluence. The medium was then removed and the culture flask was washed with 3–5 ml D-Hanks or phosphate-buffered saline (PBS). This was followed by the addition of 0.5–1 ml trypsin with 0.25% EDTA for digestion. The digestion was terminated by the addition of 3–5 ml medium, followed by centrifugaqtion at 1,000 rpm/5 min. The supernatant was then discarded and passaged according to the amount of cells.
Construction of the recombinant adenoviruses used in this study
NONMMUG014387 was inserted into the pHBAd-MCMV-GFP vector. One day prior to transfection, 293 cells were seeded in a 60 mm dish and incubated at 37°C with 5% CO2 overnight with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. The cells were transfected pHBAd-MCMV-GFP-NONMMUG014387 at 70–80% confluence. Following transfection, recombinant adenoviruses carrying NONMMUG014387 were harvested. All of the viral particles were purified by cesium chloride density gradient centrifugation and tittered by the TCID50 method.
Cell proliferation assay
The MSCs were plated in 6-well plate (cell/well), and different adenoviruses (Ad-GFP and Ad-NONMMUG014387) were added to each well at an MOI of 20. at 48 h post-transfection, the MSCs, and the MSC-GFP- and MSC-NONMMUG014387-transfected cells were digested into a single cell suspension. Each group of cells was equipped with a single cell suspension with 3×104 cells/ml concentration. Following cell adherence, 10 µl of CCK-8 (Hanheng Biotechnology Corp., Shanghai, China) were aded to the wells at 0, 24, 48, 72 and 96 h. The optical denstity (OD) was at 450 nm using a spectrophotometer.
Statistical analysis
The statistical methods employed in this study were performed using the SPSS 20.0 software packages (SPSS, Inc., Chicago, IL, USA). The statistical significance of the differential expression of various lncRNAs in the micro-array analysis and RT-qPCR validation was determined using an independent sample t-test. GO and pathway analyses were conducted using Fisher's exact test. Our data are expressed as the means ± standard deviation (SD). Statistical significance was defined as P<0.01.
Results
Assessment of sciatic nerve function index
To assess the degree of functional impairment and recovery of locomotion following sciatic nerve injury in mice, the SFI was calculated daily for 28 days. The SFI values were almost −100 immediately following PNI; after 7 days, there was rapid neural recovery; however, after 14 days, nerve function recovery had decelerated (Fig. 2A).
Figure 2.
(A) Mice exhibited severe functional impairment of the sciatic nerve. Dynamics of the sciatic nerve functional index of mice over 4 post-injury weeks (n=5, means ± SD). (B) Box plots show the symmetry of the data and the degree of dispersion. The abscissa shows the sample name. The ordinate represents the fold-change (log2) in signal values of probes. (C) Twelve samples were subjected to sample clustering. (D) Venn diagram showing the overlap of dysregulated long non-coding RNAs (lncRNAs) between gene sets at different post-injury time points (0 vs 3 days, 0 vs 7 days and 0 vs 14 days). d, day.
Differential expression of lncRNAs
The Agilent-074622 mouse lncRNA microarray consisted of 64,221 lncRNA probes collected from public databases, including GENCODE v21/Ensembl, Noncode v4 and UCSC. A total of 26,531 mRNA probes were also present on this array. Based on the SFI results, microarray experiments were performed at 0, 3, 7 and 14 days after sciatic nerve crush to identify lncRNAs that were differentially expressed during Wallerian degeneration.
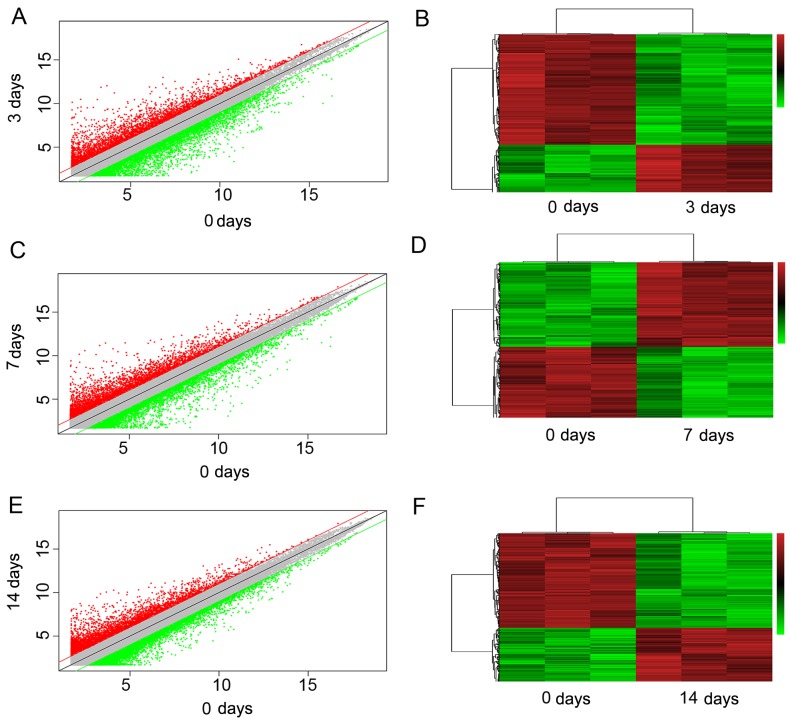
The 12 specimens shared a similar level of total characteristics, which is shown in a box plot (Fig. 2B). Sample clustering revealed that the samples from each time group were grouped into the same cluster (Fig. 2C). A total of 5,354 lncRNAs were identified as significantly differentially expressed among all groups following injury (P<0.01; fold change >2). We also compared lncRNA expression independently. A comparison between the 0 and 3 days data revealed that 3,788 lncRNAs were differentially expressed. Among these, 1,521 were upregulated, while 2,267 were downregulated. A comparison between the 0 and 7 days data revealed that 3,314 lncRNAs were differentially expressed. Among these, 1,754 were upregulated, while 1,560 were downregulated. A comparison of the data between 0 and 14 days revealed a total of 2,400 differentially expressed lncRNAs, 1,401 of which were upregulated and 999 down-regulated (Fig. 2D). The top 10 downregulated and upregulated lncRNAs between the different groups are shown in Tables II–IV. A scatter plot was generated to assess the variation in lncRNA expression among the different groups (Fig. 3A, C and E). In addition, hierarchical cluster analysis revealed lncRNA expression patterns. In hierarchical clustering analysis, we used a heatmap to indicate that differentially expressed lncRNAs at different post-injury time -points were segregated into different clusters (Fig. 3B, D and F). The results of RT-qPCR for NONMMUG018381, NONMMUG018386 and ENSMUSG00000087366 were consistent with those of the microarray analysis (Fig. 4).
Table II.
Top 10 differentially expressed lncRNAs post-injury at day 0 vs day 3.
| Upregulated lncRNAs
|
Downregulated lncRNAs
|
||||
|---|---|---|---|---|---|
| lncRNA | P-value | Fold change | lncRNA | P-value | Fold change |
| NONMMUG032014 | 0.006677 | 365.7848 | NONMMUG013007 | 6.48E-05 | 172.2607 |
| NONMMUG032012 | 0.000117 | 342.0147 | 1500009C09Rik | 0.000838 | 97.30048 |
| NONMMUG032010 | 6.50E-05 | 328.0504 | NONMMUG013007 | 0.000995 | 83.27552 |
| NONMMUG032016 | 0.000539 | 311.5205 | NONMMUG042554 | 0.001978 | 82.2234 |
| NONMMUG010768 | 0.001339 | 188.556 | NONMMUG027331 | 0.00012 | 80.19379 |
| Gm11351 | 0.000147 | 176.1802 | NONMMUG027331 | 0.000143 | 73.40636 |
| NONMMUG013417 | 1.50E-06 | 166.4564 | NONMMUG013007 | 9.24E-07 | 63.55268 |
| NONMMUG018385 | 0.000157 | 159.6788 | ENSMUSG00000101344 | 0.000463 | 56.43617 |
| NONMMUG018386 | 1.29E-05 | 138.9562 | NONMMUG025392 | 0.000777 | 55.75199 |
| NONMMUG027492 | 0.00123 | 117.741 | NONMMUG023470 | 0.000209 | 55.61781 |
Table III.
Top 10 differentially expressed lncRNAs post-injury at day 0 vs day 7.
| Upregulated lncRNAs
|
Downregulated lncRNAs
|
||||
|---|---|---|---|---|---|
| lncRNA | P-value | Fold change | lncRNA | P-value | Fold change |
| NONMMUG002067 | 0.001319 | 265.9105 | 1500009C09Rik | 0.001432 | 178.838 |
| NONMMUG032016 | 6.48E-07 | 248.2987 | NONMMUG013117 | 0.001561 | 131.4498 |
| NONMMUG032014 | 0.007602 | 225.2465 | NONMMUG042554 | 0.000353 | 109.4421 |
| NONMMUG032010 | 0.000118 | 216.8008 | NONMMUG027331 | 0.001987 | 101.779 |
| NONMMUG032012 | 0.000162 | 203.7992 | 1500009C09Rik | 0.001267 | 98.69802 |
| Bach2os | 0.000424 | 143.7661 | NONMMUG027426 | 0.00085 | 68.50972 |
| NONMMUG027492 | 0.000211 | 128.3399 | NONMMUG027331 | 0.000138 | 66.7439 |
| NONMMUG018385 | 0.000129 | 125.3089 | ENSMUSG00000101344 | 0.002055 | 63.9863 |
| NONMMUG018386 | 1.89E-05 | 103.5612 | NONMMUG019812 | 0.000344 | 60.7132 |
| NONMMUG004899 | 3.19E-06 | 102.7997 | ENSMUSG00000099137 | 5.63E-05 | 56.10263 |
Table IV.
Top 10 differentially expressed lncRNAs at post-injury day 0 vs 14.
| Upregulated lncRNAs
|
Downregulated lncRNAs
|
||||
|---|---|---|---|---|---|
| lncRNA | P-value | Fold change | lncRNA | P-value | Fold change |
| NONMMUG032016 | 4.02E-06 | 92.85173 | A330015K06Rik | 0.00014 | 36.64583 |
| NONMMUG032010 | 0.000687 | 83.81547 | 1500009C09Rik | 0.00557 | 33.69628 |
| Bach2os | 0.000716 | 79.68822 | 1500009C09Rik | 0.006163 | 28.85862 |
| NONMMUG018385 | 0.000206 | 78.50652 | NONMMUG027331 | 0.000373 | 28.00702 |
| NONMMUG032012 | 0.00025 | 76.64838 | NONMMUG025427 | 0.002157 | 26.34975 |
| NONMMUG018386 | 1.79E-05 | 73.27124 | NONMMUG043073 | 0.003822 | 24.84478 |
| NONMMUG005793 | 0.00107 | 55.83829 | NONMMUG013117 | 0.006125 | 23.96197 |
| NONMMUG010751 | 3.14E-06 | 48.46715 | ENSMUSG00000086253 | 0.001054 | 23.74235 |
| NONMMUG029505 | 0.00412 | 47.81979 | NONMMUG010960 | 0.000317 | 22.64306 |
| NONMMUG005794 | 9.66E-05 | 47.10508 | NONMMUG016086 | 0.007818 | 22.17701 |
Figure 3.
Profile of the microarray data. (A, C and E) Variations in lncRNA expression between different time-points (0 vs 3 days, 0 vs 7 days and 0 vs 14 days) are shown as a scatter plot. The x and y values on the scatter plot are the average normalized signal values, shown in a log2 scale. The red and green lines were set as fold change lines with a default change of 2.0. (B, D and F) Heatmap showing the long non-coding RNAs (lncRNAs) that were differentially expressed between the different groups. Each lncRNA is represented by a row, and each post-injury time-point is represented by a column. The color scale is used to depict the relative expression levels of lncRNAs. Red indicates increased expression, while green indicates decreased expression.
Figure 4.
The qRT-PCR results of the 3 chosen long non-coding RNAs (lncRNAs) compared with those of the microarray. (A) NONMMUG018381. (B) NONMMUG018386. (C) ENSMUSG00000087366. The heights of the columns in the chart represent the fold change (log2).
Classification of differentially expressed lncRNAs
The dysregulated lncRNAs were characterized by their lengths and chromosomal distribution (Fig. 5A and B). Several cellular biological processes, such as dedifferentiation, demyelination, redifferentiation and remyelination, are involved in Wallerian degeneration, which results from a cut or crush injury (21–23). Furthermore, lncRNA expression in response to injury may be time-dependent. We therefore performed a time series analysis to classify these dysregulated lncRNAs. Consequently, 11 lncRNA profiles had significant P-values (Fig. 5C). Recent studies have demonstrated that lncRNA transcription is often initiated simultaneously with the expression of their overlapping or interspersed sequences (20,21). This finding implies that the lncRNAs that are associated with adjacent genes may be involved inregulating gene expression. To obtain a better understanding of this association, lncRNAs were crudely classified into five categories according to their association with adjacent genes: i) sense lncRNAs overlap with the exons of coding genes and are transcribed from the same strand; ii) antisense lncRNAs overlap with the exons of coding genes and are transcribed from the antisense strand; iii) bidirectional lncRNAs are transcribed with a coding transcript that is transcribed in close proximity on the opposite strand; iv) intronic lncRNAs are derived from the introns of coding genes; and v) intergenic lncRNAs are located within the interval between two genes (24). In this study, we analyzed the categorical distribution of the 5,354 lncRNAs that were differentially expressed following sciatic nerve injury (Fig. 5D).
Figure 5.
Characteristics of differentially expressed long non-coding RNAs (lncRNAs). (A) Chromosomal distribution of differentially expressed lncRNAs indicating the variations in their chromosomal locations. (B) The length distribution of differentially expressed lncRNAs. (C) A time series analysis shows the post-injury temporal expression patterns of dysregulated lncRNAs in the sciatic nerve. Each box represents the time-dependent expression profile of a lncRNA. The upper numbers refer to the number of profiles, while the lower numbers indicate the P-value of the profiles in each box. (D) A pie chart shows the classification of a variety of differentially expressed lncRNAs based on their genomic location relative to neighboring or overlapping genes.
lncRNA target prediction and functional analysis
To further explore the dysregulated lncRNAs, we predicted the target genes that would be regulated using cis and trans mechanisms. The target genes overlapped with mRNAs that were differentially expressed at different time-points. We chose the overlapping mRNAs for further GO analysis. For further analysis, we selected 9 classes of biological processes that are closely related to nerve regeneration following PNI, including stimulation responses, inflammatory responses, immune responses, cell proliferation, cell migration, axon guidance, myelination, extracellular matrix processes, protein kinase activity and growth factor activity. The numbers of dysregulated lncRNAs that were predicted to participate in these processes are shown in Fig. 6.
Figure 6.
The number of differentially expressed long non-coding RNAs (lncRNAs) predicted to be involved in different biological processes including (A) axon guidance, (B) cell migration, (C) cell proliferation, (D) the extracellular matrix, (E) growth factor activity, (F) immune responses, (G) inflammatory responses, (H) myelination and (I) stimulus responses are shown. The x-axis represents the time after injury, and the y-axis represents the number of genes.
Functional prediction of lncRNA-mRNA pairs
Based on the classification of lncRNAs in relation to different biological processes, we predicted the lncRNA-mRNA pairs in the 9 biological processes that may be important for peripheral nerve regeneration following injury, according to the following criteria: i) the lncRNA was predicted to be involved in the 9 biological processes according to the results described above; ii) the lncRNA was predicted to use a cis or trans mechanism to target mRNAs that have been reported to be involved in peripheral nerve regeneration; iii) the mRNA has been reported to be involved in peripheral nerve regeneration; iv) both the lncRNA and mRNA were differentially expressed in the microarray experiment. A list of these lncRNA-mRNA pairs is provided in Table V.
Table V.
Dysregulated lncRNA transcripts and their potential target mRNA transcripts.
| Gene symbol | Chr | Start | End | TargetGene | Regulation type |
|---|---|---|---|---|---|
| NONMMUG036610 | Chr6 | 125479051 | 125480980 | CD9 | Intronic sense |
| NONMMUG036611 | Chr6 | 125493330 | 125507715 | CD9 | Exonic antisense |
| NONMMUG036609 | Chr6 | 125476049 | 125478755 | CD9 | Intronic sense |
| LOC100504703 | Chr10 | 127070480 | 127071101 | Cdk4 | Bidirectional |
| NONMMUG014387 | Chr15 | 39076901 | 39079473 | Cthrc1 | Exonic sense |
| NONMMUG035232 | Chr6 | 47584681 | 47585827 | Ezh2 | Intronic sense |
| ENSMUSG00000054779 | Chr3 | 37335332 | 37349740 | FGF2 | Exonic antisense |
| ENSMUSG00000097535 | Chr9 | 21034290 | 21037782 | Icam1 | Exonic antisense |
| NONMMUG042235 | Chr9 | 21034152 | 21036784 | Icam1 | Exonic antisense |
| ENSMUSG00000087366 | Chr4 | 95052951 | 95060658 | Jun | Bidirectional |
| NONMMUG020461 | Chr18 | 82575341 | 82577314 | MBP | Exonic sense |
| ENSMUSG00000100811 | Chr1 | 62714723 | 62718073 | Nrp2 | Intronic antisense |
| NONMMUG042364 | Chr9 | 29327912 | 29329715 | Ntm | Intronic sense |
| ENSMUSG00000073394 | Chr17 | 44735845 | 44737612 | Runx2 | Exonic antisense |
| NONMMUG026340 | Chr3 | 34638297 | 34680814 | Sox2 | Exonic sense |
| SOX2OT | Chr3 | 34560380 | 34677993 | Sox2 | Exonic sense |
| NONMMUG014942 | Chr15 | 79166059 | 79216796 | SOX10 | Exonic antisense |
| ENSMUSG00000075555 | Chr15 | 79166066 | 79227524 | SOX10 | Exonic antisense |
| GM10863 | Chr15 | 79166065 | 79216401 | SOX10 | Exonic antisense |
| NONMMUG027334 | Chr3 | 96559498 | 96560013 | TXNIP | Exonic sense |
| NONMMUG036332 | Chr6 | 114683284 | 114685489 | ATG7 | trans regulation |
| NONMMUG036586 | Chr6 | 125119767 | 125120865 | CHD4 | trans regulation |
| ENSMUSG00000085185 | Chr17 | 24208861 | 24221547 | FGD4 | trans regulation |
| NONMMUG026576 | Chr3 | 52349362 | 52353221 | FOXO1 | trans regulation |
| NONMMUG003088 | Chr10 | 14530916 | 14544995 | Gpr126 | trans regulation |
| NONMMUG004321 | Chr10 | 87859068 | 87862787 | IGF-I | trans regulation |
| ENSMUSG00000084785 | Chr2 | 114654425 | 114697839 | Mbp | trans regulation |
| NONMMUG022363 | Chr2 | 26498155 | 26503824 | Notch1 | trans regulation |
| NONMMUG042365 | Chr9 | 29941548 | 29963141 | Ntm | trans regulation |
| NONMMUG045895 | ChrX | 136833251 | 136834685 | Plp1 | trans regulation |
| NONMMUG010575 | Chr13 | 28949083 | 28951671 | Sox4 | trans regulation |
Validation of lncRNA-mRNA pairs by RT-qPCR
For the preliminary validation of the predicted lncRNA-mRNA pairs, we selected three lncRNA-mRNA pairs for RT-qPCR verification. NONMMUG014387 was predicted to have cis-regulating potential with Cthrc1, NONMMUG042364 was predicted to have cis-regulating potential with Ntm, and ENSMUST00000180870 was predicted to have cis-regulating potential with Icam1. The results of RT-qPCR and microarray analysis revealed that NONMMUG014387 and NONMMUG042364 were upregulated following PNI, and Cthrc1 and Ntm were also upregulated following PNI. The expression trends of the two lncRNAs at different time points were consistent with the separate trends of the two mRNAs. ENSMUSG00000097535 was downregulated following PNI, while Icam1 was upregulated following PNI, although in a different direction; the expression level of ENSMUSG00000097535 was mostly consistent with that of Icam (Fig. 7).
Figure 7.
Verification of three lncRNA-mRNA pairs by RT-qPCR and microarray analysis. (A) Comparison of the expression levels of NONMMUG014387 and Cthrc1 by microarray. (B) Comparison of the expression levels of NONMMUG014387 and Cthrc1 by RT-qPCR. (C) Comparison of the expression levels of NONMMUG042364 and Ntm by microarray. (D) Comparison of the expression levels of NONMMUG042364 and Ntm by RT-qPCR. (E) Comparison of the expression levels of ENSMUSG00000097535 and Icam1 by microarray. (F) Comparison of the expression levels of ENSMUSG00000097535 and Icam1 by RT-qPCR.
Overexpression of NONMMUG014387 enhances MSC proliferation
NONMMUG014387 was overexpressed through recombinant adenoviruses, the relative level of NONMMUG014387 was detected by RT-qPCR, and the value of the MSCs transfected with NONMMUG014387 was higher than that of the control MSCs or the MSCs + GFP group (Fig. 8A). CCK8 reavealed that the overexpression of NONMMUG014387 in the MSCs increased cell proliferation compared with the control MSCs and the MSCs + GFP group d (Fig. 8B).
Figure 8.
Overexpression of NONMMUG014387 increase mouse Schwann cell (MSC) proliferation. (A) NONMMUG014387 in MSC + lncRNA was higher than that of the control MSCs and the MSC + GFP group. (B) Overexpression of NONMMUG014387 in MSCs increased cell proliferation.
Discussion
To the best of our knowledge, this study is the first to obtain a genome-wide microarray profile of lncRNA expression at the distal end of the sciatic nerve 0, 3, 7 and 14 days following nerve crush injury and to further predict the possible lncRNA functions using bioinformatics approaches. A total of 5,354 lncRNAs were dysregulated following PNI, with 3,788, 3,314 and 2,400 lncRNAs dysregulated at 3, 7 and 14 days following nerve crush injury, respectively. Validation by RT-qPCR demonstrated that genes selected from the differentially expressed lncRNAs were dysregulated in a manner consistent with the microarray data at different time points. Therefore, many lncRNAs are predicted to target key mRNAs during peripheral nerve regeneration.
It has been demonstrated that transcriptional changes occur following sciatic nerve crush (25). Li et al demonstrated that the expression of immune response-related genes was significantly upregulated following nerve crush injury and rapidly peaked at 7 days post-crush (26). During the early stage of Wallerian degeneration, immune responses leads to debris clearance and cell death. They also found that the expression of genes associated with cell proliferation and migration showed a similar pattern to that of the immune response genes, and genes associated with guidance and regeneration were rapidly upregulated between 3 and 7 days post-crush. Li et al also demonstrated that genes related to myelination were downregulated following nerve crush injury and were then upregulated between at 7 and 14 days post-injury. In this study, SFI indicated the rapid recovery of sciatic nerve function at 7 days post-injury. However, the speed of recovery decelerated after 14 days. Overall, the gene expression data at 0, 3, 7 and 14 days following injury may reflect the process of nerve regeneration.
As mentioned above, lncRNA-mRNA pairs were predicted to be involved in peripheral nerve regeneration following injury. Several lncRNA-mRNA pairs caught our attention.
Many key genes were cis-regulated by lncRNAs. Our analysis revealed that Jun was cis-regulated by lncRNA ENSMUSG00000087366). Jun is a protein encoded by an oncogene that combines with c-Fos to form the AP-1 transcription factor. A previous study demonstrated that Jun is a central regulator of Schwann cells in response to injury (27). The knockdown of Jun has been shown to result in strikingly compromised axonal regeneration and functional repair (28,29). ENSMUSG00000075555, GM10863 and NONMMUG014942 were paired with SOX10. These three lncRNAs are located upstream of the SOX10 gene on the sense strand. Previous studies have shown that SOX10 plays an important role in the myelination of Schwann cells during development and is required for myelination maintenance in adults (30,31). Following sciatic nerve crush injury, SOX10 downregulation exerts a demyelination effect involved in the phenotypic transition of Schwann cells (30). In addition to the effects of SOX10 effects, the downregulated expression of Sox2 also has an tight association with lncRNAs. As shown by our results, Sox2 was encompassed or overlapped by two differentially expressed lncRNAs (SOX2OT and NONMMUG026340). Sox2 has been reported to be a mediator of EphB signaling, which aids in Schwann cell sorting and guides axon regeneration (32). NONMMUG042235 and ENSMUSG00000097535 were paired with intercellular adhesion molecule-1 (Icam1). It has recently been demonstrated that Icam1 is related to both inflammation and cell recruitment in peripheral nerve degeneration following injury, as well as to the function of myelinogenesis in Schwann cell (33). ENSMUSG00000054779 was paired with fibroblast growth factor-2 (FGF2). FGF2 is highly expressed in Schwann cells and may exert paracrine actions following PNI (21). FGF2 has also been found to benefit motor neuron regeneration following sciatic nerve injury and can induce Schwann cell proliferation via transforming growth factor (TGF)-β signaling (34). LOC100504703 was paired with cyclin-dependent kinase 4 (Cdk4), which has been reported to be involved in the differentiation and proliferation of Schwann cells after sciatic nerve injury (35). NONMMUG036611, NONMMUG036609 and NONMMUG036610 were paired with CD9. CD9 promotes the migration of Schwann cells in vitro. CD9 has been shown to participate in the regulation of Schwann cells in response to PNI (36,37). NONMMUG027334 was paired with thioredoxin interacting protein (TXNIP). TXNIP expression is closely associated with the process of peripheral nerve regeneration in vivo. TXNIP is required for advanced glycation end products receptor (RAGE)-induced p38 mitogen-activated protein kinase (MAPK) activation. The silencing of TXNIP affects the migration of Schwann cells and blocks interleukin (IL)-1 and fibronectin (FN) expression (38). NONMMUG042364 was paired with neurotrimin (Ntm). Ntm is a member of the neural cell adhesion molecule (NCAM) family (39). The involvement of NCAMs in cell differentiation, growth and migration is widely accepted (40). Of note, miR-182 reduces the migration ability of Schwann cells by targeting Ntm at an early stage following sciatic nerve injury (41). Whether Ntm is targeted by both lncRNAs and microRNAs simultaneously in a competing endogenous RNA network remains to be clarified.
Although overlapping or neighboring locations of mRNAs and lncRNAs could establish a tight association through cis-regulation, many lncRNAs target mRNA through trans-regulation mechanisms (42,43). In contrast to cis-regulating lncRNAs, trans-regulating lncRNAs dissociate from the primary locus of transcription and influence gene expression from a great distance. In this study, we predicted several trans-regulatory lncRNA-mRNA pairs. ENSMUSG00000084785 was paired with myelin basic protein (MBP) through a trans-regulatory mechanism. MBP has been reported to promote nerve regeneration by cleaving the neural cell adhesion molecule L1 (44). NONMMUG004321 was paired with insulin-like growth factor-I (IGF-I). IGF-I is not only an important mediator of growth hormone action, but is also a neurotrophic factor for a variety of neurons. Furthermore, IGF-I plays a key role in the development and growth of the peripheral nervous system; systemic IGF-I treatment can promote peripheral nerve regeneration (45). NONMMUG003088 was paired with G-protein-coupled receptor 126 (Gpr126). The knockdown of Gpr12 has been shown to result in limb posture abnormalities in mice, and Gpr126 is essential for peripheral nerve development in mice (46).
All the lncRNAs discussed above were closely associated with the target mRNAs. This study raises the possibility that these lncRNAs may promote nerve regeneration by targeting this set of coding genes.
This study has some limitations. First, this study was limited by the disadvantages of microarray technology, which can only detect the expression of known sequences;therefore, some unknown but crucial lncRNAs involved in axon regeneration may have been omitted. Second, due to the properties of lncRNAs and their unclear functional mechanisms involving gene expression, it is not possible to accurately predict the role of a specific lncRNA in an overt biological process based on its expression level or sequence (47). Third, the predictions of the potential function of differentially expressed lncRNAs were not fully verified; the determination of the definite roles of lncRNAs depends on further experimental validation. Despite its limitations, this study describes the lncRNA expression levels in the distal end of the sciatic nerves for the first time; predictions of lncRNA function are essential for future investigations of the important role of lncRNAs in nerve regeneration.
In conclusion, the present study, to the best of our knowledge, provides the first evidence for temporally regulated genome-wide lncRNA expression patterns in the sciatic nerve following crush injuury. We predicted the function of these altered lncRNAs based on their target mRNAs and identified lncRNAs that may play an important role in peripheral nerve regeneration.
Acknowledgments
This study was supported by the State Key Program of the National Natural Science Foundation of China (no. 81330042), the Special Program for Sino-Russian Joint Research Sponsored by the Ministry of Science and Technology, China (no. 2014DFR31210) and the Key Program Sponsored by the Tianjin Science and Technology Committee, China (nos. 13RCGFSY19000 and 14ZCZDSY00044). The authors would like to thank Xiong Hui of Tianjin Medical University (Department of Pathophysiology) and Wen-Yuan Shen of Nankai University (School of Medicine) for providing technical assistance.
References
- 1.Rochkind S. Phototherapy in peripheral nerve regeneration: From basic science to clinical study. Neurosurg Focus. 2009;26:E8. doi: 10.3171/FOC.2009.26.2.E8. [DOI] [PubMed] [Google Scholar]
- 2.Ertem K, Ceylan F, Zorludemir S, Karakoc Y, Yologlu S. Impairment of peripheral nerve healing after nerve repair in rats chronically exposed to alcohol. Arch Med Res. 2009;40:325–330. doi: 10.1016/j.arcmed.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Wang YL, Kong D, Qu B, Su XJ, Li H, Pi HY. Nerve autografts and tissue-engineered materials for the repair of peripheral nerve injuries: A 5-year bibliometric analysis. Neural Regen Res. 2015;10:1003–1008. doi: 10.4103/1673-5374.158369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas PK, King RHM. The degeneration of unmyelinated axons following nerve section: An ultrastructural study. J Neurocytol. 1974;3:497–512. doi: 10.1007/BF01098736. [DOI] [PubMed] [Google Scholar]
- 5.Court FA, Alvarez J. Local regulation of the axonal phenotype, a case of merotrophism. Biol Res. 2005;38:365–374. doi: 10.4067/S0716-97602005000400009. [DOI] [PubMed] [Google Scholar]
- 6.Gao R, Wang L, Sun J, Nie K, Jian H, Gao L, Liao X, Zhang H, Huang J, Gan S. MiR-204 promotes apoptosis in oxidative stress-induced rat Schwann cells by suppressing neuritin expression. FEBS Lett. 2014;588:3225–3232. doi: 10.1016/j.febslet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Fawcett JW, Keynes RJ. Peripheral nerve regeneration. Annu Rev Neurosci. 1990;13:43–60. doi: 10.1146/annurev.ne.13.030190.000355. [DOI] [PubMed] [Google Scholar]
- 8.Akassoglou K, Strickland S. Nervous system pathology: The fibrin perspective. Biol Chem. 2002;383:37–45. doi: 10.1515/BC.2002.004. [DOI] [PubMed] [Google Scholar]
- 9.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 10.Cao G, Zhang J, Wang M, Song X, Liu W, Mao C, Lv C. Differential expression of long non-coding RNAs in bleomycin-induced lung fibrosis. Int J Mol Med. 2013;32:355–364. doi: 10.3892/ijmm.2013.1404. [DOI] [PubMed] [Google Scholar]
- 11.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Marchese FP, Huarte M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics. 2014;9:21–26. doi: 10.4161/epi.27472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130507. doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu B, Zhou S, Hu W, Qian T, Gao R, Ding G, Ding F, Gu X. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neurosci Lett. 2013;534:117–122. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Morrison BM, Tsingalia A, Vidensky S, Lee Y, Jin L, Farah MH, Lengacher S, Magistretti PJ, Pellerin L, Rothstein JD. Deficiency in monocarboxylate transporter 1 (MCT1) in mice delays regeneration of peripheral nerves following sciatic nerve crush. Exp Neurol. 2015;263:325–338. doi: 10.1016/j.expneurol.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inserra MM, Bloch DA, Terris DJ. Functional indices for sciatic, peroneal, and posterior tibial nerve lesions in the mouse. Microsurgery. 1998;18:119–124. doi: 10.1002/(SICI)1098-2752(1998)18:2<119::AID-MICR10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Stassart RM, Fledrich R, Velanac V, Brinkmann BG, Schwab MH, Meijer D, Sereda MW, Nave KA. A role for Schwann cell-derived neuregulin-1 in remyelination. Nat Neurosci. 2013;16:48–54. doi: 10.1038/nn.3281. [DOI] [PubMed] [Google Scholar]
- 19.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tafer H, Hofacker IL. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 21.Duobles T, Lima Tde S, Levy Bde F, Chadi G. S100ß and fibroblast growth factor-2 are present in cultured Schwann cells and may exert paracrine actions on the peripheral nerve injury. Acta Cir Bras. 2008;23:555–560. doi: 10.1590/S0102-86502008000600014. [DOI] [PubMed] [Google Scholar]
- 22.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Chen G, Yang J, Fan X, Gong Y, Xu G, Cui Q, Geng B. Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS One. 2013;8:e77938. doi: 10.1371/journal.pone.0077938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang N, Li H, Sun Y, Yin D, Zhao Q, Cui S, Yao D. Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regen Res. 2014;9:1186–1194. doi: 10.4103/1673-5374.135325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Liu Q, Wang Y, Gu Y, Liu D, Wang C, Ding G, Chen J, Liu J, Gu X, Li S. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One. 2013;8:e57000. doi: 10.1371/journal.pone.0057000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson DB, Bhaskaran A, Arthur-Farraj P, Noon LA, Woodhoo A, Lloyd AC, Feltri ML, Wrabetz L, Behrens A, Mirsky R, et al. c-Jun is a negative regulator of myelination. J Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vargas ME, Barres BA. Why is Wallerian degeneration in the CNS so slow? Annu Rev Neurosci. 2007;30:153–179. doi: 10.1146/annurev.neuro.30.051606.094354. [DOI] [PubMed] [Google Scholar]
- 29.Fontana X, Hristova M, Da Costa C, Patodia S, Thei L, Makwana M, Spencer-Dene B, Latouche M, Mirsky R, Jessen KR, et al. c-Jun in Schwann cells promotes axonal regeneration and motoneuron survival via paracrine signaling. J Cell Biol. 2012;198:127–141. doi: 10.1083/jcb.201205025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glenn TD, Talbot WS. Signals regulating myelination in peripheral nerves and the Schwann cell response to injury. Curr Opin Neurobiol. 2013;23:1041–1048. doi: 10.1016/j.conb.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara S, Hoshikawa S, Ueno T, Hirata M, Saito T, Ikeda T, Kawaguchi H, Nakamura K, Tanaka S, Ogata T. SOX10 transactivates S100B to suppress Schwann cell proliferation and to promote myelination. PLoS One. 2014;9:e115400. doi: 10.1371/journal.pone.0115400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webber C, Zochodne D. The nerve regenerative microenvironment: Early behavior and partnership of axons and Schwann cells. Exp Neurol. 2010;223:51–59. doi: 10.1016/j.expneurol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Gu Y, Huang X, Shen A, Cheng C. Dynamic changes of ICAM-1 expression in peripheral nervous system following sciatic nerve injury. Neurol Res. 2011;33:75–83. doi: 10.1179/016164110X12714125204353. [DOI] [PubMed] [Google Scholar]
- 34.Allodi I, Mecollari V, González-Pérez F, Eggers R, Hoyng S, Verhaagen J, Navarro X, Udina E. Schwann cells transduced with a lentiviral vector encoding Fgf-2 promote motor neuron regeneration following sciatic nerve injury. Glia. 2014;62:1736–1746. doi: 10.1002/glia.22712. [DOI] [PubMed] [Google Scholar]
- 35.Atanasoski S, Boentert M, De Ventura L, Pohl H, Baranek C, Beier K, Young P, Barbacid M, Suter U. Postnatal Schwann cell proliferation but not myelination is strictly and uniquely dependent on cyclin-dependent kinase 4 (cdk4) Mol Cell Neurosci. 2008;37:519–527. doi: 10.1016/j.mcn.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Anton ES, Hadjiargyrou M, Patterson PH, Matthew WD. CD9 plays a role in Schwann cell migration in vitro. J Neurosci. 1995;15:584–595. doi: 10.1523/JNEUROSCI.15-01-00584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadjiargyrou M, Patterson PH. An anti-CD9 monoclonal antibody promotes adhesion and induces proliferation of Schwann cells in vitro. J Neurosci. 1995;15:574–583. doi: 10.1523/JNEUROSCI.15-01-00574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sbai O, Devi TS, Melone MA, Feron F, Khrestchatisky M, Singh LP, Perrone L. RAGE-TXNIP axis is required for S100B-promoted Schwann cell migration, fibronectin expression and cytokine secretion. J Cell Sci. 2010;123:4332–4339. doi: 10.1242/jcs.074674. [DOI] [PubMed] [Google Scholar]
- 39.Grijalva I, Li X, Marcillo A, Salzer JL, Levi AD. Expression of neurotrimin in the normal and injured adult human spinal cord. Spinal Cord. 2006;44:280–286. doi: 10.1038/sj.sc.3101842. [DOI] [PubMed] [Google Scholar]
- 40.Bachelin C, Zujovic V, Buchet D, Mallet J, Baron-Van Evercooren A. Ectopic expression of polysialylated neural cell adhesion molecule in adult macaque Schwann cells promotes their migration and remyelination potential in the central nervous system. Brain. 2010;133:406–420. doi: 10.1093/brain/awp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 43.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Lutz D, Kataria H, Kleene R, Loers G, Chaudhary H, Guseva D, Wu B, Jakovcevski I, Schachner M. Myelin basic protein cleaves cell adhesion molecule L1 and improves regeneration after injury. Mol Neurobiol. 2016;53:3360–3376. doi: 10.1007/s12035-015-9277-0. [DOI] [PubMed] [Google Scholar]
- 45.Gao WQ, Shinsky N, Ingle G, Beck K, Elias KA, Powell-Braxton L. IGF-I deficient mice show reduced peripheral nerve conduction velocities and decreased axonal diameters and respond to exogenous IGF-I treatment. J Neurobiol. 1999;39:142–152. doi: 10.1002/(SICI)1097-4695(199904)39:1<142::AID-NEU11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 46.Monk KR, Oshima K, Jörs S, Heller S, Talbot WS. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011;138:2673–2680. doi: 10.1242/dev.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]