Highlight
Combining RNA-sequencing analysis and illumina paired-end sequencing, we assembled genomic information for faba bean, extending the coverage of reads, as well as mapping organellar genome sequence variation.
Keywords: Illumina sequencing, legumes, mitochondrial genome, plastome, protein security, RNA-seq analysis.
Abstract
Grain legume improvement is currently impeded by a lack of genomic resources. The paucity of genome information for faba bean can be attributed to the intrinsic difficulties of assembling/annotating its giant (~13 Gb) genome. In order to address this challenge, RNA-sequencing analysis was performed on faba bean (cv. Wizard) leaves. Read alignment to the faba bean reference transcriptome identified 16 300 high quality unigenes. In addition, Illumina paired-end sequencing was used to establish a baseline for genomic information assembly. Genomic reads were assembled de novo into contigs with a size range of 50–5000 bp. Over 85% of sequences did not align to known genes, of which ~10% could be aligned to known repetitive genetic elements. Over 26 000 of the reference transcriptome unigenes could be aligned to DNA-sequencing (DNA-seq) reads with high confidence. Moreover, this comparison identified 56 668 potential splice points in all identified unigenes. Sequence length data were extended at 461 putative loci through alignment of DNA-seq contigs to full-length, publicly available linkage marker sequences. Reads also yielded coverages of 3466× and 650× for the chloroplast and mitochondrial genomes, respectively. Inter- and intraspecies organelle genome comparisons established core legume organelle gene sets, and revealed polymorphic regions of faba bean organelle genomes.
Introduction
Grain legumes (pulses) such as soybean are second only to cereals in terms of economic and nutritional value (O’Rourke et al., 2014). It is now becoming increasingly recognized that grain legumes have advantages over cereals, both in agriculture and in the human diet (Foyer et al., 2016). Pulses are rich in protein, starch, fibre, and other essential nutrients required in the human diet, and they are also valuable in the production of foodstuffs and animal feed. The protein content of pulses greatly exceeds that of cereals, and their amino acid composition is complementary (Boye et al., 2010; Roy et al., 2010). The fixation of atmospheric nitrogen in leguminous plants (Leguminosae), which occurs through the symbiotic union with soil bacteria (Barton et al., 2014; Lassaletta et al., 2014), is used as a natural means of soil nitrogen fertilization in many parts of the world. The increasing organic market, in which farmers rely on legumes to provide much of the nitrogen needed to support other crops, would favour intensification of legume-based agriculture. Although the global demand for legumes for bioenergy purposes as well as food and feed is increasing (Foyer et al., 2016), the yields of many legumes are currently unstable (Cernay et al., 2015). Crucially, the essential underpinning genetic improvement of many potentially important legumes such as faba beans (Vicia faba) is currently limited by a lack of genetic resources. Despite some advances, the development of gene-based resources in legumes such as faba bean has not kept pace with cereal crops (O’Rourke et al., 2014; Foyer et al., 2016).
According to FAOSTAT (2016), faba beans, also called broad beans, are the fourth most widely grown cool season legume after pea (Pisum sativum), chickpea (Cicer arietinum), and lentil (Lens culinaris). They have a protein content that is higher than that of many other common food legumes (Griffiths and Lawes, 1978; Burstin et al., 2011). Moreover, the total grain yield of faba bean is not sacrificed in favour of high seed protein contents. In cool climates, faba beans have advantages over legumes such as soybean because they are adapted to growth under low temperatures. As such, they are well suited to sustainable farming practices, particularly in developing countries that are routinely subject to chilling temperatures (Temesgen et al., 2015). However, faba bean yield remains unstable, as is the case with many other major legumes (Cernay et al., 2015).
The Mendelian inheritance traits for faba bean were first characterized in the 1930s (Erith, 1930); however, faba bean genetics received only intermittent interest in the following years (O’Sullivan and Angra, 2016). In the 1970s, asynaptic mutants were identified, from which a series of trisomic lines was developed and analysed (Sjodin, 1970, 1971). Subsequently, genetic markers could be assigned to physical chromosomes (vaz Patto et al., 1999). In addition, genes controlling rhizobial symbiosis and pigment composition were identified (Duc and Picard, 1986; Duc et al., 1999), with segregating recombinant inbred lines (RILs) being used to determine the Mendelian inheritance of a seed dormancy gene (Ramsay, 1997). Parental germplasm resources from early faba bean research are still available (O’Sullivan and Angra, 2016).
Use of RILs and molecular markers has allowed the production of high resolution linkage maps and the identification of quantitative trait loci (QTLs) (Torres et al., 2010; O’Sullivan and Angra, 2016). The high resolution linkage-based sequences have been used to produce maps that are syntentic with other legumes (Webb et al., 2016). For example, in a map of 687 V. faba single nucleotide polymorphism (SNP)-based linkage markers, each was matched to an orthologue in Medicago truncatula and assigned a linkage group, analogous to one of the six faba bean chromosomes, allowing a putative alignment of M. truncatula genomic regions and sequences to corresponding V. faba chromosomes (Webb et al., 2016).
Although faba bean transcriptome data have increased in recent years, surprisingly little DNA sequence data are available in public databases (Ray and Georges, 2010; Kaur et al., 2012; Arun-Chinnappa and McCurdy, 2015; Zhang et al., 2015; O’Sullivan and Angra, 2016). The only public data set of genomic DNA sequences was reported by Yang et al. (2012), who performed 454 sequencing on the pooled genomic DNA of 247 accessions in order to identify simple sequence repeat (SSR) markers. The current lack of publicly available genomic sequence data for faba bean may be attributed to the intrinsic difficulties of assembling and annotating the giant (~13 Gb) genome. However, the faba bean mitochondrial genome, which was annotated using plasmid/fosmid libraries, shows extensive repetitive content and a low degree of homology to other sequenced mitochondrial genomes (Negruck, 2013). A study on the inverted-repeat-lacking clade of legumes showed that L. culinaris and V. faba have plastomes that are smaller in size than other chloroplast genomes, but they have similar levels (6.7%) of repetitive DNA (Sabir et al., 2014). However, a blastn analysis of all intergenic spacer (IGS) regions from the sequenced plastomes failed to produce any hits for the V. faba sequence in the nucleotide database in GenBank (Sabir et al., 2014). In the following analysis, genomic and transcriptome sequencing were performed on faba bean, providing essential underpinning foundation data that can be used to improve gene identification, which is a prerequisite for future studies on the improvement of key traits of agronomic importance.
Materials and methods
The faba bean seeds (V. faba cultivar Wizard) used in the following studies were provided by Wherry and Sons Ltd, Rippingale, Lincolnshire, UK. Seeds were sown and plants were self-pollinated in the University of Leeds glasshouse facilities to produce first-generation inbred seeds, which were used in all the following experiments.
Chromosome identification
Faba bean seeds were imbibed for 3 d until germination. Germinated seeds were stored at 4 ºC for a 24 h period to allow cold-induced metaphase synchronization. The root apex was then excised, fixed, and stained for DNA, followed by chromosome imaging under a light microscope.
DNA extraction, sequencing, and assembly
Faba bean seeds were imbibed for 48 h in distilled water and the embryonic axes were excised. DNA was extracted from excised embryos using the Qiagen DNeasy plant mini kit (Qiagen Ltd, Manchester, UK). Extracted DNA was quantified by nanodrop, and sequencing was performed at the Leeds Institute of Molecular Medicine (University of Leeds, UK). An Illumina HiSeq2500 platform (Illumina, 2014) was used, running a single paired-end library on two flow cell lanes. FastQC analysis (Arun-Chinnappa and McCurdy, 2015) was performed on raw sequenced reads, and ‘Trim Galore!’ (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, last accessed 4 April 2017) was used to remove adaptor sequences using an average Phred quality score of 30. Trimmed reads were aligned to both the Cool Season Food Legume V. faba transcriptome database (CSFL; Humann et al., 2016), and the M. truncatula reference transcriptome (Benedito et al., 2008). ABySS (Simpson et al., 2009) was used for de novo paired-end sequence assembly, with an empirically determined k-mer size of 32. Alignment against full-length contigs, corresponding to those used by Webb et al. (2016) (BioProject PRJNA225873), was then performed using NCBI BLAST+ (v2.5; Camacho et al., 2009). DNA sequence reads for the Wizard cultivar are available at the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra, last accessed 4 April 2017) under the accession numbers SRR5015739 and SRR5015740.
RNA extraction, sequencing, and assembly
Faba bean (Wizard cultivar) were grown for 24 d in a controlled-environment chamber under a 12 h day/night cycle with an irradiance of 200–250 μmol m–2 s–1 and day/night temperatures of 25 °C/20 °C, respectively. Leaf discs (2 cm) were taken from the youngest mature leaves of nine plants. Discs from three biological replicates were combined and a single extraction was performed. A total of three RNA samples (representing nine individual plants) were produced. The mRNA samples were sequenced at the Leeds Institute of Molecular Medicine using an Illumina HiSeq 3000 platform producing paired-end 150 bp reads. FastQC analysis (Andrews, 2010) was performed on raw sequenced reads, and ‘Trim Galore!’ was used to remove adaptor sequences using an average Phred quality score of 30. Trimmed mRNA reads were aligned to the CSFL database (Humann et al., 2016) using Bowtie 2.3 (Langmead and Salzberg, 2012). Aligned sequences were analysed using bedtools 2.26.0 Quinlan and Hall, 2010). Trimmed sequence reads are available at the NCBI SRA (https://www.ncbi.nlm.nih.gov/sra, last accessed 4 April 2017) under the BioProject accession PRJNA369531. Single nucleotide variants (SNVs) were identified between RNA alignments and the reference using a samtools (Li, 2011) mpileup-based pipeline.
Unigene verification
Alignment data files for the RNA sequencing (RNA-seq) versus CSFL and DNA sequencing (DNA-seq) (de novo contigs of >1 kbp) were processed using a bedtools-based pipeline to identify contiguous regions of coverage within the CSFL unigenes. The DNA-seq alignments were subsequently separately processed to identify potential splice/breaks in the alignment of unspliced DNA-seq reads versus the spliced RNA reference, as indicated by peaks of bowtie softclips within a 2 bp window.
Linkage-enhanced genome contigs
Linkage marker sequences were obtained from the BioProject associated with Webb et al. (2016), with marker sequences being identified based on work performed by Khazaei et al. (2014) and Ellwood et al. (2008). Full-length sequences corresponding to these linkage markers were searched with de novo contig sequences using NCBI BLAST+.
Organellar genome comparisons
Chloroplast and mitochondrial reference sequences for V. faba and other species were downloaded from NCBI. The DNA-seq reads were aligned to the relevant reference genomes for chloroplasts and mitochondria using Bowtie2 (Langmead and Salzberg, 2012). These were used to align the DNA-seq reads and the relevant reference genome for chloroplasts and mitochondria. Samtools mpileup was used to identify SNVs. A core set of chloroplast and mitochondrial genes was aligned by BLAST between pairs of species, where one or more genes were present in annotated reference sequences from all species. SNVs in DNA-seq reads were visualized using Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013). The effect of SNVs on coding sequence changes was determined using provean (Choi et al., 2012).
Results
Genomic imaging and read assembly
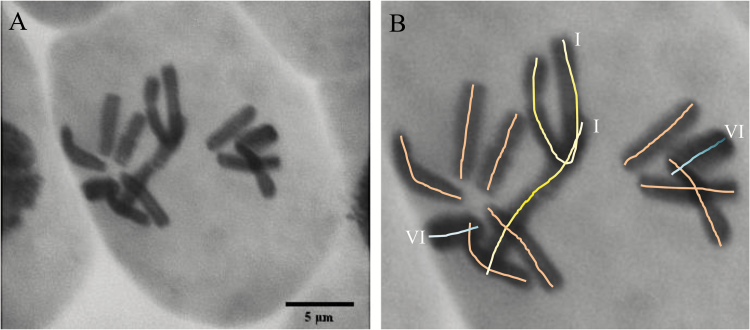
Cytogenetic analysis showed that the faba bean chromosome number was 2n=12 (Fig. 1A). The largest chromosome pair (I) and the smallest chromosome pair (VI) were identified (Fig. 1B), with chromosome I appearing to be considerably larger than the other chromosomes.
Fig. 1.
Light microscope image (magnification ×40) of Vicia faba chromosomes in metaphase. (A) Feulgen staining of DNA against a FastGreen cytoplasm stain; (B) chromosome labels with roman numerals denote the largest (I) and smallest (VI) chromosome pairs. Scale bars=5 µm.
DNA was sequenced on an Illumina HiSeq 2500 platform using 100 bp paired-end reads of a single library across two lanes. Following quality trimming, raw reads were aligned to both chloroplast and mitochondrial reference sequences (Table 1). Nearly 13% of reads mapped to the established CSFL reference coding DNA sequence (CDS) of V. faba with a total depth of 30×. A small percentage (0.95%) of the reads aligned to the chloroplast reference genome with a coverage of 3466× (Sabir et al., 2014). Similarly, 1.07% of reads aligned to the mitochondrial reference genome with a coverage of 650× (Negruk, 2013). However, 85.1% of reads could not be aligned to either the organellar reference genomes or the cool season food legume database (Table 1; Humann et al., 2016). Of the reads that could not be aligned, only ~10% were identified in a repetitive element database (Bao et al., 2015). The Gypsy class of long terminal repeat (LTR) retrotransposons formed the majority of the repetitive elements, accounting for 3.61% of the total reads. The VicSatellite, LTR (uncharacterized), and Copia class LTR elements comprised 2.49, 2.09, and 0.99% of the genome reads, respectively (Table 2).
Table 1.
Quality-trimmed genomic read data for DNA extracted from Vicia faba (cv. Wizard) detailing read number and coverage, and depth and coverage over the faba bean (CSFL) and Medicago v4 reference transcriptomes
| No. of paired reads | Reads accounted for (%) | Coverage | |
|---|---|---|---|
| Total number of reads | 812 092 660 | 100.00 | 6× |
| Reads aligned to chloroplast | 7 712 928 | 0.95 | 3466× |
| Reads aligned to mitochondria | 8 657 582 | 1.07 | 650× |
| Reads aligned to faba bean transcriptome database | 104 668 765 | 12.89 | 30× |
| Unaligned reads | 691 053 385 | 85.10 | <6× |
Table 2.
Quality-trimmed genomic read data aligned against repeat element databases, showing repetitive element identities, the number of paired reads mapped to these repetitive sequences, and the total number of reads expressed as a percentage
| Repetitive element | No. of paired reads | Total reads accounted for (%) |
|---|---|---|
| LTR/Gypsy | 29 358 977 | 3.615225 |
| VicSatellite | 20 274 464 | 2.496570 |
| LTR (uncharacterized) | 16 974 274 | 2.090189 |
| LTR/Copia | 8 090 787 | 0.996289 |
| rRNA | 1 986 302 | 0.244591 |
| Ogre | 1 708 252 | 0.210352 |
| Other/simple | 1 199 173 | 0.147665 |
| Uncharacterized | 1 054 668 | 0.129870 |
| Mobile element | 411 832 | 0.050712 |
| DNA/En-Spm | 332 062 | 0.040890 |
| Retroelement | 92 278 | 0.011363 |
| Non-LTR | 53 835 | 0.006629 |
| Other | 45 904 | 0.005653 |
| DNA | 40 982 | 0.005046 |
| DNA/MuDR | 36 393 | 0.004481 |
| DNA/hAT | 34 576 | 0.004258 |
| LINE | 21 923 | 0.002700 |
| Satellite | 9917 | 0.001221 |
| DNA/Harbinger | 8519 | 0.001049 |
| tRNA | 7049 | 0.000868 |
| DNA/TcMar | 6118 | 0.000753 |
| DNA/Mite | 3643 | 0.000449 |
| RC/Helitron | 1874 | 0.000231 |
| SINE | 883 | 0.000109 |
| DNA/hAT-Ac | 815 | 0.000100 |
| DNA/Stowaway | 344 | 0.000042 |
| DNA/Tourist | 55 | 0.000007 |
| Other/Centromeric | 52 | 0.000006 |
| DNA/TcMar-Pogo | 6 | 0.000001 |
| Repetitive element total | 81 755 957 | 10.07 |
| Genome uncharacterized | 609 297 428 | 75.03 |
Unigene model verification
To enhance the unigenes in the CSFL reference transcriptome, aligned RNA-seq data were generated (96.2 million reads, 14.4 Gbp), aligned, and analysed for breadth and depth of coverage (Table 3). RNA-seq reads showed a large number of patterns of alignment; however, we identified a core set of 16 300 unigenes which showed a single peak of alignments over 95% of the unigene, the majority of which showed an average coverage of at least 10 reads per base. We also observed a similar number with lower breadth, but with a consequent decrease in depth of coverage, as well as decreasing amounts of unigenes showing patterns of alignment peaks.
Table 3.
Alignment of RNA-seq reads from the Wizard cultivar to the CSFL faba bean transcriptome database
Peaks show independent read clustering. Breadth shows the percentage of the unigene model covered, and coverage shows the number of transcripts for which coverages of <10, 10–100, and >100 were obtained. Total shows the total number of unigenes that were present.
| No. of peaks | Breadth | Coverage | Total | ||
|---|---|---|---|---|---|
| <10 | 10–100 | >100 | |||
| 1 | >95% | 3067 | 9169 | 4064 | 16 300 |
| 50–95% | 9677 | 5338 | 1094 | 16 109 | |
| <50% | 4280 | 47 | 2 | 4329 | |
| 2–3 | >95% | 326 | 1515 | 271 | 2112 |
| 50–95% | 4339 | 2976 | 268 | 7583 | |
| <50% | 1198 | 18 | 2 | 1218 | |
| 4–10 | >95% | 14 | 70 | 5 | 89 |
| 50–95% | 1176 | 429 | 17 | 1622 | |
| <50% | 282 | 8 | 0 | 290 | |
| 0% | 0 | ||||
| >10 | >95% | 1 | 0 | 0 | 1 |
| 50–95% | 15 | 1 | 0 | 16 | |
| <50% | 3 | 0 | 0 | 3 | |
In addition, to identify potential genomic structure and putative splice points, we aligned the DNA-seq reads to the CSFL unigenes, and searched the alignment files for indications of localized DNA-seq read alignment breaks (see Supplementary Table S1 at JXB online). This uncovered 25 535 unigenes with at least one potential splice site (total 56 668).
De novo assembly and linkage map alignment
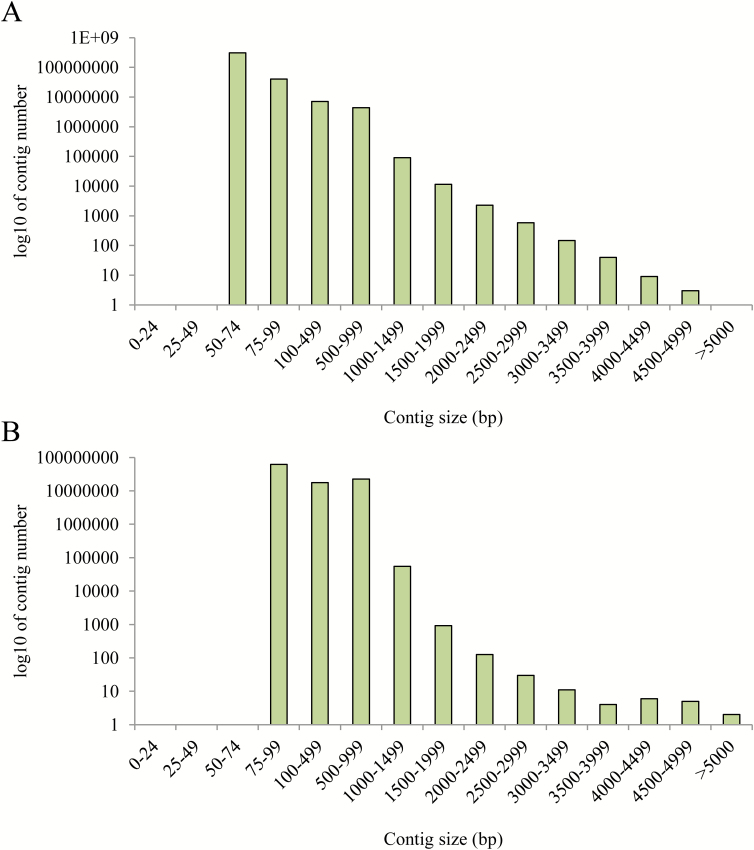
ABySS assembly of short reads resulted in the generation of >361 million contigs, which varied in length between 50 bp and 5 kb (Fig. 2A, B). The abundance of contigs that were generated was highly dependent on k-mer size (Fig. 2A), with 32 bp k-mers showing a wider range of contig sizes, together with a higher number of contigs compared with those produced using a k-mer size of 64 bp (Fig. 2B). The contigs shown in Fig. 2 were filtered to a subset of 14 682 with a size of ≥1 kbp and aligned against the raw contigs used in the synteny-based SNP linkage map produced by Webb et al. (2016). Using a criterion of >95% sequence identity over >95% of a contig, this alignment confirmed the assembled sequence of 94 of the de novo contigs, which were smaller than those from Webb et al. (2016). The alignment with the existing contigs has also produced an increase in the length of another 461 of these raw loci contigs. A complete list of the syntenic marker-related contigs and the aligned de novo contig sequences can be found in Supplementary Table S2. Existing loci contigs were increased by an average of 874 bp (in a range of 55–2995 bp).
Fig. 2.
(A) The log10 contig abundance over a range of contig sizes, produced from 32 bp k-mers; (B) the log10 contig abundance over a range of contig sizes, produced from 64 bp k-mers.
Single nucleotide polymorphisms in the organellar genome of Vicia faba
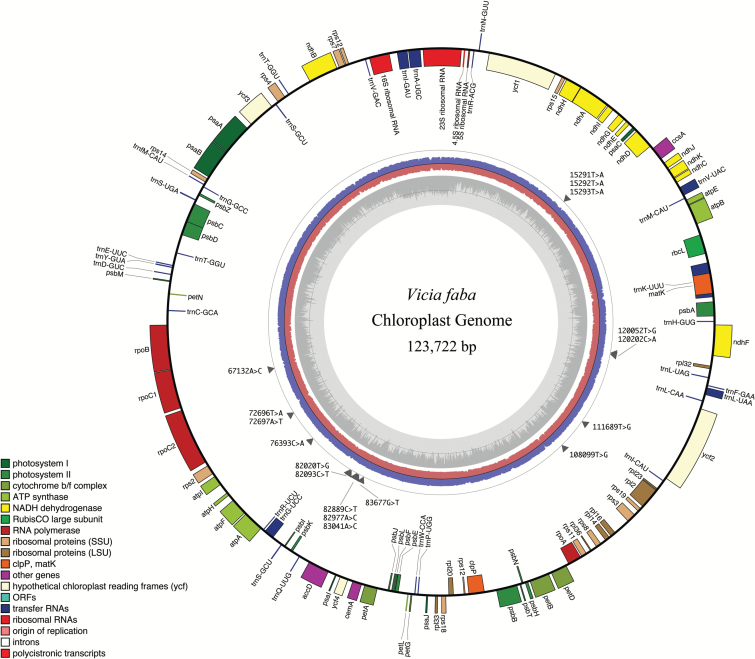
Species-level variation within the organellar genomes of V. faba raw DNA sequence reads was investigated by mapping against the reference sequences for the chloroplast and mitochondria (Negruk, 2013; Sabir et al., 2014). The locations of gene-based variations within the V. faba chloroplast genome (Fig. 3) were obtained by aligning the raw DNA sequence reads to the published faba bean chloroplast sequence (Sabir et al., 2014). While a high level of homology was found between the published reference sequence and the chloroplast genome reported here, a number of SNPs were identified (Table 4). Four genes encoding the RNA polymerase β chain, ATPase subunit I, tRNA Q, and ribosomal protein S3 had a single polymorphism (Table 4). The gene encoding the acetyl co-A carboxylase subunit (accD) showed three SNPs. The data in Table 4 show whether the SNPs are synonymous or non-synonymous.
Fig. 3.
Vicia faba chloroplast genome map. The outermost circle shows gene identities and read direction, with outward-facing genes being in the positive direction and inward-facing genes being in the negative direction. SNPs are denoted by grey triangles with the location and nature of polymorphism detailed. Read depth is shown for the forward reads (blue ring) and reverse reads (red ring). GC content (%) is shown by the innermost grey circle, with 50% being denoted by the dark grey line. Gene mapping and annotation were performed in OGDRAW.
Table 4.
Single nucleotide polymorphisms in the chloroplast genome of Vicia faba, comparing the Wizard cultivar with the reference sequence published by Sabir et al. (2014)
The amino acid change is provided for non-synonymous mutations including the predicted effect on the protein calculated with Provean. SYN is a synonymous effect.
| Map identifier | Gene identity | SNP position | Reference base | Sequenced base | Effect | Provean score |
|---|---|---|---|---|---|---|
| rpoC1 | RNA polymerase β chain | 67 132 | A | C | M→L | –1.687 |
| atpF | ATPase subunit I | 76 393 | C | A | – | – |
| trnQ-UUG | tRNA | 82 093 | G | A | – | – |
| accD | Acetyl co-A carboxylase subunit | 82 977 | A | C | Y→S | 0.775 |
| accD | Acetyl co-A carboxylase subunit | 83 041 | A | C | L→F | –0.77 |
| accD | Acetyl co-A carboxylase subunit | 83 677 | G | T | SYN | – |
| rps3 | Ribosomal protein S3 | 108 099 | T | G | K→T | –2.802 |
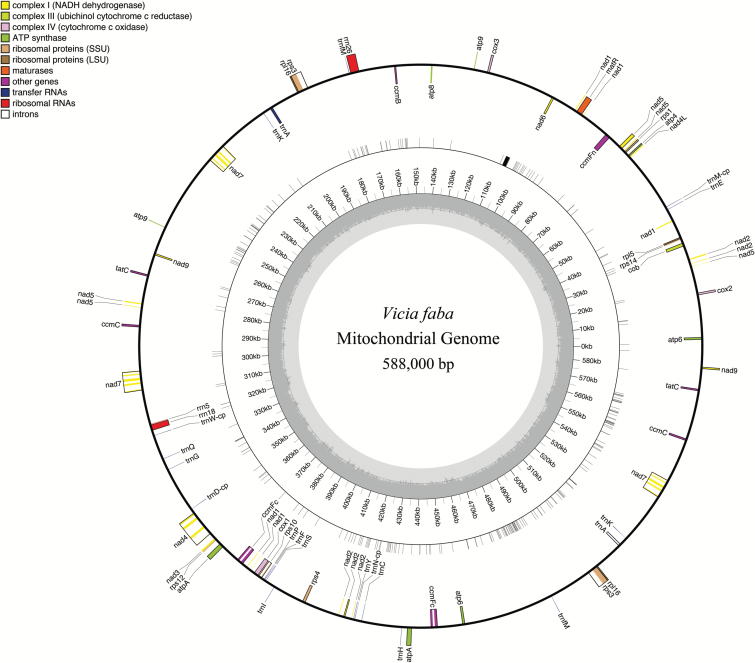
A faba bean mitochondrial genome reference map (Fig. 4) based on genomic read alignment was constructed relative to the cultivar Broad Windsor (Negruk, 2013). While a high level of homology was found between the gene space published by Negruk (2013) and that assembled from raw genomic reads of the Wizard cultivar, seven genes were found to have polymorphisms (Table 5). Analysis of within-species variation for the mitochondrial genome (Fig. 5) revealed a high number of SNPs in the gene coding and intergenic regions. The number of polymorphisms present varied between genes, with the genes encoding 26S rRNA, 18S rRNA, and NADH dehydrogenase subunit 4 having one SNP and the genes encoding maturase R and NADH dehydrogenase subunit 6 having two SNPs. The genes encoding ribosomal protein S14 and cytochrome c oxidase subunit II had six SNPs. We checked whether the SNPs found in the chloroplast and mitochondrial genomes will cause an amino acid change (i.e. non-synonymous SNPs). The data in Tables 4 and 5 show that a number of the observed SNPs will change the amino acid and thus they have the potential to have an effect on the functioning of the protein.
Fig. 4.
Vicia faba mitochondrial genome map. The outermost circle shows gene identities and read direction, with outward-facing genes being in the positive direction and inward-facing genes being in the negative direction. SNPs are denoted by grey bars on the second concentric circle. Base pair positioning is detailed in the third circle. GC content (%) is shown by the innermost grey circle, with 50% being demarked by the dark grey line. The key provides details of mitochondrial gene families. Gene mapping and annotation were performed in OGDRAW.
Table 5.
Single nucleotide polymorphisms in the mitochondrial genome of Vicia faba, comparing the Wizard cultivar with the reference sequence published by Negruk (2013)
The amino acid change is provided for non-synonymous mutations including the predicted effect on the protein calculated with Provean. SYN is a synonymous effect.
| Map identifier | Gene identity | SNP position | Reference base | Sequenced base | Effect | Provean score |
|---|---|---|---|---|---|---|
| rrn26.r01 | 26S rRNA | 167 803 | T | A | – | – |
| rrn18.r01 | 18S rRNA | 321 111 | T | G | – | – |
| rps14 | Ribosomal protein S14 | 36 460 | T | G | SYN | – |
| rps14 | Ribosomal protein S14 | 36 461 | C | G | G→A | –0.404 |
| rps14 | Ribosomal protein S14 | 36 462 | C | A | G→STOP | – |
| rps14 | Ribosomal protein S14 | 36 463 | A | T | SYN | – |
| rps14 | Ribosomal protein S14 | 36 490 | G | T | F→L | –1.059 |
| rps14 | Ribosomal protein S14 | 36 616 | A | G | SYN | – |
| nad6 | NADH dehydrogenase subunit 6 | 101 407 | A | T | SYN | – |
| nad6 | NADH dehydrogenase subunit 6 | 101 508 | G | T | L→I | –0.026 |
| nad4.CDS.3 | NADH dehydrogenase subunit 4 | 361 807 | T | G | F→L | 4.0 |
| matR | Maturase R | 90 908 | G | T | L→F | 1.622 |
| matR | Maturase R | 92 082 | A | C | K→Q | –0.223 |
| ORF106 | Hypothetical protein | 25 588 | C | A | H→N | –7.0 |
| ORF295 | Putative succinate dehydrogenase | 25 760 | C | A | H→N | 0.6 |
| ORF295 | Putative succinate dehydrogenase | 25 862 | T | A | F→I | 0.044 |
| ORF295 | Putative succinate dehydrogenase | 25 863 | T | A | F→Y | –0.103 |
Fig. 5.
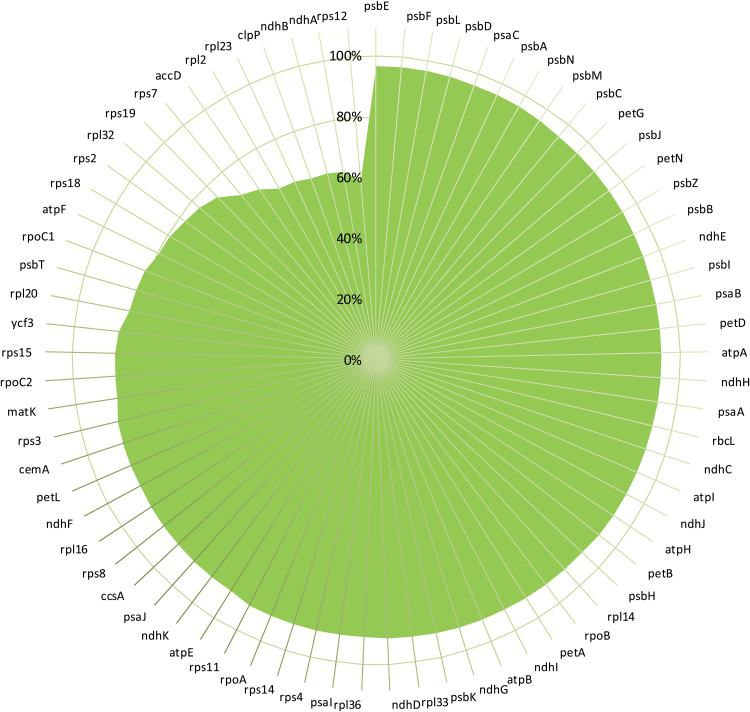
The percentage sequence identity for chloroplast-encoded genes, showing the average data for a 55-way alignment for individual chloroplast genes of 11 species. Gene identities are given in the outermost circle. The scale shows the homology percentage (0–100%). Species comparisons were performed for Glycine max (G.m), Lotus japonicus (L.j), Vigna unguiculata (V.u), Lens culinaris (L.c), Cicer arietinum (C.a), Vigna radiata (V.r), Pisum sativum (P.s), Medicago truncatula (M.t), Arachis hypogaea (A.h), Phaseolus vulgaris (P.v), and Vicia faba (V.f).
Comparison of legume organelle genome sequences
The coding regions of the faba bean chloroplast and mitochondrial genomes obtained from the resources provided by Sabir et al. (2014) and Negruk (2013) were compared with those of other legume organelle genomes that are available on the NCBI database (Benson et al., 2013). Genome sequence alignment of 11 reference chloroplast sequences available for legumes (Glycine max, Vicia faba, Lotus japonicus, Vigna unguiculata, L. culinaris, C. arietinum, Vigna radiata, P. sativum; M. truncatula; Arachis hypogaea, and Phaseolus vulgaris) allowed for the identification of a core set of chloroplast genes that are conserved across all legumes (Fig. 5). Chloroplast sequences were well conserved (>80% for most genes encoding photosynthetic electron transport proteins). However, the percentage homology was lower in genes encoding ribosomal proteins. Between the plastid genomes, the greatest sequence variations were found in clpP, ndhA, and rps12, which showed <60% homology. The least variable genes were psbE, psbF, nd psbD, all of which had a homology >95%. The psb genes encode proteins that are part of PSII and therefore may be strongly conserved between species.
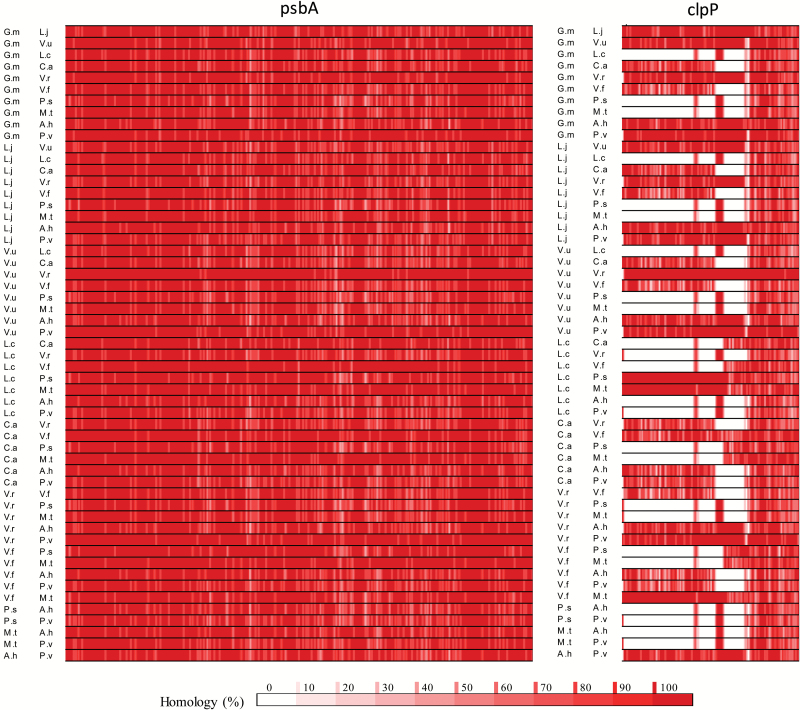
Chloroplast genome variation was further analysed in homology maps in a pairwise manner for each gene and species. While psbA shows a very high homology across all species, clpP genes show far more interspecies variation, with large regions being absent from the coding sequences of L. culinaris, M. truncatula, and P. sativum (Fig. 6). Comparisons for all genes can be found in Supplementary Table S3. However, for simplicity, representative examples are shown in Fig. 7 for the psbA and clpP genes.
Fig. 6.
Representative examples of sequence identity maps drawn for the highly homologous psbA gene and the clpP gene with low homology. Maps were constructed from a multiway homology comparison between 11 legume species; Glycine max (G.m), Lotus japonicus (L.j), Vigna unguiculata (V.u), Lens culinaris (L.c), Cicer arietinum (C.a), Vigna radiata (V.r), Pisum sativum (P.s), Medicago truncatula (M.t), Arachis hypogaea (A.h), Phaseolus vulgaris (P.v), and Vicia faba (V.f). High degrees of homology between compared species are shown in dark red, while a lack of detectable matches is shown in white. Quantitation is as shown in the scale bar. Homology was determined across a 10 bp frame (i.e. a single polymorphism would give 90% homology).
Fig. 7.
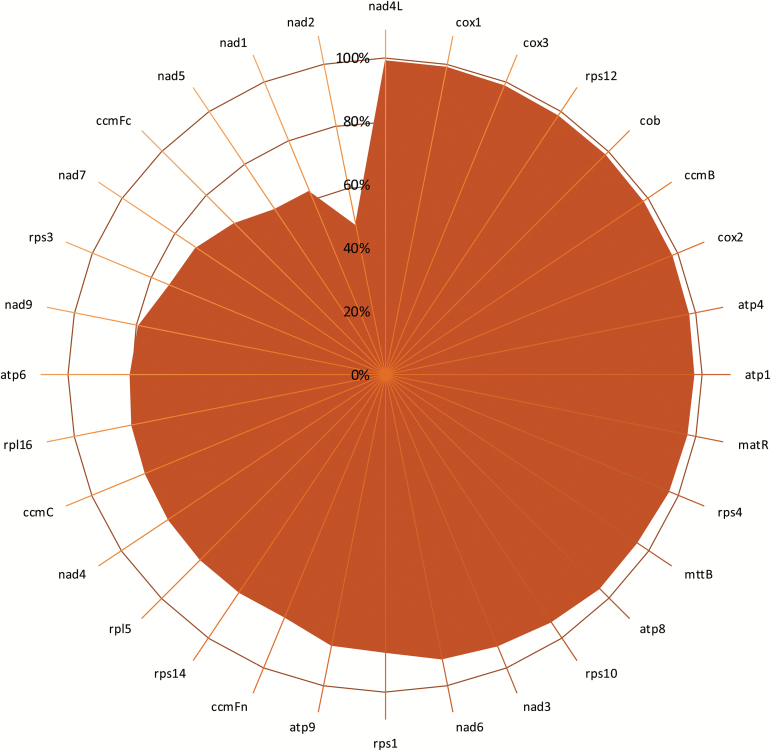
The percentage sequence identity for genes encoded in the mitochondrial genome. Average data for individual mitochondrial genes from six species were subjected to a 15-way alignment. Gene identities are given in the outermost circle. The scale shows the percentage sequence identity (0–100%). Species comparisons were performed for Medicago truncatula (M.t), Lotus japonicus (L.j), Glycine max (G.m), Vigna angularis (V.a), Vigna radiata (V.r), and Vicia faba (V.f).
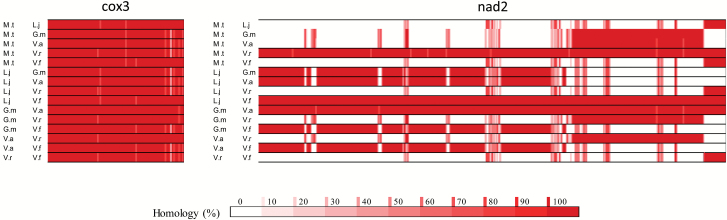
Reference mitochondrial genomes were available for six legume species on the NCBI database (Benson et al., 2013). These sequences were compared to define a core set of conserved mitochondrial genes. Mitochondrial genome sequences are well conserved (Fig. 7), with all of the cytochrome-associated genes having an average sequence homology of >90%. However, genes encoding ribosomal proteins and the NADH dehydrogenase complex had a lower average sequence homology (Fig. 7). The most variable genes across the mitochondrial genomes were nad5, nad1, and nad2 which showed <60% homology across the legume sequences analysed. The least variable genes were nad4L, cox1, and cox3, all of which had a homology >95%. Examples of the most (nad2) and least variable genes (cox3) are shown in Fig. 8.
Fig. 8.
Percentage homology maps for cox3 and nad2. Maps were constructed from a multiway homology comparison of six legume species: Medicago truncatula (M.t), Lotus japonicus (L.j), Glycine max (G.m), Vigna angularis (V.a), Vigna radiata (V.r), and Vicia faba (V.f). High degrees of homology are shown in dark red, and a lack of detectable match is shown in white, with quantitation as shown in the scale bar.
Discussion
While most plant species have small genomes (Kelly and Leitch, 2011), the data presented here confirm that faba bean belongs to those lineages that have ‘giant genomes’. This is rather surprising given that faba bean is diploid (2n=12 chromosomes). Chromosome fusion is known to have occurred during faba bean evolution (Fuchs et al., 1998). As with other large genomes, genome size expansion can be accounted for by repetitive DNA sequences that consist of different types of transposable elements. In the data presented here, only 9.64% of reads could be identified in a repetitive element database (Bao et al., 2015). However, many of the repetitive elements in the faba bean genome remain to be annotated and do not yet have a homologue in this database. Large plant nuclear genomes, such as that of maize, can contain 85% repetitive DNA made up of intact and simply organized retrotransposons (RTs; Schnable et al., 2009). Many RTs that use element-encoded mRNAs as transposition intermediates can rapidly proliferate in copy number, resulting in large differences in genome sizes between related species. The action of these mobile elements can restructure the genome, resulting in a high level of genetic variability. Moreover, these RT-induced mutations are usually stable and can be used as molecular tools to study gene tagging and functional analysis (Bennetzen, 2000). The Gypsy and Copia type RTs belong to the LTR family and are referred to as Metaviridae and Pseudoviridae elements, respectively (Grandbastien, 2015). LTR replication via an RNA template is reminiscent of retroviral proliferation, and interestingly these are the predominant RTs in plants. Genome fluidity arising from the activities of these mobile elements has long been recognized as an opportunity for the genome to evolve in response to environmental challenges (Lisch, 2002; Wicker et al., 2007). The expression of LTR RTs is important in host functional plasticity, with regulation being co-ordinated in response to a diverse array of external stresses (Reichmann et al., 2012; Grandbastien, 2015; Giordani et al., 2016).
RNA-seq data were used in the present study to identify a set of high confidence unigenes through alignment to the faba bean CSFL reference transcriptome. This analysis yielded a total of 16 300 unigenes with full depth coverage identified in CSFL. This subset was more likely to have an alignment to known genes than the full set of unigenes (71.2% versus 53.2%), but the alignment was no more likely to be longer (21.5 bp versus 21.0 bp) or to have a higher identity (82.2% versus 79.6%). The data were also more likely to yield fewer aligned DNA-seq read clips, indicative of transcriptome assembly breaks compared with the genomic architecture from which they originate. By combining the genomic sequence data presented here with the uncropped linkage marker data produced by Webb et al. (2016), sequence length data were increased at 461 chromosomal loci, extending current CDS sequence data into the genomic space.
Although the present characterization of the nuclear genome is challenging given the length and coverage of genomic reads, the mapping of the organellar genomes was much more informative. By comparing the plastome sequence reported here with the publicly available sequences (Sabir et al., 2014), we identified relatively few variants in the coding regions. However, SNPs were identified in the genes encoding the RNA polymerase subunit C1 (rpoC1), the ATpase β subunit (atpF), glutamine tRNA (trnQ-UUG), and the β-carboxyl transferase subunit of acetyl co-A carboxylase (accD). Among these, accD contained the most SNPs, with three detectable polymorphisms. While the tobacco accD knockout mutants are lethal (Kode et al., 2005), the accD gene was shown to be highly variable across 24 M. truncatula ecotypes (Csanad and Pal, 2014). Ecotype-specific accD gene sequences ranging from 650 to 796 amino acids in length were reported (Csanad and Pal, 2014). Despite this variability, a conserved carboxylase domain was always present, and gene length expansion and contraction was attributed to repetitive sequence integration in intronic regions (Csanad and Pal, 2014). The accD gene has been lost during evolution in a number of species (Rousseau-Gueutin et al., 2013), but in such situations it has been functionally replaced by a nuclear-encoded analogue (Rousseau-Gueutin et al., 2013).
Intraspecies comparisons of the faba bean chloroplast genes reported here may form the basis for future cultivar genotyping. A low level of plastome gene sequence variation was found amongst the 11 legumes analysed in the present study. There were notable exceptions however, particularly in the rps2, rpl32, rps19, rps7, accD, rpl2, rpl23, clpP, ndhB, ndhA, and rps12 genes, which showed sequence homologies of only between 60% and 79%. These findings are consistent with the literature view that most photosynthesis-related genes are conserved to a higher level than ribosomal genes, with chloroplast genomes showing far higher degrees of conservation than the nuclear and mitochondrial genomes (Xu et al., 2015). Despite the high level of variation in the rps12 gene, loss of rps12 function might be predicted to lead to impaired chloroplast translation (Ramundo et al., 2013).
The number of polymorphisms in the mitochondrial genome was greater than those detected in the plastome, with six genes showing single base pair mutations compared with the reference sequence (Negruk, 2013). Two SNPs were identified in NADH dehydrogenase subunit 6 and maturase R, with ribosomal protein S14 and cytochrome c oxidase subunit II having six SNPs. In comparison with the chloroplasts, the mitochondria were found to be more prone to the accumulation of SNPs, although this was restricted to the intergenic spaces. Plant mitochondrial genomes are characterized by their very large size, ranging from 200 kb to 2500 kb, with many introns and repeated elements (Kitazaki et al., 2010). Mitochondrial genome recombination is key to within- and between-species variation (Galtier, 2011). However, the rate of nucleotide substitution is low, mitochondrial genome variation within species being derived from dsDNA breakage and repair via highly efficient recombination of asymmetric sequences (Davila et al., 2011).
In conclusion, the novel sequence data presented here provide an essential foundation for the generation of a draft nuclear genome sequence and further analysis of organellar genomes. The transcriptome reference for faba bean in the CSFL database is comprised of data from multiple cultivars. The data reported here for Wizard match this genomic resource but have advantages over the existing CSFL faba bean resource because they provide increased accuracy (Table 3). Given the rapid advances in sequencing technologies, it is feasible that a high quality faba bean reference genome will be produced in the near future by intense long-read sequencing, accompanied by innovative bioinformatics approaches. Unravelling the nuclear and organellar genomes of faba bean, their plasticity, and regulation in response to environmental challenges will not only increase our understanding of how stress tolerance is regulated but will also accelerate breeding progress. These data also provide an essential underpinning resource required for the genetic improvement of faba bean, an important cold season legume, paving the way for future realization of its considerable potential for improvement.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of putative splice points identified by aligning DNA-seq reads to the CSFL.
Table S2. List of full length markers upon which the publication of Webb et al. (2016) was based, with aligned DNA contigs and sequence extension data.
Table S3. Pairwise alignments of sequences of identified genes in the chloroplast and mitochondrial genomes.
Supplementary Material
Acknowledgements
This work was funded by a BBSRC CASE studentship (BB/K501839/1) in association with Wherry and Sons Ltd, The Old School, High Street, Rippingale, Lincolnshire PE10 0SR, UK. We are indebted to A. Cuming and Y. Kamisugi for their guidance in the production of Fig. 1. We thank I. Carr and his group at the Leeds Institute of Molecular Medicine, University of Leeds, UK for the sequencing of DNA and RNA samples. This work was undertaken on MARC1, part of the High Performance Computing facilities at the University of Leeds, UK.
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Retrieved on 4 April 2017.
- Arun-Chinnappa KS, McCurdy DW. 2015. De novo assembly of a genome-wide transcriptome map of Vicia faba (L.) for transfer cell research. Frontiers in Plant Science 6, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L, Thamo T, Engelbrecht D, Biswas WK. 2014. Does growing grain legumes or applying lime cost effectively lower greenhouse gas emissions from wheat production in a semi-arid climate? Journal of Cleaner Production 83, 194–203. [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, et al. 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. 2000. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology 42, 251–269. [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Research 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye J, Zare F, Pletch A. 2010. Pulse proteins: processing, characterization, functional properties and applications in food and feed. Food Research International 43, 414–431. [Google Scholar]
- Burstin J, Gallardo K, Mir RR, Varshney RK, Duc G. 2011. Improving protein content and nutrition quality. In: Pratap A, Kumar J, eds. Biology and breeding of food legumes. Wallingford, UK, CAB International, 314–328. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernay C, Ben-Ari T, Pelzer E, Meynard JM, Makowski D. 2015. Estimating variability in grain legume yields across Europe and the Americas. Scientific Reports 5, 11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanad G, Pal M. 2014. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Research 21, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, Arrieta-Montiel MP, Wamboldt Y, Cao J, Hagmann J, Shedge V, Xu YZ, Weigel D, Mackenzie SA. 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biology 9, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc G, Moussy F, Zong X, Ding G. 1999. Single gene mutation for green cotyledons as a marker for the embryonic genotype in faba bean, Vicia faba. Plant Breeding 118, 577–578. [Google Scholar]
- Duc G, Picard J. 1986. Note on the presence of the sym-1 gene in Vicia faba hampering its symbiosis with Rhizobium leguminosarum. Euphytica 35, 61–64. [Google Scholar]
- Ellwood SR, Phan HT, Jordan M, Hane J, Torres AM, Avila CM, Cruz-Izquierdo S, Oliver RP. 2008. Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 9, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erith AG. 1930. The inheritance of colour, size and form of seeds, and of flower colour in Vicia faba L. Genetica 12, 477–510. [Google Scholar]
- FAOSTAT 2016. United Nations Food and Agriculture Organisation Website http://www.fao.org/faostat/en/ Retrieved on 4 April 2017.
- Foyer CH, Lam HM, Nguyen HT, et al. 2016. Neglecting legumes has compromised human health and sustainable food production. Nature Plants 2, 16112. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Strehl S, Brandes A, Schweizer D, Schubert I. 1998. Molecular–cytogenetic characterization of the Vicia faba genome—heterochromatin differentiation, replication patterns and sequence localization. Chromosome Research 6, 219–230. [DOI] [PubMed] [Google Scholar]
- Galtier N. 2011. The intriguing evolutionary dynamics of plant mitochondrial DNA. BMC Biology 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani T, Cossu RM, Mascagni F, Marroni F, Morgante M, Cavallini A, Natali L. 2016. Genome-wide analysis of LTR-retrotransposon expression in leaves of Populus × canadensis water-deprived plants. Tree Genetics and Genomes 12, 75. [Google Scholar]
- Grandbastien MA. 2015. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochimica et Biophysica Acta 1849, 403–416. [DOI] [PubMed] [Google Scholar]
- Griffiths DW, Lawes DA. 1978. Variation in the crude protein content of field beans (Vicia faba L.) in relation to the possible improvement of the protein content of the crop. Euphytica 27, 487–495. [Google Scholar]
- Humann JL, Jung S, Zheng P, et al. 2016. Cool season food legume genome database: an up-to-date resource enabling genetics, genomics and breeding research in pea, lentil, faba bean and chickpea. Proceedings of the International Plant and Animal Genome Conference San Diego, CA, USA. [Google Scholar]
- Illumina 2014. Sequencing methods review. A review of publications featuring Illumina Technology. Illumina Protocols 199, 27. [Google Scholar]
- Kaur S, Pembleton LW, Cogan NO, Savin KW, Leonforte T, Paull J, Materne M, Forster JW. 2012. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, Leitch IJ. 2011. Exploring giant plant genomes with next-generation sequencing characterization of the Vicia faba genome. Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Research 6, 219–230. [DOI] [PubMed] [Google Scholar]
- Khazaei H, O’Sullivan DM, Sillanpää MJ, Stoddard FL. 2014. Use of synteny to identify candidate genes underlying QTL controlling stomatal traits in faba bean (Vicia faba L.). Theoretical and Applied Genetics 127, 2371–2385. [DOI] [PubMed] [Google Scholar]
- Kitazaki K, Kubo T, Kitazaki K, Kubo T. 2010. Cost of having the largest mitochondrial genome: evolutionary mechanism of plant mitochondrial genome. Journal of Botany 2010, 1–12. [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. 2005. The tobacco plastid accD gene is essential and is required for leaf development. The Plant Journal 44, 237–244. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaletta L, Billen G, Grizzetti B, Anglade J, Garnier J. 2014. 50 year trends in nitrogen use efficiency of world cropping systems: the relationship between yield and nitrogen input to cropland. Environmental Research Letters 9, 105011. [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2002. Mutator transposons. Trends in Plant Science 7, 498–504. [DOI] [PubMed] [Google Scholar]
- Negruk V. 2013. Mitochondrial genome sequence of the legume Vicia faba. Frontiers in Plant Science 4, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JA, Bolon YT, Bucciarelli B, Vance CP. 2014. Legume genomics: understanding biology through DNA and RNA sequencing. Annals of Botany 113, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan DM, Angra D. 2016. Advances in Faba bean genetics and genomics. Frontiers in Genetics 7, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G. 1997. Inheritance and linkage of a gene for testa-imposed seed dormancy in faba bean (Vicia faba L.). Plant Breeding 116, 287–289. [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix JD. 2013. Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. The Plant Cell 25, 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H, Georges F. 2010. A genomic approach to nutritional, pharmacological and genetic issues of faba bean (Vicia faba): prospects for genetic modifications. GM Crops 1, 99–106. [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Huang X, Higginson E, Ayliffe M, Day A, Timmis JN. 2013. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiology 161, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, Meehab RR, Adams IR. 2012 Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS Computational Biology 8, e1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy F, Boye JI, Simpson BK. 2010. Bioactive proteins and peptides in pulse crops: pea, chickpea and lentil. Food Research International 43, 432–442. [Google Scholar]
- Sabir J, Schwarz E, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen R, Ruhlman T. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnology Journal 12, 743–754. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Research 19, 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin J. 1970. Induced asynaptic mutants in Vicia faba L. Hereditas 66, 215–232. [Google Scholar]
- Sjodin J. 1971. Induced morphological variation in Vicia faba L. Hereditas 67, 155–180. [Google Scholar]
- Temesgen T, Keneni G, Sefera T, Jarso M. 2015. Yield stability and relationships among stability parameters in faba bean (Vicia faba L.) genotypes. Crop Journal 3, 258–268. [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AM, Avila CM, Gutierrez N, Palomino C, Moreno MT, Cubero JI. 2010. Marker-assisted selection in faba bean (Vicia faba L.). Field Crops Research 115, 243–252. [Google Scholar]
- vaz Patto MC, Torres AM, Koblizkova A, Macas J, Cubero JI. 1999. Development of a genetic composite map of Vicia faba using F2 populations derived from trisomic plants. Theoretical and Applied Genetics 98, 736–743. [Google Scholar]
- Webb A, Cottage A, Wood T, et al. 2016. A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.). Plant Biotechnology Journal 14, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. 2007. A unified classification system for eukaryotic transposable elements. Nature Reviews. Genetics 8, 973–982. [DOI] [PubMed] [Google Scholar]
- Xu JH, Liu Q, Hu W, Wang T, Xue Q, Messing J. 2015. Dynamics of chloroplast genomes in green plants. Genomics 106, 221–231. [DOI] [PubMed] [Google Scholar]
- Yang T, Bao SY, Ford R, et al. 2012. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics 13, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Wheeler S, Xia X, Radchuk R, Weber H, Offler CE, Patrick JW. 2015. Differential transcriptional networks associated with key phases of ingrowth wall construction in trans-differentiating epidermal transfer cells of Vicia faba cotyledons. BMC Plant Biology 15, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.