Abstract
Sequence similarity tools like Basic Local Alignment Search Tool (BLAST) are essential components of many functional genetic, genomic, phylogenetic and bioinformatic studies. Many modern analysis pipelines use significant sequence similarity scores (p- or E-values) and the ranked order of BLAST matches to test a wide range of hypotheses concerning homology, orthology, the timing of de novo gene birth/death and gene family expansion/contraction. Despite significant contrary findings, many of these tests still implicitly assume that stronger or higher-ranked E-value scores imply closer phylogenetic relationships between sequences. Here, we demonstrate that even though a general relationship does exist between the phylogenetic distance of two sequences and their E-value, significant and misleading errors occur in both the completeness and the order of results under realistic evolutionary scenarios. These results provide additional details to past evidence showing that studies should avoid drawing direct inferences of evolutionary relatedness from measures of sequence similarity alone, and should instead, where possible, use more rigorous phylogeny-based methods.
Keywords: phylogenetics, sequence similarity, BLAST, rate heterogeneity, phylostratigraphy, compositional bias
Introduction
Local sequence alignment tools are central to many molecular comparative analyses and informatics pipelines. The Basic Local Alignment Search Tool (BLAST) [1] revolutionized the speed with which sequences could be compared with large databases. As such, BLAST has become essential in many analyses ranging from assessment of gene homology, orthology and annotation to large-scale phylogenetics, phylogenomics and phylostratigraphy [2–13]. While BLAST is ubiquitously used to address questions in these areas, its specific uses and interpretations vary widely.
Unlike exact search approaches like Smith-Waterman that guarantee optimal local alignments [14], BLAST uses a heuristic method to quickly produce significant local alignments and provide several similarity scores. Alignments that have scores above a specified threshold are presented in ranked order by significance. The significance score often used by BLAST users is the E-value, which is interpreted as the expected number of random alignments with at least the same quality as the alignment calculated by BLAST between the query and subject sequence. The smaller the E-value, the fewer random alignments are expected for the given parameters.
While the BLAST algorithm was intended for simple sequence or motif similarity searches, modern usages make significantly broader assumptions. One common application of sequence similarity programs is the interpretation of reciprocal best hits (RBH) between species as homologous or orthologous [15, 16]. However, homology and orthology are hypotheses concerning common descent (rather than mere similarity) and are therefore phylogenetic in nature [9]. Using sequence similarity alone has been considered insufficient evidence for identifying common ancestry (i.e. orthology or homology) [17, 18] and imposes a potentially unsupported evolutionary interpretation on sequence similarity scores. Other applications that rely on the rank-order of results from BLAST make the same assumption. Many phylogenomic and phylogenetic analysis pipelines use either BLAST or other related similarity scores for homolog and ortholog identification at an early stage, including eggNOG, OrthoMCL, OMA, HaMStR and OrthoFinder [2, 3, 19–22]. While alternatives that incorporate phylogeny exist (e.g. [9, 23, 24]), rank-based BLAST analyses are still common.
Phylostratigraphy, another research approach that relies on sequence similarity searches, attempts to determine the age of a gene based on the phylogenetic completeness of sequence similarity hits [4, 5, 25, 26]. Phylostratigraphy assumes sequence similarity searches will return unbiased and complete (or nearly complete) significant hits from a sequence database. Recent examinations have demonstrated that differences in molecular rates of evolution and gene length can bias phylostratigraphy results [12], but these analyses primarily focus on biases related to molecular properties of the sequences themselves rather than phylogenetic bias.
Several factors may cause BLAST results not to reflect phylogenetic relationships (Figure 1). In general, sequence similarity measures presumably suffer from the same problems that complicate all distance-based phylogenetic measures and methods [27]. In addition, the specific challenges that affect phylogenetic reconstruction should also affect BLAST results (Figure 1), including lineage-specific rate heterogeneity, saturation and non-stationarity of composition [8, 27–30]. Even sophisticated molecular evolutionary models that accommodate for these processes can still have difficulty reconstructing phylogenies [8, 31], and therefore these factors are expected to complicate BLAST results as well.
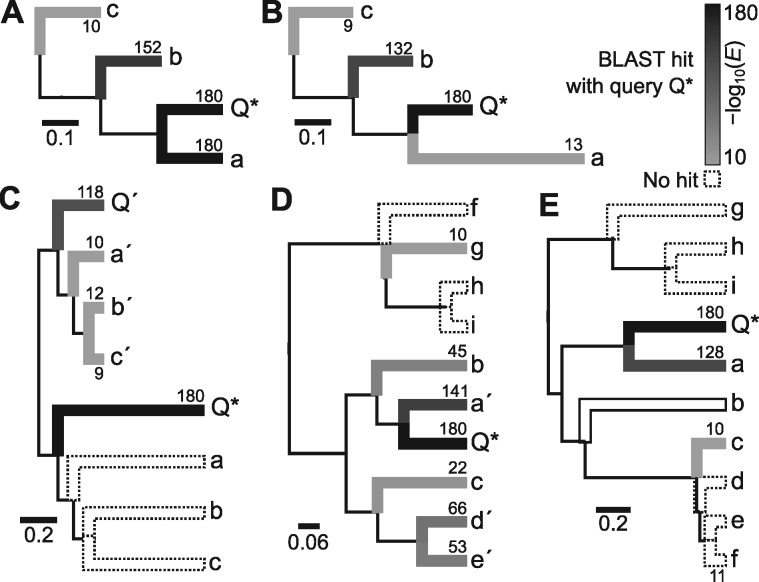
Figure 1.
Examples of phylogenetic scenarios that pose potential problems for BLAST analyses. Each scenario shows the ‘true tree’ over which sequences have evolved. Branch intensities and labels show –(E) for BLAST hits using query sequence against all other sequences in the tree, where higher values indicate a stronger match and white with dotted lines indicates no BLAST hit. (A) A simple example where BLAST of results in E-values that reflect phylogenetic distances. All branch lengths are 0.125. (B) An example where lineage-specific rate heterogeneity will mislead in the order of results. The branch length of a is twice that of . The first hit for is b and not a. (C) An example of a gene duplication where each ortholog ( and ) has a different local rate of molecular evolution. A BLAST of in the faster gene hits only the slow ortholog and incorrectly hits first. (D) An example of compositional bias where have biased and similar nucleotide composition and other sequences have equal composition. A BLAST of hits, in order, . (E) A simple example demonstrating the problem of saturation. The height of this tree is such that BLAST of does not hit each sequence and instead only hits, in order, a, c, f.
Understanding the biases and expectations for sequence similarity analyses and how these relate to phylogenetics is important for a number of reasons. As described above, many phylogenetic and phylogenomics analyses interpret the significance scores or the order of search hit completeness as a proxy for evolutionary relatedness (i.e. phylogeny). If results from sequence similarity analyses are to be used to make decisions confidently about further evolutionary analyses, then they should reflect phylogeny as accurately as possible. But if BLAST results do not accurately reflect relatedness, then BLAST should not be used instead of phylogenies where approximation of phylogenetic relationships is needed. The suggestion to use phylogenies instead of BLAST to increase the accuracy of inference in functional genomics is far from a recent, with the first findings on this subject appearing over 15 years ago [32] and related debates over distance-based phylogenetic methods stretching back even further (e.g. [33]). Despite these past findings and clear arguments against the use of BLAST to approximate phylogenetic relationships, this practice still persists, perhaps in part because this relationship has been under-examined statistically.
Here, we detail some of the potential problems for using BLAST (or other similarity-based measures) to address questions of sequence relatedness (Figure 1) and examine these issues through simulations (Figure 2; see also Supplementary Figure S1A). Specifically, we explore lineage-specific rate heterogeneity, compositional bias and saturation in relation to both the ranked order and completeness of BLAST results. These realistic scenarios using BLAST represent only a few of the many possible parameters, problems and approaches that complicate similarity score results, but already demonstrate the danger of presuming phylogenetic relationships based on E-values in situations where even simple phylogenetic methods would be more appropriate.
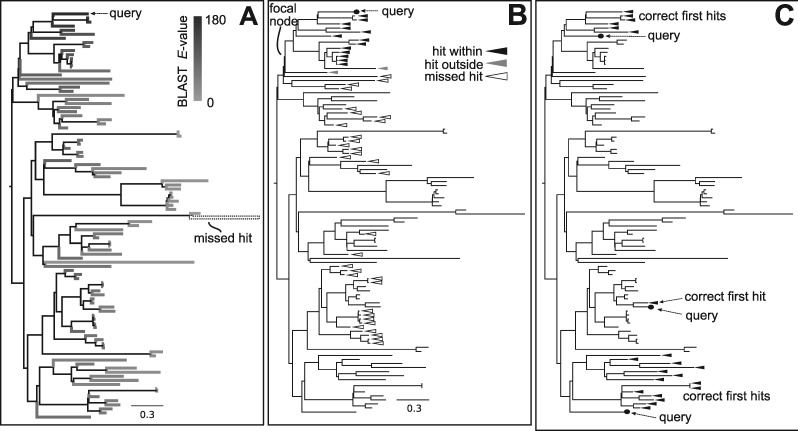
Figure 2.
Analyses of BLAST results examined here. (A) Shows a query taxon and the BLAST –(E)-values as distributed on the phylogeny. (B) Shows a query taxon and a focal node identifying a clade and the missed hits within the clade, hits outside the focal clade and hits within the clade. (C) Three sets of query taxa and the identification of the set of hits that would be correct phylogenetically.
Methods
Phylogenies and molecular sequence data were simulated under several scenarios. Pure birth trees were simulated with 100 and 1000 taxa with a standard molecular model (JC for 2000 nucleotide sites and WAG for 400 amino acid sites), including indels, rate heterogeneity (low = RH+, high = RH ++) and biased base composition (CB+). Tree heights (in substitutions per site) varied for each of these scenarios from 0.5, 1, 2, 5 and 10. For each scenario, 100 replicates were performed and summarized. More detailed description of the simulation scenarios can be found in the Supplementary Methods and Supplementary Results.
Pairwise alignment analyses were conducted using blastn for nucleotides and blastp for amino acids. BLAST and associated programs are heuristic and so the alignments are not guaranteed to be the best possible hits. Therefore, pairwise alignments were also conducted using the exact Smith-Waterman algorithm as implemented in SWIPE [34]. The parameters used for blastn and blastp included an E-value cutoff of and word sizes of four for blastn and three for blastp. The state space of amino acids and nucleotides differs such that the E-value may perform differently. However, here we are interested in the bias and so E-values were not corrected for this inherent nucleotide-amino acid difference. While BLAST does not guarantee that all best hits will be returned, SWIPE guarantees that all best hits will be reported given a particular E-value cutoff. SWIPE reports exact best hits and therefore served as more complete all-by-all results. The SWIPE E-value cutoff used was for both amino acids and nucleotides.
The resulting BLAST results were compared with the phylogenies used to generate the sequences in a number of different ways (Figure 2). The – transformation of the E-values were used in all comparisons primarily because they are the most frequently used statistic in phylogenomics. Briefly, the E-value is the number of hits with the same or better score expected to randomly be hit given the particular parameters of the search. They are closely related to p-value as . The – transformation of the E-value is the commonly used statistic in homology/orthology assessment and phylogenetic studies because this transformation allows for higher E-values to be associated with lower p-values. To determine the rough correlation of E-values with phylogeny, we compared pairwise E-values with phylogenetic distance as measured by the sum of the branch lengths (i.e. substitutions per site) separating the subject and query sequence in the pairwise alignment. To examine the completeness of the returned hits, the results returned from BLAST were compared with those returned by SWIPE. Those results from SWIPE that were not recorded in BLAST were recorded, including the E-values (as recorded from SWIPE) and the phylogenetic distance of the sequences involved. To determine how complete results are from BLAST in relation to phylogeny, the proportion of BLAST hits that were recorded within a clade along with the number of hits recorded outside of each clade was recorded (Figure 3C, see also Supplementary Figures S3 and S4). To facilitate the comparison of results across different simulations, a statistic that we name was calculated for each simulation.
Figure 3.
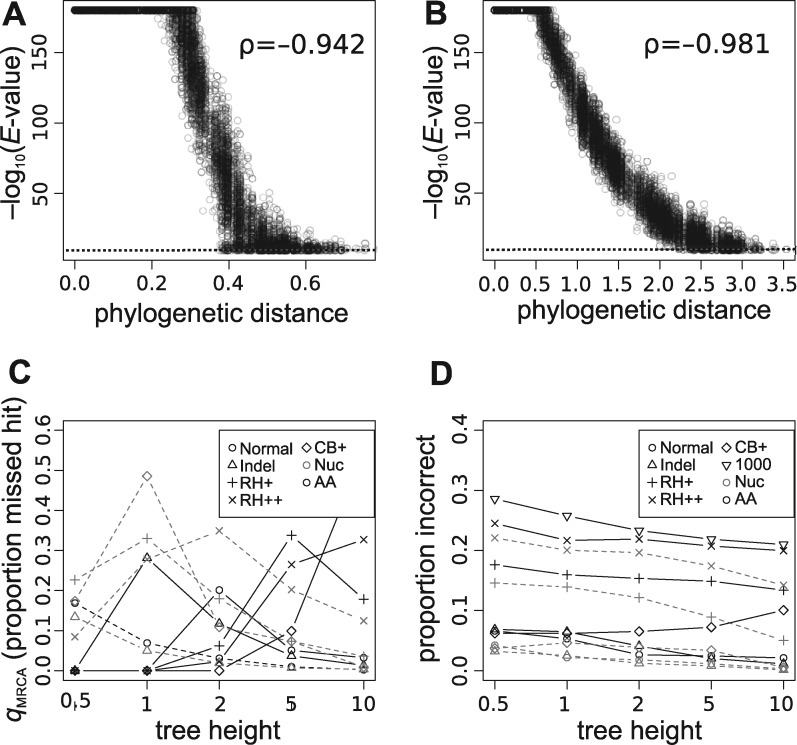
Correlation of phylogenetic distance and E-values for both (A) nucleotides and (B) amino acid results, shown for tree height of 1 and 10, respectively. The dashed line identifies an E-value cutoff of , and BLAST has an implicit maximum of . Because of the density of points, a random sample of 10 000 points for each plot is shown. Spearman’s rank correlation (ρ) is shown on each plot. See also Supplementary Figure S2. (C) The mean of the (i.e. hits missed among taxa sharing the same MRCA) statistic as calculated across each simulated tree (see Methods for details). (D) Examination of the errors in the first hits. When the first hit is not phylogenetically sister, it is recorded as ‘false’. The proportion of false sister BLAST hits are presented here. Results from BLAST for both nucleotide and amino acids are presented. See also Supplementary Figures S3 and S4.
When a BLAST search is conducted using a given query sequence (xq) on a given tree (T), each potential subject sequence (xs) in T that is hit by the BLAST search must also share a most recent common ancestor (MRCA) with xq. All other sequences in T that also descend from the MRCA of xs and xq can be defined as the set of sequences Xqs. If xs is a BLAST hit of xq and similarity scores are correlated with phylogenetic relatedness, then all sequences in Xqs are expected to also be BLAST hits of xq. We define the measure (for a given xq and xs) as the proportion of sequences in Xqs that do not have a BLAST hit for xq (i.e. are ‘missed hits’). If one or more hits are missed among these equally or more related sequences, then . Alternatively, if BLAST hits all sequences in Xqs, then this would result in an optimum score of . The mean of was calculated across the set of sequences for a tree, and the mean of these tree-wide values was calculated across each simulated sequence set.
In addition to the phylogenetic pattern of missed hit, the order of hits can be important for certain phylogenetic and phylogenomic analyses. We also calculated the phylogenetic error in first BLAST hits. This is useful not only because BLAST first hits are used in some analyses, but also as a general proxy for errors in order.
Results and discussion
Sequence similarity correlates broadly but variably with phylogenetic distance
To demonstrate potential problems with using similarity scores to infer phylogenetic relatedness, we first need to characterize the relationship between sequence similarity and phylogenetic relatedness. We found BLAST E-values (expressed as –) were correlated (Spearman’s ρ) with phylogenetic distance for both nucleotide and amino acid sequences (Figure 3A–B, Supplementary Figure S2 and Table S1). The relationship between the E-value and phylogenetic distance remained with simulations including indels (Supplementary Figure S2 and Table S1) or rate heterogeneity (Supplementary Figure S2 and Table S1). No significant difference was shown with rate heterogeneity runs including 1000 taxa. Simulations that included biases in base composition are better compared with the same data sets without biased composition (‘CB + ’ versus ‘CB0’) as these use a single tree on which to simulate data and both use the p4 simulation engine (instead of indelible, Supplementary Figure S2 and Table S1). Collectively, these results demonstrate that, in general, sequence similarity and phylogenetic distance are grossly correlated though composition bias and rate heterogeneity somewhat weaken this correlation.
Missing and misordered hits
One common way BLAST scores are used in a broad phylogenomic comparative and phylostratigraphy analyses is to examine phylogenetic patterns of the presence or absence of BLAST hits. When a given query sequence is used to conduct a BLAST search, the expectation might be that it will hit all most closely related sequences until reaching a most distantly related sequence that is still a BLAST hit. We can then define a new measure, called , as the proportion of sequences missed by BLAST that share the same MRCA as the query and most-distant-hit (i.e. are members of the clade defined by these sequences). For amino acids and relatively small tree height (i.e. low substitutions), BLAST performs well and generally hits all sequences within a clade before hitting outside of a clade (, see Methods for an extended definition). However, for tree height = 2 and no other molecular processes present, on average 20% of the sequences within the MRCA of the hits for a sequence were missing (Figure 3C). For nucleotides and large tree height (i.e. complete saturation), BLAST also performed generally well. However, for tree height = 0.5, on average 18% of the hits within the MRCA were missing. In the presence of lineage-specific rate heterogeneity or biased base composition, the missing hit percentages rose sharply to >30% and >50%, respectively. While the composition bias examined here is extreme, it demonstrates the potential for errors (though probably at lower rates).
While completeness can be important for some analyses, the exact order can be important for many others (e.g. RBH analyses). To examine error in the order of hits, the first significant BLAST hit was examined. This serves not only to address procedures that specifically use the first BLAST hit, but also gives a simple measure to describe errors in the order of hits. The lowest error rates, 0.3% for nucleotides and 2% for amino acids, was with tree height equal to 10 as generally only closely related sequences would have successful hits (Figure 3D). As with the other measures, introducing indels, lineage-specific rate heterogeneity and biased composition increased the error. The highest error rates were found with lower root heights, but this could simply be owing to high sequence similarity at lower root heights. Data sets with extreme rate heterogeneity in amino acids produced error rates of 20–24%. The source of the error in the order of BLAST hits can be demonstrated on a small simulated data set (Figures 1 and 2). These findings highlight the first hit often is not the most phylogenetically related sequence and should not be used to identify orthologs. Orthology is fundamentally a phylogenetic question, and therefore a phylogeny should be constructed to infer orthology (e.g. [9]).
Interpretation of E-values
The issues examined here may be somewhat alleviated by more focused and precise interpretations of BLAST results. For example, instead of using the resulting p- or E-values as a quantitative measure of homology, these scores could be interpreted as a Boolean (i.e. true or false). In this way, p- and E-values would be used—as intended—as frequentist significance test statistics. The null hypothesis in a BLAST analysis is that the proposed alignment between the query and subject sequences is random and follows a null distribution. A significantly small p-value, as used for BLAST, is evidence against the null hypothesis that the alignment could have been generated by random sequences, given the sequence database and search parameters. The E-value is interpreted as the number of random hits expected with an alignment score equal to or better than the score obtained between the query and subject. As with any frequentist statistic, an insufficiently small p-value does not necessarily mean that the alignment is random, but rather lacks sufficient information to distinguish it from a random alignment. Furthermore, sufficiently small p- and E-values do not necessarily mean that the alignment is the result of common descent. First, a small p-value offers evidence that the null hypothesis does not adequately explain the observation. However, the alternative to the null hypothesis for a BLAST analysis is a non-random alignment and not a homologous one. A smaller p-value more strongly refutes the null hypothesis of random sequences, but does not more strongly support homology. This more conservative usage of BLAST not only is a more accurate representation of the measures themselves, but also avoids explicitly addressing phylogenetic questions.
Sequence similarity, phylogenetic relatedness and suggestions
Many of the potential pitfalls discussed and examined here will not be surprising to phylogeneticists, and significant research has demonstrated the failings of particular methods for phylogenetic reconstruction methods, including those relying solely on sequence distance [27, 35]. Fundamentally, sequence distance alone has limited ability to reflect phylogeny. As expected and shown here, a rough correspondence exists between E-value and phylogenetic distance. However, this does not imply that distance matrices alone can replace construction of a phylogeny. Also, lineage-specific rate heterogeneity, saturation and compositional bias exacerbate errors in these types of interpretations of BLAST results. Unfortunately for BLAST analyses, these problematic molecular patterns can occur in common scenarios. For example, aside from the lineage-specific rate heterogeneity that is common throughout the tree of life [36, 37], few data sets (even small ones) conform to a strict molecular clock, which has spurred extensive research in relaxed molecular clock models [38–41]. Even though phylogenies may better model these processes, they can still pose problems for phylogeny reconstruction [8, 28, 31].
While in this study we have focused examining how aspects of similarity-based searches impact phylogenetic conclusions, our results are also relevant to ongoing community efforts to codify methods and assessment of ortholog prediction ([42, 43], and reviewed in [44]). Some studies in this area have found mixed performance for some tree-based prediction of orthologs and assignment of gene function (e.g. [45]). In particular, these studies cite weaker performance of phylogenetic ortholog prediction methods compared with similarity-based ones, particularly more rigid tree topology-based reconciliation methods that do not incorporate gene tree discordance owing to biological processes (i.e. incomplete lineage sorting, introgression). While our study does not advocate a particular practical solution for ortholog prediction or gene functional annotation, we would agree with these studies that improvements to phylogenetic ortholog prediction methods are needed, specifically in resolving both phylogenetic discordance (produced by both error and biological forces) and heterogeneity of both molecular rates and compositions that we describe. However, we would also caution that similarity-based searches, as we have demonstrated here, suffer from major biases in relation to these common molecular evolutionary processes.
BLAST and other sequence similarity tools will continue to be essential for bioinformatics, phylogenetic, genomic and phylogenomic analyses. However, the lessons from decades of phylogenetic method development need to be integrated into the culture of homolog identification, phylostratigraphy and other analyses. As expected from the large phylogenetic literature on distance-based methods, significant biases exist in how BLAST similarity corresponds to phylogenetic relatedness, and indicate that applications of BLAST that presume to estimate phylogenetic relationships are misguided. Instead of a particular approach, we advocate caution when using and interpreting sequence similarity results, especially as they are more frequently applied to phylogenetic questions or act as inputs for more complex analysis methods. Finally, where a phylogenetic relationship is needed, a phylogeny will likely produce more accurate results than the order of BLAST results.
Key Points
BLAST is often incorrectly used to infer evolutionary relatedness of sequences.
Reciprocal best hits from BLAST are often not the closest related phylogenetically under common scenarios.
Phylogenetic methods should be used to infer orthology instead of similarity-based methods.
Supplementary Data
Supplementary data are available online at http://bib.oxfordjournals.org/.
Supplementary Material
Acknowledgments
We are grateful to Joseph Brown, Bryan Moyers, Caroline Parins-Fukuchi, Jeet Sukumaran, Joseph Walker and Ya Yang for helpful discussions.
Funding
This work was supported by the National Science Foundation [1354048 to S.A.S., 1338694 to J.B.P.].
Stephen A. Smith is an assistant professor in Evolutionary Biology at the University of Michigan, Ann Arbor. His research focuses on large-scale phylogenetics, transcriptomics and molecular evolution.
James B. Pease is a postdoctoral fellow in Evolutionary Biology at the University of Michigan, Ann Arbor. His research focuses on genomic and transcriptomic patterns of molecular evolution.
References
- 1. Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 2. Li L, Stoeckert CJ, Roos DS.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13:2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen F, Mackey AJ, Stoeckert CJ, et al. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 2006;34:D363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Domazet-Lošo T, Brajković J, Tautz D.. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 2007;23:533–9. [DOI] [PubMed] [Google Scholar]
- 5. Domazet-Lošo T, Tautz D.. An ancient evolutionary origin of genes associated with human genetic diseases. Mol Biol Evol 2008;25:2699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parfrey LW, Grant J, Tekle YI, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 2010;59:518–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee EK, Cibrian-Jaramillo A, Kolokotronis SO, et al. A functional phylogenomic view of the seed plants. PLoS Genet 2011;7:e1002411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jarvis ED, Mirarab S, Aberer AJ, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014;346:1320–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Y, Smith SA.. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol 2014;31:3081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wickett NJ, Mirarab S, Nguyen N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 2014;111:E4859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katz LA, Grant JR.. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol 2015;64:406–15. [DOI] [PubMed] [Google Scholar]
- 12. Moyers BA, Zhang J.. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol 2015;32:258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y, Moore MJ, Brockington SF, et al. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol Biol Evol 2015;32(8):2001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith TF, Waterman MS.. Identification of common molecular subsequences. J Mol Biol 1981;147:195–7. [DOI] [PubMed] [Google Scholar]
- 15. Wall D, Fraser H, Hirsh A.. Detecting putative orthologs. Bioinformatics 2003;19:1710–11. [DOI] [PubMed] [Google Scholar]
- 16. Moreno-Hagelsieb G, Latimer K.. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 2008;24:319–24. [DOI] [PubMed] [Google Scholar]
- 17. Theobald DL. A formal test of the theory of universal common ancestry. Nature 2010;465:219–22. [DOI] [PubMed] [Google Scholar]
- 18. Theobald DL. On universal common ancestry, sequence similarity, and phylogenetic structure: the sins of p-values and the virtues of Bayesian evidence. Biol Direct 2011;6:60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ebersberger I, Strauss S, von Haeseler A.. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol 2009;9:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altenhoff AM, Schneider A, Gonnet GH, et al. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res 2011;39:D289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Powell S, Forslund K, Szklarczyk D, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 2014;42:D231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emms DM, Kelly S.. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 2015;16:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van der Heijden RT, Snel B, Van Noort V, et al. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics 2007;8:83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dunn CW, Howison M, Zapata F.. Agalma: an automated phylogenomics workflow. BMC Bioinformatics 2013;14:330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Domazet-Lošo T, Tautz D.. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol 2010;8:66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Šestak MS, Božičević V, Bakarić R, et al. Phylostratigraphic profiles reveal a deep evolutionary history of the vertebrate head sensory systems. Front Zool 2013;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holder MT, Zwickl DJ, Dessimoz C.. Evaluating the robustness of phylogenetic methods to among-site variability in substitution processes. Philos Trans Roy Soc B 2008;363:4013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foster PG. Modeling compositional heterogeneity. Syst Biol 2004;53:485–95. [DOI] [PubMed] [Google Scholar]
- 29. Heath TA, Hedtke SM, Hillis DM.. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol 2008;46:239–57. [Google Scholar]
- 30. Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol 2011;9:e1000602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morgan CC, Foster PG, Webb AE, et al. Heterogeneous models place the root of the placental mammal phylogeny. Mol Biol Evol 2013;30:2145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res 1998;8:163–7. [DOI] [PubMed] [Google Scholar]
- 33. Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool 1970;19:99–113. [PubMed] [Google Scholar]
- 34. Rognes T. Faster Smith-Waterman database searches with inter-sequence SIMD parallelisation. BMC Bioinformatics 2011;12:221.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thornton JW, DeSalle R.. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet 2000;1:41–73. [DOI] [PubMed] [Google Scholar]
- 36. Smith SA, Donoghue MJ.. Rates of molecular evolution are linked to life history in flowering plants. Science 2008;322:86–9. [DOI] [PubMed] [Google Scholar]
- 37. Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009;459:1122–5. [DOI] [PubMed] [Google Scholar]
- 38. Yoder AD, Yang Z.. Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol 2000;17:1081–90. [DOI] [PubMed] [Google Scholar]
- 39. Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol 2002;19:101–9. [DOI] [PubMed] [Google Scholar]
- 40. Drummond AJ, Ho SY, Phillips MJ, et al. Relaxed phylogenetics and dating with confidence. PLoS Biol 2006;4:699.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Drummond AJ, Suchard MA.. Bayesian random local clocks, or one rate to rule them all. BMC Biol 2010;8:114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kriventseva EV, Tegenfeldt F, Petty TJ, et al. Orthodb v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res 2015;43:D250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonnhammer EL, Östlund G.. Inparanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 2015;43:D234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sonnhammer EL, Gabaldn T, Sousa da Silva AW, et al. Big data and other challenges in the quest for orthologs. Bioinformatics 2014;30:2993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dalquen DA, Altenhoff AM, Gonnet GH, et al. The impact of gene duplication, insertion, deletion, lateral gene transfer and sequencing error on orthology inference: a simulation study. PloS One 2013;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.