Highlight
A new genome wide scan method using signals of positive selection identifies C4 candidate genes in the grasses independent of a priori knowledge of C4 biochemistry.
Keywords: Adaptation, C4 photosynthesis, cross-species selection scans, gene discovery, grasses, parallel evolution.
Abstract
C4 photosynthesis is perhaps one of the best examples of convergent adaptive evolution with over 25 independent origins in the grasses (Poaceae) alone. The availability of high quality grass genome sequences presents new opportunities to explore the mechanisms underlying this complex trait using evolutionary biology-based approaches. In this study, we performed genome-wide cross-species selection scans in C4 lineages to facilitate discovery of C4 genes. The study was enabled by the well conserved collinearity of grass genomes and the recently sequenced genome of a C3 panicoid grass, Dichanthelium oligosanthes. This method, in contrast to previous studies, does not rely on any a priori knowledge of the genes that contribute to biochemical or anatomical innovations associated with C4 photosynthesis. We identified a list of 88 candidate genes that include both known and potentially novel components of the C4 pathway. This set includes the carbon shuttle enzymes pyruvate, phosphate dikinase, phosphoenolpyruvate carboxylase and NADP malic enzyme as well as several predicted transporter proteins that likely play an essential role in promoting the flux of metabolites between the bundle sheath and mesophyll cells. Importantly, this approach demonstrates the application of fundamental molecular evolution principles to dissect the genetic basis of a complex photosynthetic adaptation in plants. Furthermore, we demonstrate how the output of the selection scans can be combined with expression data to provide additional power to prioritize candidate gene lists and suggest novel opportunities for pathway engineering.
Introduction
C4 photosynthesis evolved multiple times coincident with a steep decline in global CO2 levels approximately 30–40 mya (Giussani et al., 2001; Sage, 2004; Vicentini et al., 2008; Edwards and Smith, 2010; Sage et al., 2011, 2012). This correlation suggests that C4 adaptively evolved as a mechanism to concentrate carbon in the vicinity of ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco), thus significantly reducing energetic losses associated with photorespiration (Sage, 2004; Sage et al., 2011, 2012). The majority of C4 plants utilize two dimorphic cell types to fix CO2. Bundle sheath (BS) cells perform most of the reactions required for the Calvin cycle and some cyclic electron transport while the surrounding mesophyll (M) cells serve as the initial site of carbon capture and perform linear electron transport to drive the production of NADPH and ATP. BS and M cells form a wreath-like structure surrounding vasculature tissues known as Kranz anatomy. This is most often associated with C4 photosynthesis (Brown, 1975; Giussani et al., 2001; Sage et al., 2011, 2012). This morphological adaptation and associated division of biochemical activities serves to pump C4 acids into the BS that are later decarboxylated in the BS plastid where most Calvin cycle enzymes are localized. These innovations have resulted in some of the most productive plants on the planet, accounting for an estimated 25% of global primary production, despite including only 3% of all angiosperm species (Still et al., 2003). Traditionally, C4 species have been classified into three major subtypes based on the primary decarboxylating enzyme present in the BS (Sage, 2004; Furbank, 2011; Sage et al., 2011): NADP malic enzyme (NADP-ME), NAD malic enzyme (NAD-ME) and phosphoenolpyruvate carboxykinase (PCK).
The evolution of the C4 carbon pump involves a number of dramatic changes: increased vein density, increased photosynthetic capacity of the BS cells, repositioning of organelles, changes in photosynthetic membranes, and the redistribution of enzymes into subcellular compartments. In many cases, genes encoding proteins that perform other functions in C3 plants have been co-opted into new roles in C4 photosynthesis (Sage et al., 2011, 2012). Molecular approaches to dissect the regulatory networks guiding C4 differentiation have focused on profiling or co-expression studies that often yield hundreds to thousands of candidate genes (Li et al., 2010; Chang et al., 2012; John et al., 2014; Wang et al., 2014a; Huang and Brutnell, 2016), with little evidence for prioritization. Reverse genetic screens have largely been limited to known components such as carbon shuttle enzymes (Bailey et al., 2000; Cousins et al., 2006; Studer et al., 2014) and have not yielded insights into networks regulating the differentiation process. Comparative studies of molecular evolution, on the other hand have shown core C4 genes such as phosphoenolpyruvate carboxylase (PEPC) (Christin et al., 2007), NADP-ME (Christin et al., 2010) and PCK (Christin et al., 2009) to be adaptively evolving in C4 clades. However, like reverse genetic screens, these studies relied on a priori information on the biochemistry of the C4 carbon shuttle pathway to first identify gene candidates.
In this report we describe a novel method to use signals of adaptive evolution to identify candidate genes required for C4. The method conducts an automated genome-wide scan and does not rely on a priori information to define candidates. Rather, putative C4 genes are identified based strictly on the ratio of rates of nucleotide substitutions. We focus our study on the grasses (Poaceae), as C4 has originated in grasses at least 25 times and they include some of the most ecologically successful C4 species (Giussani et al., 2001; Sage et al., 2011; Grass Phylogeny Working Group II, 2012). We identify 88 genes that show potential adaptive evolution in C4 lineages. These genes include both known components of the C4 pathway and several suspected and novel components. When coupled with expression profiling, this approach provides a powerful tool for gene discovery and potentially for engineering alternative forms of C4 photosynthesis.
Materials and methods
Obtaining syntenic orthologs and quality control
Reference primary coding DNA sequences (CDSs) of rice, Brachypodium distachyon, Setaria italica, sorghum and maize were downloaded from Phytozome 10 (http://phytozome.jgi.doe.gov). The CDSs of Dichanthelium oligosanthes were obtained from CoGe (http://genomevolution.org, genome ID no. 20291) (A. J. Studer, J. C. Schnable, S. Weissmann et al., unpublished data). Lists of known syntenic orthologs were obtained from (Schnable et al., 2012), and S. italica syntenic orthologs were identified using the same method as described in (Schnable et al., 2012). Ortholog groups that were duplicated in the maize whole genome duplication event (Schnable et al., 2009) were merged, and BLASTN (Camacho et al., 2009) was used to identify the closest D. oligosanthes homolog to the S. italica ortholog. This yielded 16 943 ortholog groups. We then considered four patterns of gene relationship: (i) one ortholog in all six species (8143); (ii) two orthologs in maize (homeologs) and one ortholog in the other five species (3262); (iii) rice ortholog missing and one ortholog in the other five species (1029), and (iv) B. distachyon ortholog missing and one ortholog in the other five species (604). Blast hits without gene annotation were considered missing. These patterns were specifically considered because C4 branches can be unambiguously assigned. Collectively these occasions accounted for about 77% (13 038 out of 16 934) of ortholog groups. Codon-based alignment was performed using ProGraphMSA (Szalkowski, 2012), and the resulting alignments were trimmed using Gblocks (Castresana, 2000) and short alignments (less than 30% coverage) discarded. A maximum likelihood (ML) phylogenetic tree was constructed using RaxML (Stamatakis, 2014) using all sites and MEGA-CC (Kumar et al., 2012) using only the third position of codons. The GTR+gamma+I mutation model was used in both analyses. Resulting trees were then compared with the species phylogeny and tested for topological congruence using qdist (Mailund and Pedersen, 2004). Failing both phylogenetic congruence tests resulted in exclusion from further analysis. Finally 6784 ortholog groups were obtained.
Test for potential selection and identification of candidate genes
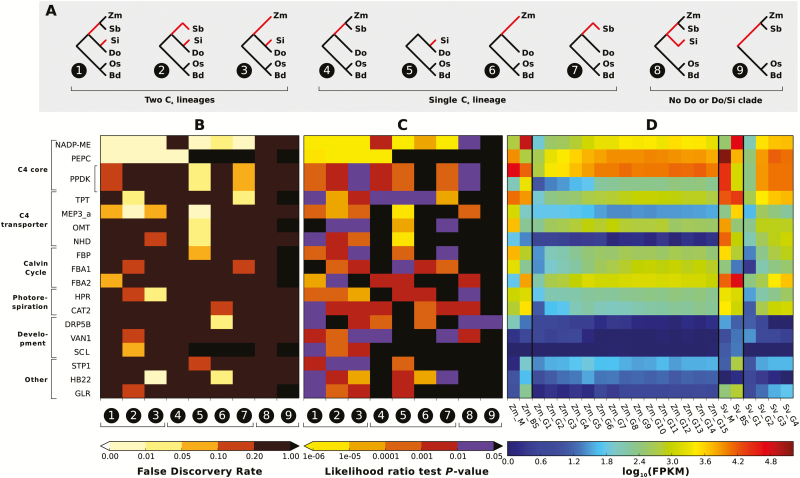
The branch model of PAML 4.2 (Yang, 2007) was used to calculate likelihoods of the data given the null hypothesis (H0) assuming all branches shared the same ratio of dN/dS, and the alternative hypothesis (Ha) assuming C4 branches had a dN/dS ratio independent from all other branches (Fig. 1A). A likelihood ratio test was used to evaluate the significance of Ha over H0 (Yang, 2007). The full phylogeny with all six species (condition 1) theoretically requires an ortholog group to be under positive selection in all three C4 species. In order to account for possible selection that only occurred in specific subsets of C4 lineages, additional tests under six conditions with one or two C4 lineages manually removed were considered. These conditions include maize removed (condition 2), sorghum removed (condition 3), S. italica removed (condition 4), the maize–sorghum clade removed (condition 5), sorghum and S. italica removed (condition 6), and maize and S. italica removed (condition 7). To determine the importance of the Setaria–Dichanthelium clade, two additional conditions were also considered in which Dichanthelium (condition 8) and the Setaria–Dichanthelium clade (condition 9) were removed manually (Fig. 1A). The testing topologies under these conditions were further modified in cases when maize duplication and rice/Brachypodium gene loss needed to be accounted for, and tests were not conducted if there were less than four taxa available (for final testing topologies see Supplementary Table S1 at JXB online).
Fig. 1.
Phylogenies used for selection scan, statistical significance and tissue-specific expression data for top 18 candidate C4 ortholog groups. (A) Nine phylogenetic conditions used for selection scan. Red branches are branches where the C3 to C4 transitions are inferred to have occurred (C4 branches). Zm: maize; Sb: sorghum; Si: S. italica; Do: D. oligosanthes; Os: rice; Bd: B. distachyon. (B) False discovery rates from selection scan. Each column represents tests under the same phylogenetic condition corresponding to (A), and each row (or two rows in the case of maize, which has two homeologs) represent one ortholog group. Lighter color indicates higher significance. Ortholog groups are grouped according to their functional relevance to C4, specified on the left. (C) P-values of likelihood ratio tests from selection scan. These are single test statistics and not multi-tests corrected. (D) Tissue specific expression profile of corresponding ortholog groups in maize and Setaria, shown on log scale. Zm_M/BS: maize mesophyll/bundle sheath; Zm_G1-15: maize leaf gradient. Sv_M/BS: Setaria viridis mesophyll/bundle sheath; Sv_G1-4: S. viridis leaf gradient. The BS/M original data are downloaded from John et al. (2014) and were originally generated by John et al. (2014) and Chang et al. (2012). The maize leaf gradient data are obtained from Wang et al. (2014a). The S. viridis gradient data are obtained from (A. J. Studer, J. C. Schnable, S. Weissmann et al., unpublished data).
A multi-test correction was performed under each phylogenetic condition to obtain the false discovery rate (FDR) using the R package fdrtools (Strimmer, 2008). Ortholog groups with FDR<0.2 in at least one test (indicating an elevated dN/dS ratio in at least one C4 branch) were merged to generate a final candidate gene list, grouping by their putative relationship to C4. The cell-type and leaf gradient expression profile measured in fragments per kilobase of exon per million fragments mapped (FPKM) of these candidates in maize and Setaria were extracted from previous studies (Li et al., 2010; Chang et al., 2012; John et al., 2014; Wang et al., 2014a; A. J. Studer, J. C. Schnable, S. Weissmann et al., unpublished data). A gene ontology enrichment analysis was also performed using the GO annotations of the closest Arabidopsis thaliana homolog using AgriGO (Du et al., 2010) with the background as the non-redundant A. thaliana homolog of 6784 ortholog groups. Finally, we manually examined ten homolog groups that were putatively involved in C4 photosynthesis (Chang et al., 2012; John et al., 2014) but were filtered out from the automated workflow (Supplementary Table S2). Homologs were identified using BLASTN when syntenic orthologs are not available. Case-specific phylogenies were used to determine orthology and account for the complexities involved in these situations.
Results
An overview of the candidates and the automated workflow
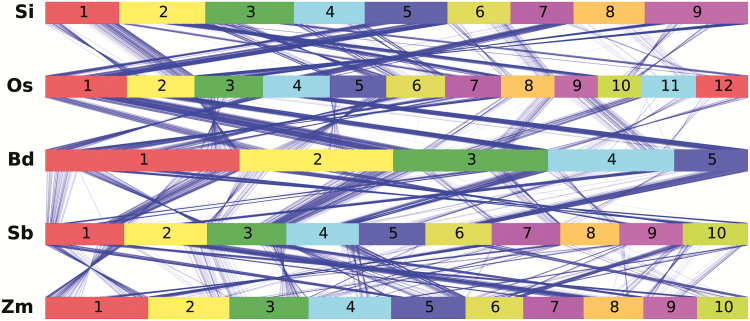
Although phylogenetic relationships have been the subject of intense study in the grasses and several branches of C3 to C4 transitions defined (simplified as ‘C4 branches’ hereafter) (Christin et al., 2007, 2009), gene duplication, loss and polyploidization confound attempts to streamline genome-wide scans (Wang et al., 2009; Christin et al., 2013). Thus, we have employed a set of orthologous relationships among five grass species based on syntenic conservation (Schnable et al., 2012) (Fig. 2). These species are Oryza sativa (rice; Ouyang et al., 2007), Brachypodium distachyon (Vogel et al., 2010), Setaria italica (Bennetzen et al., 2012), Sorghum bicolor (sorghum; Paterson et al., 2009) and Zea mays (maize; Schnable et al., 2009). Rice and B. distachyon employ C3 photosynthesis while the other three employ C4 photosynthesis. Among the three C4 species, maize and sorghum share a common origin of C4 while S. italica has evolved C4 photosynthesis independently (Giussani et al., 2001; Edwards and Smith, 2010; Grass Phylogeny Working Group II, 2012). The relationships of genes present at syntenic locations in the genomes of multiple species strictly follow the phylogeny of the species themselves, meaning a uniform phylogeny can be applied to all genes for analysis. This makes it possible to conduct a cross-species genome scan in an automated fashion.
Fig. 2.
Gene synteny across five grass species (a random set of 1400 ortholog groups are shown). Si: Setaria italica; Os: Oryza sativa (rice), Bd: Brachypodium distachyon; Sb: Sorghum bicolor (sorghum); Zm: Zea mays (maize). Each colored segment represents one chromosome in one species, and the blue lines between species denote position of a pair of syntenic orthologs. Genome lengths of all species are normalized to be equal to each other.
The most recent common ancestor of the C3 and C4 lineages included in the set of five grasses with complete assembled genomes represents at least ~50 million years of evolutionary divergence (Christin et al., 2014). Accordingly, signals of positive selection may be obscured by many other randomly fixed changes along the long branches separating these grass lineages. Furthermore, while two independent origins of C4 (Setaria and maize–sorghum) are available, the C3 clades between these two C4 clades are not represented (Giussani et al., 2001; Grass Phylogeny Working Group II, 2012). Thus the two independent origins of C4 are not distinguishable on the phylogeny of these five species. The recently published draft genome of Dichanthelium oligosanthes, a C3 panicoid grass, mitigates both issues described above (A. J. Studer, J. C. Schnable, S. Weissmann et al., unpublished data). D. oligosanthes is more closely related to Setaria than it is to the maize–sorghum clade. Thus, inclusion of D. oligosanthes in our analysis greatly reduces divergence time between C3 and C4 lineages, and also phylogenetically separates Setaria from the maize–sorghum clade.
The genetic unit employed in this study is the pan-grass syntenic orthologous gene or ortholog group (Fig. 2). We define an ortholog group to be a set of genes that are syntenically orthologous across maize, sorghum, Setaria, rice, and Brachypodium, together with their putative Dichanthelium ortholog. A total of 13 038 ortholog groups were considered after grouping together syntenic homologs due to whole genome duplication in maize (Schnable et al., 2009) and controlling for taxon coverage. Among them, 6784 ortholog groups passed both alignment quality and phylogenetic congruence tests, and were tested for potential positive selection using nonsynonymous to synonymous substitution rates ratio (dN/dS) based methods (see Materials and methods for details). Because elevated dN/dS is a strong indication of positive selection or relaxed negative selection (Yang, 2007), we effectively conducted a cross-species selection scan. To account for the potential that different genes were co-opted in independent evolutionary origins of C4, an analysis was performed using the full phylogeny (condition 1), together with phylogenies with one (conditions 2–4) or two C4 species manually removed (conditions 5–7; Fig 1A). In total 88 ortholog groups were identified that show elevated dN/dS in at least one C4 lineage after multi-test corrections (FDR<0.2, see Discussion), and 18 ortholog groups were prioritized based on their test significance and putative functions (Fig. 1; Supplementary Table S3). We also extracted expression data from published datasets (Chang et al., 2012; John et al., 2014; Wang et al., 2014a) for further comparisons (Fig. 1).
Core C4 genes
Of the five genes encoding the enzymes of the NADP-ME subtype carbon shuttle, three were among the resulting list of the automated workflow (Fig. 1). They include NADP-ME (Si000645m; for simplicity only the Setaria CDS is used unless otherwise necessary; for corresponding orthologs across all six species, see Supplementary Table S4), pyruvate, phosphate dikinase (PPDK; Si021174m) and PEPC (Si005789m). In both Setaria and maize these genes are highly expressed (fragments per kilobase of exon per million fragments mapped, FPKM>500) (Chang et al., 2012; John et al., 2014) in photosynthetic tissues, and thus are likely to be functional for photosynthesis (Fig. 1B, C, D). Another core C4 gene, NADP malate dehydrogenase (NADP-MDH; Si013632m) did not show evidence of adaptive evolution. A separate manual test for carbonic anhydrase (CA; Si003882m) was conducted because gene duplication and fusion resulted in its exclusion from the automated workflow (Studer et al., 2014; A. J. Studer, J. C. Schnable, S. Weissmann et al., unpublished data). Tests of CA based only on the putative photosynthetically active homologs (highly expressed homologs; Supplementary Table S2) failed to provide signals of adaptive evolution.
A proposed PCK pathway in maize (Wingler et al., 1999) utilizes aspartate to shuttle carbon between M and BS. This pathway is maize specific, and thus was not included in the automated workflow. Manual examination of two syntenic orthologs of PCK, however, did reveal a signal of elevated dN/dS in only one of the two (GRMZM2G001696; P=0.000000012; FDR for manual tests are not calculated because manual tests are case-specific; Supplementary Table S2). This ortholog shows high and biased expression in maize BS, consistent with a functional role in the PCK C4 pathway (Chang et al., 2012). The two aspartate amino transferases (AspAT1 and AspAT2) did not show signals of adaptive evolution.
Putative C4-related transporters
Of the six putative C4-related transport proteins (Kinoshita et al., 2011; Furumoto et al., 2011; Chang et al., 2012; John et al., 2014) that were included in the automated workflow, four were identified as targets of potential adaptive evolution (Fig. 1B, C; Supplementary Table S3). They include a dicarboxylate translocator (OMT, Si024403m), a putative pyruvate transporter (MEP3_a, Si024315m), an H+/Na+ antiporter relating to pyruvate transportation (NHD, Si029362m) and a triose-phosphate transporter (TPT; Si001693m). Another dicarboxylate translocator (DCT2, Si013503m) showed significance in a few single tests, but failed the corresponding multi-test corrections. Manual examinations of the other six ortholog groups, which were not included in the automated workflow due to our inability to unambiguously define orthology relationships, showed single test level significance in a dicarboxylate transporter (DCT4, Si035016m), a putative pyruvate transporter (sodium bile acid symporter BASS2, Si001591m) and a phosphoenolpyruvate/phosphate translocator (PPT1, Si013874m) (Supplementary Table S2). Tests for MEP3_c (Si005376m) were not conducted because a corresponding Dichanthelium homolog was not found. Among the three ortholog groups that did not show any signal of positive selection (MEP3_b, Si000451m; DCT1, Si029415m; PPT2, Si005351m), two showed low levels of expression in leaf tissue of Setaria and maize. In contrast, the ortholog groups that appear to have similar functions and show potential evidence of selection were all highly expressed in at least one C4 species (Supplementary Tables S2 and S3).
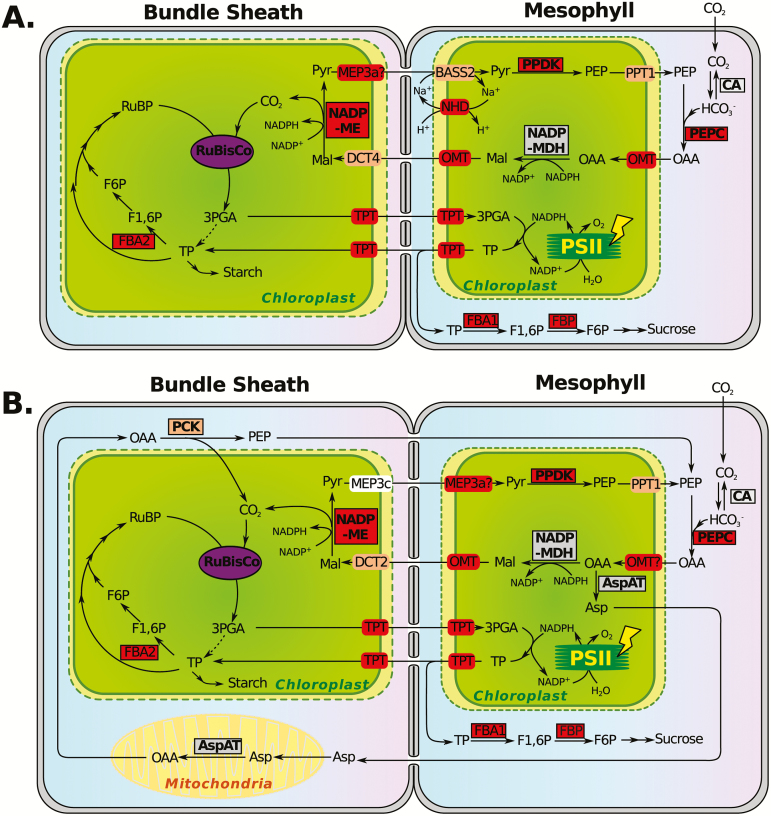
Combining our results with bundle sheath/mesophyll (BS/M) expression profiles, proteomics and models of metabolite flow from previous studies (Aoki et al., 1992; Majeran and van Wijk, 2009; Kinoshita et al., 2011; Furumoto et al., 2011; Chang et al., 2012; John et al., 2014), we generated a hypothesized overview of the adaptively evolving C4-related enzymes and transporters in maize and Setaria (Fig. 3). Although some uncertainties remain, an important observation for the C4 transporters is that the homolog groups showing potential evidence of selection collectively cover most plastidial transport roles needed for the NADP-ME subtype of C4 based on their putative function (Fig. 3). These results suggest that plastid membrane transporters in general are key components of C4 adaptive evolution, in addition to core C4 enzymes. Unlike the core C4 genes in which the same ortholog groups have been recruited in parallel, Setaria and the maize–sorghum lineages sometimes adopt transporters from different ortholog groups to achieve similar functions. This result reflects the great flexibility in biochemistry of the parallel C4 origins.
Fig. 3.
Hypothesized metabolite flow in (A) Setaria italica/viridis and (B) maize. Enzymes are enclosed in rectangles, and transporters are located on plastid membranes. The enzyme/transporter names correspond to those listed in Supplementary Tables S3 and S4. Enzymes and transporters colored in red show significant signal of positive selection (FDR<0.2) in at least one C4 lineage by the automated workflow. Those colored in orange are significant only at the single test level (P<0.01) in the automated workflow or manually, those colored in grey show no signal of positive selection in any test performed, and MEP3_c colored in white means meaningful tests could not be performed. 3PGA: 3-phosphoglycerate; Asp: aspartate; F1,6P: fructose-1,6-bisphosphate; F6P: fructose-6-phosphate; Mal: malate; OAA: oxaloacetate; PEP: phosphoenolpyruvate; Pyr: pyruvate; RuBP: ribulose bisphosphate; TP: triose phosphate.
Calvin–Benson–Basham cycle and photorespiration-related genes
Both the Calvin–Benson–Basham (CBB) cycle and photorespiration are processes that are predominantly BS-localized in C4 photosynthesis. As shown in Fig. 1 and Supplementary Table S3, two fructose-1,6-bisphosphate aldolases (FBAs) and one fructose-1,6-bisphosphate phosphatase (FBP) appear to have potential C4-specific activities. Among them, FBA2 (Si026480m) shows BS-preferential expression and is likely required for CBB function. FBA and FBP show M-preferential expression and are putatively involved in downstream sugar metabolism. The automated workflow also identified two ortholog groups with putative roles in the photorespiratory pathway (Fig. 1B, C), a catalase (CAT2, Si035374m) and a hydroxypyruvate reductase (HPR, Si017480m).
Novel C4 candidate genes
In addition to the genes mentioned above, many candidate genes that had not been previously considered as C4 related (Fig. 1B, C and Supplementary Table S3) were identified by this method. They include three ortholog groups implicated in leaf development. Ortholog group Si028928m encodes an ADP-ribosylation factor-GTPase activating protein. Disruptions in the closest homolog from Arabidopsis thaliana (AT5G13300, VASCULAR NETWORK DEFECTIVE 1, VAN1) result in leaf vein patterning defects (Sieburth, 2006). Ortholog group SCL (Si026111m) is a GRAS family transcription factor and a homolog to SCARECROW-like 14 in A. thaliana. SCARECROW-like genes are known to be involved in endodermis pattern specification in roots in A. thaliana, and recently have been suspected of playing a key role in vasculature/BS/M patterning in leaves of C4 plants (Slewinski et al., 2012). Another ortholog group with a potential link to leaf development is DRP5B (Si009435m), a dynamin-related family protein homologous to A. thaliana DRP5B, which is known to be involved in chloroplast division and development (Pyke and Leech, 1994).
Several potential C4-related transcription factors were also identified. Among them, a zinc finger homeodomain transcription factor (HB22, Si032496m) is of particular interest. It is homologous to a homeodomain transcription factor that has been shown to bind the promoter region of PEPC in dicot C4Flaveria species, but not to bind the promoter region of PEPC in C3Flaveria species (Windhövel et al., 2001). The previously discussed SCL ortholog group (Si026111m) is also a transcription factor.
A gene ontology enrichment analysis using the GO annotations of homologous genes in A. thaliana showed a significant enrichment in molecular functions related to transporter activities (GO:0005215, FDR<0.05; Supplementary Table S5) among the 88 orthologous groups identified. In addition to the C4-related transporters described above, at least 12 other ortholog groups in our candidate list have predicted transporter functions. One of them is a putative sugar transporter (STP1, Si035219m), which shows preferential BS expression in both maize and Setaria. Many ortholog groups in the candidate list have never been linked to C4 photosynthesis, but some showed high significance in certain tests as well as BS/M differential expression profiles in maize and Setaria (Supplementary Table S3). One example is a glutamate receptor-like (GLR, ortholog group Si005804m) protein. Its homolog in A. thaliana, GLR3.4, has recently been shown to affect lateral root primordium formation through Ca2+ signaling pathways (Vincill et al., 2013). As root development modules have been implicated in driving BS/M differentiation in C4 grasses (Slewinski et al., 2012), we speculate that this gene may also have been co-opted from lateral root development in vein patterning of C4 grass leaves.
Discussion
Overview of the cross-species selection scans
In this study we have developed a genome-wide (6784 ortholog groups) unbiased survey for signals of positive selection or relaxed negative selection to discover genes related to C4 photosynthesis in six grass species. We used a relaxed FDR of <0.2 to capture a broad list of C4 candidate genes and identified a list of 88 candidate genes that have likely been co-opted into a C4 differentiation process (Supplementary Table S3). To develop a test for enrichment of C4-related genes identified in the selection scan, we compared the frequencies of known ‘C4 genes’ (carbon shuttle enzymes and transporters) in the set of 88 prioritized candidates with the total tested 6784 genes. Seven of the 11 known C4 genes were detected in the automated workflow. Thus, a significant enrichment in C4 genes was achieved using the automated workflow (Fisher’s exact test, P= 2.3×10–9). There are three major advantages of this evolutionary based approach for gene discovery. First, it does not require any a priori knowledge of C4 biochemistry or development to identify candidate genes, and is completely independent from expression and proteomics data (Huang and Brutnell, 2016). Second, it provides a much smaller list of candidate genes, defined by a robust statistical test, than other, ‘guilt by association’ techniques such as cell-type specific expression analysis and coexpression network clusters (Li et al., 2010; Wang et al., 2014a). Third, the automated nature of this cross-species selection scan workflow is also quite flexible. It may be expanded with new genomes/transcriptomes, and adopted for other traits under strong adaptive evolution in taxa of interest.
An important validation of this approach was revealed in the identification of known C4-associated genes including PEPC, PPDK, NADP-ME and OMT. However, as with most computationally based gene discovery platforms, the workflow suffers from both type I and type II errors. False positives can be caused by genes under selection due to other causes, relaxed negative selection rather than positive selection (e.g. pseudogenes in C4 lineages), or random fluctuations of dN/dS (Yang, 2007). In the long run, these problems can be largely overcome through increasing species sampling, especially through increasing the number of phylogenetically independent C3–C4 comparisons (Christin et al., 2007, 2009). This approach is feasible for grasses in particular, because C4 has originated in grasses at least 25 times (Grass Phylogeny Working Group II, 2012). New draft genomes/transcriptomes also provide more robust phylogenies for the tests performed and increase the specificity of detecting C4-related genes. More independent C4 lineages can also help with identifying genes under lineage-specific positive selection.
False negatives will not be as easily resolved through the inclusion of data from additional species. In addition to a large number of genes not recovered in synteny analysis, many ortholog groups are not considered due to complicated duplication/loss and mis-annotation, failing the multiple sequences alignment threshold, and/or failing the phylogeny congruence test (10 148 out of 16 934, ~59.9%), as a necessary sacrifice to ensure conservative predictions and automation of the workflow. As shown here, our false negatives included one core C4 carbon shuttle enzyme (CA) and four putative C4-related transporters. A key to solve this problem is to improve annotations of all genomes. It greatly reduces false gene losses (when a syntenic ortholog exists in one species but is not annotated), improves the quality of multispecies alignments and increases the chance of reconstructing the correct gene phylogeny. For example, probable candidate ortholog groups that are significant in manual tests could have been included in the automated workflow (e.g. BASS2 and PPT1; Supplementary Table S2) with improved genome annotations and/or alignments. Gene orthology calls based on gene synteny, if applied across a broader range of species, would improve existing gene annotations (Schnable et al., 2012). Additionally, topology-based congruence tests for orthology may be substituted by a Bayesian statistical framework to test if an alignment-based gene tree significantly deviates from the expected (genome wide estimated) species tree to allow some more flexibility accounting for errors introduced by a small species sample and short alignments.
It is also likely that the protein sequences of some genes co-opted into C4 photosynthesis are simply not subject to positive selection. This could include proteins involved in non-rate limiting steps of metabolic networks (NADP-MDH is a potential example), or genes where adaption to a role in C4 photosynthesis occurs through mechanisms other than amino acid substitutions (e.g. copy number variation and/or cis-element-induced expression level changes). Accordingly, the method presented here is not comprehensive in identifying all C4-related genes in a group of species, but it does represent a novel and complementary approach to gene discovery based on biochemical or transcriptional characterizations.
The Setaria–Dichanthelium clade is a key for C4 gene discoveries in grasses
Two additional phylogenetic conditions (condition 8, phylogeny without Dichanthelium, and condition 9, phylogeny without the Setaria–Dichanthelium clade; Fig. 1A and Supplementary Table S3) were used to determine the importance of the Setaria–Dichanthelium clade for our results. Clearly, the power to detect C4-related genes dramatically decreases under these two conditions (Fig. 1B, C). None of the three core C4 genes (PEPC, NADP-ME and PPDK) shows statistical significance at the FDR<0.2 level. Excluding the Dichanthelium branch alone is slightly better than excluding the entire Setaria–Dichanthelium clade, under which the detection power is lost almost completely. The lack of detection power is most likely due to the small number of sampled species and long divergence time between the panicoid and pooid lineages.
This result clearly shows the inclusion of the Setaria–Dichanthelium clade, a recently diverged C3–C4 species pair, is crucial for identifying C4-related genes using our approach. In the absence of such closely related C3–C4 pairs, it is often necessary to employ simple pairwise comparisons, frequently between long-diverged lineages such as rice vs. maize (Wang et al., 2014a). This more recent C3–C4 comparison affords a dramatic increase of power in detecting signals of selection, suggesting that other methods such as expression profiling and proteomics could benefit from such comparisons as well. It also indicates that the inclusion of additional recently diverged C3–C4 comparisons will increase both the power and the specificity in revealing novelties associated with C4 gene evolution.
Adaptive evolution in C4-related genes and its implications for engineering
As discussed above, signals of elevated dN/dS were observed in many carbon shuttle enzymes and key transporters (Fig. 1), indicating changes in protein function that act to increase metabolic flux within the C4 cycle. These findings suggest that movement of metabolites between BS and M cells are potential rate limiting steps in C4 metabolism networks, consistent with prior metabolic modeling studies (Pick et al., 2011; Wang et al., 2014b). When considering the engineering of C4 photosynthesis into C3 plants, our findings point to ‘lessons learned’ from the evolutionary trajectories of C4 plants and reveal which enzymes and transporters may be necessary for insertion into C3 plants (Heckmann et al., 2013; Wang et al., 2014b). One example of such a component is the putative triose phosphate transporter (TPT), which is responsible for plastidial membrane transport of triose phosphate and 3-phosphoglycerate. While little engineering attention has been paid to this gene relative to core C4 genes such as PEPC and NADP-ME, recent modeling work has shown that the TPT is a critical component for the efficiency of C4 photosynthesis (Wang et al., 2014b). Our findings support the conclusion that TPT is a good target for engineering. Furthermore, as TPT is functional in both BS and M, it is unlikely to be detected from BS/M differential expression analysis without a priori knowledge of the biochemistry (John et al., 2014).
Another important finding with potential engineering significance is that while some C4 core enzymes are recruited in parallel, others are differentially recruited in different lineages. Such parallelism versus divergence is evident when considering the three C4 subtypes, which are named after the primary decarboxylases expressed in BS cells (Furbank, 2011). This might indicate an evolutionary trajectory in which the shared genes are more constrained in enzymatic activities (e.g. PEPC and PPDK), whereas decarboxylase recruitment was more flexible. In maize, for instance it appears that both NADP-ME and PEPCK pathways are both utilized (Wingler et al., 1999; Pick et al., 2011). The divergence in C4 transporters creates fascinating opportunities for cross-species engineering. One example is the NHD-BASS2 system in Setaria. Early physiological work indicated two types of M plastidial pyruvate uptake systems in C4 species: the maize–sorghum clade uses an H+-dependent pyruvate transport system, while Setaria, Panicum and many other non-Andropogoneae species rely on a Na+-dependent pyruvate transport system (Aoki et al., 1992). In the C4 eudicot Flaveria, the homologous NHD-BASS2 system has been suggested to be responsible for pyruvate uptake in a Na+-dependent fashion in M cells (Furumoto et al., 2011). We find that both NHD (Si029362m, automated workflow) and BASS2 (Si001591m, manual) orthologs are likely under strong selection pressure in Setaria, but not in maize and/or sorghum (Fig. 1; Supplementary Tables S2 and S3). In addition, both NHD and BASS2 are highly expressed in M of Setaria but not in maize (Chang et al., 2012; John et al., 2014). The combined results strongly indicate NHD-BASS2 is a Setaria-specific pyruvate transport system that is not operational in maize. Accordingly, insertion of the NHD-BASS2 complex into maize could facilitate pyruvate flux into M, and ultimately increase overall photosynthetic assimilation efficiency.
Conclusions
C4 photosynthesis drives productivity in some of the most ecologically and agronomically important species on the planet, but a genetic dissection of C4 has been limited by the lack of resolution of available tools. Here we demonstrated the potential of cross-species selection scans, based on the concept of adaptive molecular evolution, as a powerful new method to identify candidate genes for C4 photosynthesis. Unlike current -omics based approaches for gene discovery, our method is independent of a priori knowledge of C4 biochemistry and results in a small list of candidate genes. Using this method, we have identified 88 candidate C4-related genes, including both known and novel genes. These candidates, along with the method, provide new insight into engineering plants with better photosynthetic efficiency, and engineering C4 photosynthesis into C3 plants. This approach can also be broadly applied to other traits under adaptive evolution and represents a powerful new approach to gene discovery.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Phylogenies used for positive selection test given maize duplication, gene loss in rice or Brachypodium, under different phylogenetic conditions.
Table S2. Manually conducted tests.
Table S3. Candidates from automated workflow.
Table S4. Gene names and syntenic ortholog group correspondence for six grass species.
Table S5. Gene ontology enrichment analysis using Arabidospsis thaliana homologs.
Supplementary Material
Acknowledgements
This research was supported by a National Science Foundation Plant Genome Research Program award (IOS-1127017) to T.P.B., a Department of Energy – Division of Biosciences Fellowship of the Life Sciences Research Foundation to A.J.S. (DE-FG02-12ER16337), a National Science Foundation Plant Genome Postdoctoral Fellowship to J.C.S. (IOS-1306738) and funding from the Donald Danforth Plant Science Center. The authors thank Indrajit Kumar and other Brutnell lab members for helpful suggestions.
References
- Aoki N, Ohnishi J, Kanai R. 1992. Two different mechanisms for transport of pyruvate into mesophyll chloroplasts of C4 plants—a comparative study. Plant and cell Physiology 33, 805–809. [Google Scholar]
- Bailey KJ, Battistelli A, Dever LV, Lea PJ, Leegood RC. 2000. Control of C4 photosynthesis: effects of reduced activities of phosphoenolpyruvate carboxylase on CO2 assimilation in Amaranthus edulis L. Journal of Experimental Botany 51, 339–346. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria. Nature Biotechnology 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Brown WV. 1975. Variations in anatomy, associations, and origins of Kranz tissue. American Journal of Botany 62, 395–402. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552. [DOI] [PubMed] [Google Scholar]
- Chang Y-M, Liu W-Y, Shih AC-C, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiology 160, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution 5, 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Petitpierre B, Salamin N, Buchi L, Besnard G. 2009. Evolution of C4 phosphoenolpyruvate carboxykinase in grasses, from genotype to phenotype. Molecular Biology and Evolution 26, 357–365. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Christin PA, Samaritani E, Petitpierre B, Salamin N, Besnard G. 2010. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biology and Evolution 1, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Spriggs E, Osborne CP, Stromberg CAE, Salamin N, Edwards EJ. 2014. Molecular dating, evolutionary rates, and the age of the grasses. Systematic Biology 63, 153–165. [DOI] [PubMed] [Google Scholar]
- Cousins AB, Badger MR, von Caemmerer S. 2006. Carbonic anhydrase and its influence on carbon isotope discrimination during C4 photosynthesis. Insights from antisense RNA in Flaveria bidentis. Plant Physiology 141, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Smith SA. 2010. Phylogenetic analyses reveal the shady history of C4 grasses. Proceedings of the National Academy of Sciences of the United States of America 107, 2532–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Yamaguchi T, Ohshima-Ichie Y, et al. 2011. A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. [DOI] [PubMed] [Google Scholar]
- Giussani LM, Cota-Sánchez JH, Zuloaga FO, Kellogg EA. 2001. A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. American Journal of Botany 88, 1993–2012. [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist 193, 304–312. [DOI] [PubMed] [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber AP, Lercher MJ. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. [DOI] [PubMed] [Google Scholar]
- Huang P, Brutnell TP. 2016. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Current Opinion in Plant Biology 31, 91–99. [DOI] [PubMed] [Google Scholar]
- John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM. 2014. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiology 165, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita H, Nagasaki J, Yoshikawa N, Yamamoto A, Takito S, Kawasaki M, Sugiyama T, Miyake H, Weber APM, Taniguchi M. 2011. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism: OMT in malate valve and C/N interaction. The Plant Journal 65, 15–26. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Peterson D, Tamura K. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28, 2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Mailund T, Pedersen CNS. 2004. QDist—quartet distance between evolutionary trees. Bioinformatics 20, 1636–1637. [DOI] [PubMed] [Google Scholar]
- Majeran W, van Wijk KJ. 2009. Cell-type-specific differentiation of chloroplasts in C4 plants. Trends in Plant Science 14, 100–109. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. 2007. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Research 35, D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. [DOI] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. 1994. A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiology 104, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. New Phytologist 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Review of Plant Biology 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E. 2012. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biology and Evolution 4, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sieburth LE. 2006. SCARFACE Encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. The Plant Cell 18, 1396–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski TL, Anderson AA, Zhang C, Turgeon R. 2012. Scarecrow plays a role in establishing kranz anatomy in maize leaves. Plant and Cell Physiology 53, 2030–2037. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still CJ, Berry JA, Collatz GJ, DeFries RS. 2003. Global distribution of C3 and C4 vegetation: Carbon cycle implications. Global Biogeochemical Cycles 17, 6-1–6-14. [Google Scholar]
- Strimmer K. 2008. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462. [DOI] [PubMed] [Google Scholar]
- Studer AJ, Gandin A, Kolbe AR, Wang L, Cousins AB, Brutnell TP. 2014. A limited role for carbonic anhydrase in C4 photosynthesis as revealed by a ca1ca2 double mutant in maize. Plant Physiology 165, 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalkowski AM. 2012. Fast and robust multiple sequence alignment with phylogeny-aware gap placement. BMC Bioinformatics 13, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. 2008. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biology 14, 2963–2977. [Google Scholar]
- Vincill ED, Clarin AE, Molenda JN, Spalding EP. 2013. Interacting glutamate receptor-like proteins in phloem regulate lateral root initiation in Arabidopsis. The Plant Cell 25, 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, et al. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. [DOI] [PubMed] [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, et al. 2014a Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nature Biotechnology 32, 1158–1165. [DOI] [PubMed] [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. 2009. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biology 10, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Long SP, Zhu X-G. 2014b Elements required for an efficient NADP-malic enzyme type C4 photosynthesis. Plant Physiology 164, 2231–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhövel A, Hein I, Dabrowa R, Stockhaus J. 2001. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Molecular Biology 45, 201–214. [DOI] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen Z-H, Leegood RC. 1999. Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiology 120, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.