Abstract
Genetic variants in the purinergic receptors P2RX4 and P2RX7 have been shown to affect susceptibility to multiple sclerosis (MS). In this study we set out to evaluate whether rare coding variants of major effect could also be identified in these purinergic receptors. Sequencing analysis of P2RX4 and P2RX7 in 193 MS patients and 100 controls led to the identification of a rare three variant haplotype (P2RX7 rs140915863:C>T (p.T205M), P2RX7 rs201921967:A>G (p.N361S) and P2RX4 rs765866317:G>A (p.G135S)) segregating with disease in a multi-incident family with six family members diagnosed with MS (LOD=3.07). Functional analysis of this haplotype in HEK293 cells revealed impaired P2X7 surface expression (p<0.01), resulting in over 95% inhibition of ATP-induced pore function (p<0.001) and a marked reduction in phagocytic ability (p<0.05). In addition, transfected cells showed 40% increased peak ATP-induced inward current (p<0.01), and a greater Ca2+ response to the P2X4 135S variant compared to wild type (p<0.0001). Our study nominates rare genetic variants in P2RX4 and P2RX7 as major genetic contributors to disease, further supporting a role for these purinergic receptors in MS and suggesting the disruption of transmembrane cation channels leading to impairment of phagocytosis as the pathological mechanisms of disease.
Keywords: Multiple sclerosis, mutation, variant, familial, Mendelian, P2RX4, P2RX7, P2X4, P2X7
INTRODUCTION
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system (CNS) characterized by myelin loss, axonal pathology and progressive neurological dysfunction. The etiology of MS is complex with both genetic and environmental factors implicated in disease susceptibility, with the human leucocyte antigen (HLA)-DRB1 region providing the highest known attributable risk (Traboulsee et al. 2014). Recently, functional variants in genes for the purinergic receptors P2RX7 (MIM# 602566) and P2RX4 (MIM# 600846) have been shown to modulate MS susceptibility (Gu et al. 2015). P2RX7 variants resulting in loss-of-function were found to be protective against MS, particularly rs28360457:G>A (p.R307Q) which has a dominant negative effect on risk of MS; whereas a P2RX7 rs1718119:G>A (p.A348T) gain-of-function variant increased risk of disease. In addition, a rare P2RX4 rs28360472:A>G (p.Y315C) loss-of-function mutation showed a trend towards association suggesting it increases the risk of MS (Gu et al. 2015; Stokes et al. 2011; Wiley et al. 2011).
The purinergic receptors P2X4 and P2X7 are non-selective transmembrane cation channels with critical roles in immunity and inflammation (Bours et al. 2006; Burnstock 2016; Khakh and North 2006). Activation of P2X7 by extracellular adenosine triphosphate (ATP) leads to pore formation in cells of monocyte-macrophage lineage and triggers a pro-inflammatory response, which includes activation of the NALP3 inflammasome, proliferation of B and T cells, and cytokine release (Feske et al. 2012; Junger 2011). However, in the absence of ATP, the P2X7 receptor has an alternative function in the recognition and regulation of apoptotic cell removal which has an important role in early human neurogenesis (Gu et al. 2011; Lovelace et al. 2015). Interestingly, postmortem studies have shown a greater density of P2X7 receptors in microglia cells from MS patients compared to controls (Yiangou et al. 2006); further supporting a role for P2X receptors in the mechanism of disease. Given the role of purinergic receptors in the innate and adaptive immune response, and the genetic and biological associations described in MS patients, we studied genetic variants in the P2RX7-P2RX4 loci by mining exome sequencing data from MS patients and healthy controls. A unique P2RX7-P2RX4 haplotype, which impairs the phagocytic function of the P2X7 receptor, was found to co-segregate with disease in one family with multiple affected members. While the P2X7 receptor is only one of many scavenger receptors in the CNS, our data show for the first time that an inherited defect in a scavenger receptor contributes to the pathogenesis of MS.
MATERIAL AND METHODS
Participants
A Caucasian case-control series consisting of 2,211 MS patients and 880 unrelated healthy control subjects from Canada were included in this study. Patients were diagnosed with MS according to Poser criteria prior to 2001 (Poser et al. 1983), or McDonald criteria thereafter (McDonald et al. 2001; Polman et al. 2005). The mean age at blood collection was 46.7 ± 11.8 years (SD) for MS patients and 67.2 ± 9.8 years (SD) for controls, with a male to female ratio of 1:2.75 and 1:0.96 respectively. The mean age at MS onset was 31.0 ± 9.7 years (SD), with a median Expanded Disability Status Scale (EDSS) score of 3.5 and an average of 4.05 ± 2.59 (SD). Exome sequencing data from 193 unrelated MS patients (51 male and 141 female) and 100 healthy controls (26 male and 74 female) was available for analysis. All samples were collected through the longitudinal Canadian Collaborative Project on the Genetic Susceptibility to Multiple Sclerosis (CCPGSMS) (Sadovnick et al. 1998; Traboulsee et al. 2014). The ethical review board at the University of British Columbia approved the study, and all participants provided informed consent.
Exome Sequencing
Exonic regions were enriched using an Ion AmpliSeq exome kit (57.7Mb) and sequenced in an Ion Proton sequencer (Life Technologies, Carlsbad, CA, USA) with a minimum average coverage of 70 reads per base and an average read length of 150 bases. The Ion Torrent Server was used to map reads to NCBI Build 37.1 reference genome and identify variants differing from the reference (Forwell et al. 2016). Annotation of variants was performed with ANNOVAR (Wang et al. 2010), and damage prediction on protein function assessed using Combined Annotation Dependent Depletion (CADD v1.3) (http://cadd.gs.washington.edu) (Kircher et al. 2014). All variants of interest were confirmed by Sanger sequencing as previously described (Trinh et al. 2013).
Linkage Analysis
Two-point logarithm of odds (LOD) score were obtained with MLINK assuming a dominant model, with a fully penetrant mutation and without phenocopies, as previously described (Chartier-Harlin et al. 2011; Ott 1989; Wang et al. 2016). The deleterious allele was defined with a 0.0001 frequency and the marker-allele frequencies determined empirically from genotyped individuals within the family.
Ethidium uptake
ATP-induced ethidium uptake assays were performed on HEK293 cells transfected with 4 μg P2X7-AcGFP plasmid DNA and/or 0.5 μg P2X4-AcGFP plasmid DNA. The pAcGFP-N1 vector (0.2 μg) was used as the mock transfected control, using linear polyethylenimine as described previously (Gu et al. 2015). Cell suspensions (1.0 ml) in HEPES-buffered KCl medium (150 mM KCl, 10 mM HEPES, pH 7.5, plus 5 mM D-glucose, 0.1% BSA) were added to the cuvette of a Time Zero module and gently stirred with temperature maintained at 37°C. Ethidium (25 μM) was added, followed 40 s later by addition of 1.0 mM ATP. Cells were analyzed at 1000 events/seconds on a FACSCalibur flow cytometer and were gated by forward and side scatter. The linear mean channel of fluorescence intensity (0–255 channel) of AcGFP+ cells over successive 5-second intervals was analyzed by WinMDI software and plotted against time as previously described (Gu et al. 2009). Paired Student’s t-test of the area under ATP-induced ethidium uptake curves were used to calculate statistical differences between P2X7 mutant constructs and wild-type P2X7 or P2X4-P2X7 constructs and wild type P2X4-P2X7.
P2X7 surface expression
Surface expression of the P2X7 receptor was measured by equilibrium binding of the Alexa 647 - conjugated P2X7 monoclonal antibody to transfected HEK293 cells. Transfected cells were incubated with Alexa 647 monoclonal anti-P2X7 antibody (Clone L4, 6 μg/ml) for 20 min at 4°C, washed once and analyzed by two-color flow cytometry with gating on AcGFP+ cells. Statistical differences between mutant constructs and equivalent wild-type transfections were assessed using Student’s t-test.
Phagocytosis of YO beads by transfected HEK293 cells
Transfected HEK293 cells (~2x106/mL) were resuspended in HEPES-buffered NaCl medium (145 mM NaCl, 5 mM KCl, 10 mM HEPES, pH 7.5, plus 5 mM D-glucose, 0.1% BSA, 0.1 mM CaCl2). For phagocytosis assay, 5 μL 1.0 μm carboxylated yellow-orange (YO) beads (Polyscience, USA) were added at zero time. The linear mean channel of fluorescence intensity for gated AcGFP+ HEK293 cells over successive 10-s intervals was analyzed by WinMDI software and plotted against time, as described previously (Gu et al. 2014). Paired student’s t-test was used to assess differences between mutant and wild-type constructs.
Ca2+ influx assay
HEK293 cells were transfected with pAcGFP-N1 (mock), P2X4-WT-AcGFP and P2X4-135S-AcGFP constructs. Cells (2x106) were incubated in Ca2+ free Na medium (145 mM NaCl, 5 mM KCl, 10 mM Hepes, pH 7.5, supplemented with 0.1% BSA and 5mM glucose) with Fura-Red acetoxymethyl ester (1 μg/mL) for 30 min at 37°C. After washed once, cells were kept at 37°C for another 30 min followed by the second wash. Cells were resuspended in 1 mL Na medium and analysed by a FACSCalibur flow cytometer with a Time Zero module (Cytek, USA). CaCl2 (3 mM) was added and the FL3 voltage is adjusted to give a linear mean channel fluorescence intensity of ~700 for the gated AcGFP+ population. ATPγS (100 μM) was added 40 sec later. The mean fluorescence intensity of Fura Red was recorded in every 2 seconds and plotted against time. The data from each run (about 250 sec) is saved into a listmode file and analyzed with WinMDI software (The Scripps Research Institute, CA, USA). The area above Ca2+ influx curve 60 seconds after addition of agonist was calculated as the arbitrary units for quantitation.
In vitro electrophysiology
HEK293 cells were transfected with pAcGFP-N1 (mock), P2X4-WT-AcGFP and P2X4-135S-AcGFP constructs. Forty-eight hours post-transfection the cells were detached using 0.5 mL Accutase® Cell Detachment Solution (Innovative Cell Technologies Inc., San Diego, CA, USA) and resuspended at a density of 1x106 to 5x107 per mL in 50 % serum-free media and 50 % external recording solution v/v. The external recording solution comprised (mmol/L): 145 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 13 D-glucose, 10 HEPES (pH 7.4 with NaOH; ~298 mOsm). The internal recording solution comprised (mmol/L): 85 NaCl, 60 NaF, 10 EGTA, 10 HEPES (pH 7.2 with NaOH; ~285 mOsm). Solutions were filtered using a 0.2 μm membrane filter (Minisart; Sartorius Stedim Biotech, Goettingen, Germany). Cells were kept in suspension by gentle automatic pipetting. Currents generated by P2X4 receptors were recorded using the Patchliner® (Nanion Technologies, Munich, Germany) in the whole-cell configuration. Medium single-hole planar NPC-16 chips with an average resistance of ~2.5 M3 were used. Pipette and whole cell capacitance were fully compensated. Recordings were acquired at 1 kHz with the low pass filter set to 0.33 kHz in PATCHMASTER (HEKA Instruments Inc., NY, USA) and performed at room temperature. P2X4 receptor-dependent currents were induced by application of 100 μM ATP. The cells were held at −50 mV. Offline analysis was performed using MATLAB R2015a (The Math Works Inc., Natick, Massachusetts, United States) and GraphPad Prism 6 (Molecular Devices). Data are shown as means ± standard error of the mean (SEM). Statistical analysis was performed using Student’s t-test. Statistical differences were considered significant when p<0.05.
RESULTS
Genetic analysis of P2RX7 and P2RX4 in MS patients
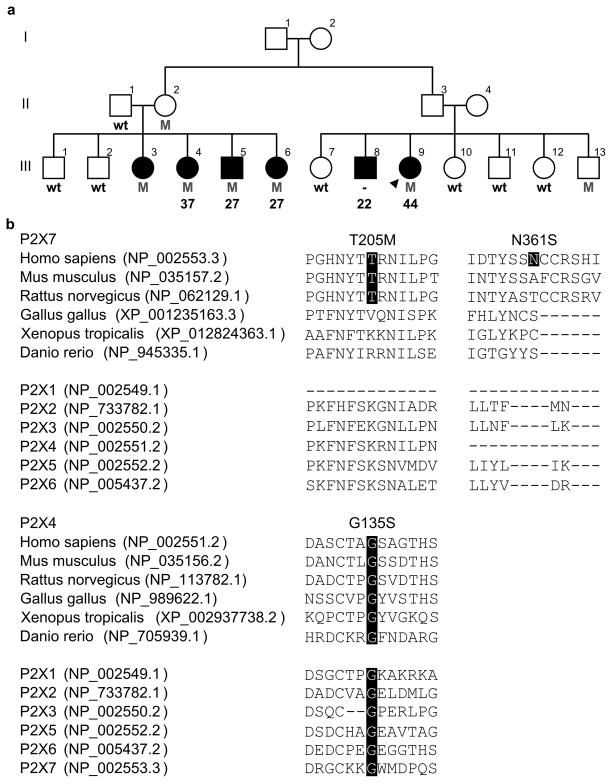
The human P2RX7 and P2RX4 genes are located 23 kb apart on chromosome 12q24.31. P2RX7 (NM_002562.5) contains 13 exons which encode P2X7, a protein 595 amino acids in length. P2RX4 (NM_002560.2) has 12 exons and encodes a 388 amino acid protein, P2X4. Exome sequencing in 193 MS probands and 100 controls resulted in an average sequencing depth of 156.9 ± 90.2 reads (SD) for P2RX7 and 133.2 ± 86.7 (SD) for P2RX4. All coding exons were comprehensively sequenced with the exception of P2RX7 exon 12 and P2RX4 exon 1, which presented an average depth of 9.4 ± 5.2 (SD) and 6.0 ± 7.8 (SD) respectively. Exome analysis identified 23 missense substitutions in P2RX7 and four in P2RX4 (Table 1). Of those, 12 were exclusively identified in MS patients and were assessed for segregation with disease in additional family members when available (Supp. Figure S1). Seven variants were considered not to segregate with disease as they were present in at least two unaffected blood-related family members (excluding obligate carriers), or were not observed in at least two family members diagnosed with MS. All remaining variants (P2RX7 rs140915863:C>T (p.T205M), rs146725537:T>C (p.Y288H), rs201921967:A>G (p.N361S), rs34219304:G>A (p.V522I) and P2RX4 rs765866317:G>A (p.G135S)) were genotyped in a case control series consisting of 2,211 MS patients and 880 healthy controls. P2X7 p.Y288H and p.V522I were identified at similar frequencies in MS patients and controls (Supp. Table S1), thus suggesting they are both rare polymorphisms of no effect towards the pathogenesis or susceptibility to MS. P2X7 p.T205M, p.N361S and P2X4 p.G135S, which are found on a single haplotype, were not observed in any additional subjects. This P2RX7-P2RX4 three variant haplotype was identified in five family members diagnosed with MS, one obligate carrier and only one out of seven seemingly healthy blood-relatives from a single family (Fig. 1A). One additional family member diagnosed with MS was known to exist (III-8), but was deceased at the time of collection.
Table 1.
P2RX7 and P2RX4 variants identified in MS patients and healthy controls. Genomic coordinates are given from NCBI Build 37.1; RNA and protein position are based on GenBank RefSeq NM_002562.5 (P2RX7) and NM_002560.2 (P2RX4); dbSNP IDs are provided from build 149; minor allele frequencies (MAF) were obtained from The Exome Aggregation Consortium (ExAC) database.
| Chromosome: Position | cDNA change | Aminoacid change | dbSNP rs ID | ExAC MAF | Controls MAF | MS MAF |
|---|---|---|---|---|---|---|
| P2RX7 | ||||||
| 12:121592689 | c.227T>C | p.V76A | rs17525809 | 0.0597 | 0.081 | 0.078 |
| 12:121593936 | c.349C>T | p.R117W | rs28360445 | 0.0013 | 0.005 | 0.010 |
| 12:121598715 | c.374G>T | p.R125L | rs201668926 | 2.47x10−5 | 0 | 0.003* |
| 12:121600233 | c.443A>G | p.Q148R | rs150235326 | 2.49x10−5 | 0 | 0.003* |
| 12:121600238 | c.448G>A | p.G150R | rs28360447 | 0.0127 | 0 | 0.016* |
| 12:121600253 | c.463C>T | p.H155Y | rs208294 | 0.4757 | 0.409 | 0.422 |
| 12:121603240 | c.614C>T | p.T205M | rs140915863 | 0.0005 | 0 | 0.003* |
| 12:121605337 | c.791G>A | p.R264H | rs149639375 | 0.0002 | 0 | 0.003* |
| 12:121605355 | c.809G>A | p.R270H | rs7958311 | 0.2448 | 0.278 | 0.313 |
| 12:121605373 | c.827G>A | p.R276H | rs7958316 | 0.0177 | 0.010 | 0.016 |
| 12:121605408 | c.862T>C | p.Y288H | rs146725537 | 0.0019 | 0 | 0.003* |
| 12:121613229 | c.920G>A | p.R307Q | rs28360457 | 0.0081 | 0.030 | 0.021 |
| 12:121615103 | c.1042G>A | p.A348T | rs1718119 | 0.3582 | 0.354 | 0.298 |
| 12:121615131 | c.1070C>G | p.T357S | rs2230911 | 0.1323 | 0.081 | 0.078 |
| 12:121615143 | c.1082A>G | p.N361S | rs201921967 | 8.24x10−6 | 0 | 0.003* |
| 12:121622115 | c.1298C>T | p.A433V | rs28360459 | 0.0109 | 0.005 | 0.003 |
| 12:121622196 | c.1379A>G | p.Q460R | rs2230912 | 0.1318 | 0.136 | 0.132 |
| 12:121622304 | c.1487A>C | p.E496A | rs3751143 | 0.1903 | 0.162 | 0.189 |
| 12:121622380 | c.1563C>G | p.H521Q | rs2230913 | 0.0132 | 0.005 | 0.003 |
| 12:121622381 | c.1564G>A | p.V522I | rs34219304 | 0.0035 | 0 | 0.003* |
| 12:121622421 | c.1604C>T | p.A535V | rs201256156 | 0.0004 | 0 | 0.003* |
| 12:121622448 | c.1631G>A | p.R544Q | rs34567077 | 3.04x10−5 | 0 | 0.003* |
| 12:121622520 | c.1703T>A | p.I568N | rs1653624 | 0.0149 | 0.040 | 0.023 |
| P2RX4 | ||||||
| 12:121647974 | c.7G>T | p.G3C | rs200492184 | 0.0089 | 0.005 | 0 |
| 12:121659945 | c.403G>A | p.G135S | rs765866317 | 3.32x10−5 | 0 | 0.003* |
| 12:121666646 | c.724A>G | p.S242G | rs25644 | 0.1566 | 0.136 | 0.106 |
| 12:121670276 | c.944A>G | p.Y315C | rs28360472 | 0.0091 | 0 | 0.018* |
Variants assessed for segregation are indicated with an asterisk.
Figure 1. Pedigree presenting the P2X7 p.T205M, P2X7 p.N361S and P2X4 p.G135S haplotype and protein conservation.
A) Males are represented by squares and females by circles with the proband indicated win an arrow head. Patients diagnosed with MS have black filled symbols with corresponding age at onset of disease when available. Heterozygote carriers for all three mutations (M) and wild type (wt) haplotypes are indicated. B) Conservation of P2X7 and P2X4 variants in orthologs and human paralogs are highlighted in black. Protein homologs were aligned via ClustalO, and RefSeq accession numbers are provided.
To determine whether any other variant could be ascribed to the high prevalence of disease observed in this family, we performed exome sequencing analysis in all additional family members diagnosed with MS for whom a DNA sample was available (III-3, 4, 5 and 6; Fig. 1A). The pattern of disease inheritance in this family is suggestive of an autosomal dominant trait with reduced penetrance, therefore only rare heterozygote coding substitutions identified in all affected individuals were considered potentially pathogenic; detailed methodology has been previously described (Wang et al. 2016). Exome analysis identified a total of 82,073 variants across all five affected subjects, with 3,548 autosomal heterozygote genotypes being observed in all family members examined. After excluding non-coding and silent substitutions, as well as variants with a reported minor allele frequency over 1% in proprietary and public databases (Exome Aggregation Consortium et al. 2015), only the three variants on the P2RX7-P2RX4 haplotype were found to be present in all five family members diagnosed with MS. Parametric linkage analysis resulted in a maximum LOD score of 3.07, confirming the co-segregation of this haplotype with disease; thus indicating the variants in this haplotype as the most likely pathogenic mechanism of disease for this family. In silico analysis of these variants found P2X7 p.T205M to be evolutionarily conserved in mammals (Fig. 1B) and predicted likely damaging to protein function with a phred-scaled CADD score of 25.8; whereas P2X7 p.N361S is neither evolutionarily conserved nor predicted damaging (CADD = 0.003). P2X4 p.G135S is predicted deleterious to protein function (CADD = 28.0) and shows the highest level of conservation amongst these three mutations, with the glycine residue being observed not only in vertebrates but also all human paralogs.
Clinical phenotype for P2RX7-P2RX4 patients
The family harboring the P2RX7-P2RX4 disease haplotype is of mixed European/Acadian ancestry. The mean age at the onset of MS was 31.4 ± 8.9 years (SD) with a range of 22 to 44 (Fig. 1A). Two family members were deceased by age 48 (III-6) and 57 (III-8), 21 and 35 years after the onset of MS respectively. Detailed clinical phenotype was available for three family members (III-4, III-6 and III-9), and was consistent with a relapsing-remitting course which became progressive for III-6 and III-9 within 15 and 13 years from onset, respectively. The initial symptoms for III-4 and III-6 were sensory disturbances whereas III-9 presented slowness of movements at the onset of MS. Two seemingly unaffected mutation carriers were also identified (II-2 and III-13). Biological samples from these two family members were collected at 76 (II-2) and 51 (III-13) years of age, and MS symptoms were not reported at collection thus indicating that the P2RX7-P2RX4 three mutation haplotype has reduced penetrance.
A unique P2RX7-P2RX4 haplotype confers major inhibition on P2X7 pore function
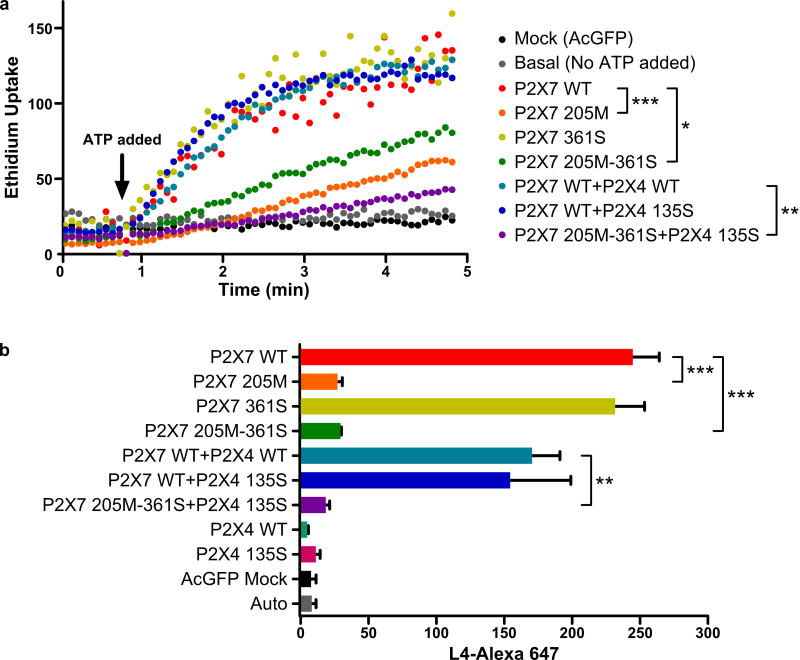
Activation of P2X7 by ATP induces pore formation in cells of monocytic/microglia lineage, which has downstream effects including formation of the NALP3 inflammasone and secretion of proinflammatory cytokines (Mariathasan et al. 2006). We examined P2X7 pore formation by measuring ATP induced ethidium uptake in HEK293 cells transfected with wild type or mutated P2X7 and/or P2X4 constructs. The P2X7 205M variant conferred greater than 80% inhibition of P2X7 mediated ethidium uptake (p=0.0003) whereas P2X7 361S constructs gave the same ethidium uptake as cells transfected with wild type P2X7 (Fig. 2A). Transfection of HEK293 cells with P2X7 205M plus 361S double mutant constructs resulted in approximately 70% inhibition of P2X7 pore function compared to wild type (p=0.011). The greatest inhibition of pore function (>95%) was observed when this double P2X7 mutant was co-transfected with P2X4 135S to reproduce the full P2RX7-P2RX4 haplotype observed in MS patients (p=0.008). Our data show that the P2X7 205M variant is the major contributor to the inhibition of P2X7 pore function, an inhibition that is observed in the absence or presence of variant P2X4 135S. In contrast P2X7 361S appears to have little or no effect on P2X7 pore function.
Figure 2. P2X7 functional assay.
A) ATP induced ethidium uptake was used to measure P2X7 pore formation in HEK293 cells transfected with various P2X7 and/or P2X4 constructs. The mean fluorescent intensity of ethidium uptake into cells was measured at 5 sec intervals by two-color flow-cytometry. B) P2X7 expression on the surface of HEK 293 cells transfected with AcGFP-tagged P2X7 and/or P2X4 constructs. Bars represent mean values and SEM of three replicate experiments. WT = wild type. *p<0.05, **p<0.01, ***p<0.001.
The P2X7 205M-361S variant fails to express on the surface of transfected HEK293 cells
We measured the surface expression of the P2X7 receptor in transfected HEK293 cells by the binding of Alexa-647 conjugated monoclonal antibody (Clone L4) specific for the extracellular domain of P2X7 (Buell et al. 1998). Identical expression of wild type P2X7 either with or without the 361S mutation was observed in HEK293 cells and there was a trend to lower values in cells co-transfected with the P2X4 constructs, either wild type or 135S mutant (Fig. 2B). A major reduction in surface expression was found in cells expressing the 205M mutation of P2X7. An identical major reduction in P2X7 expression was observed in cells harboring 205M either with or without the 361S mutation or in P2X7 205M 361S cells co-transfected with P2X4 135S (p=0.002–0.0004). Confocal microscopy confirmed surface expression of P2X7 in wild-type and 361S transfected cells, whereas P2X7 constructs containing the 205M variant did not show detectable surface expression (Supp. Figure S2). This data shows that the P2X4 135S and P2X7 361S mutations have no significant effect on P2X7 expression and that the P2X7 205M variant has by far the greatest influence on surface expression of this receptor.
Phagocytosis mediated by the P2X7 receptor is abolished by the T205M variant
Phagocytosis mediated by P2X7 requires surface expression of this receptor in a membrane complex which contains non-muscle myosin IIA and other cytoskeletal proteins (Gu et al. 2009) which allow recognition and engulfment of apoptotic cells or latex beads in the absence of extracellular ATP (Gu et al. 2010; Gu et al. 2011). We assayed this scavenger role of P2X7 by measuring the phagocytosis of non-opsonized YO fluorescent beads in HEK293 cells transfected with the same P2X7 and P2X4 constructs previously described. Wild type P2X7 transfected HEK293 cells, as well as P2X7 361S and all P2X4 constructs, showed brisk engulfment of beads at 37°C, while cells transfected with any combination of constructs containing P2X7 205M showed significantly impaired bead uptake (p<0.05) (Supp. Figure S3 & S4).
Functional studies on the P2X4 135S variant
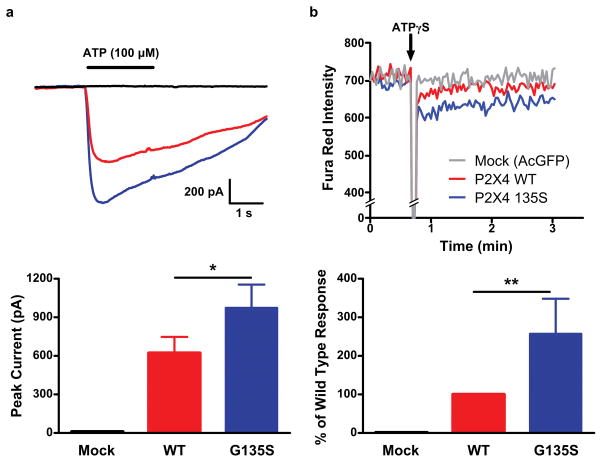
P2X4 and P2X7 receptors have been reported to interact on the surface of immune cells eliciting changes in function, but not expression levels of P2X7 (Guo et al. 2007). The p.G135S mutation was introduced into a human P2X4 plasmid and expressed in HEK293 cells to assess changes in functional responses. Whole-cell patch clamp recordings showed approximately 40% increased peak ATP-induced inward current for P2X4 135S compared to wild type (p<0.01, Fig. 3A). The functional response of the P2X4 135S variant was also assessed by a second technique in which HEK293 cells transfected with P2X4 wild type or 135S were loaded with the Ca2+-sensitive dye Fura Red. The fluorescence response to activation of P2X4 by its specific ligand, ATPγS (100 μM) was measured by real-time flow cytometry, which again showed a greater Ca2+ response to the P2X4 135S variant compared to wild type (p<0.0001, Fig. 3B). These data show that the P2X4 135S variant retains its high permeability to Ca2+ ions at wild type or greater levels.
Figure 3. P2X4 functional assay using HEK293 cells transfected with AcGFP, wild type P2X4-AcGFP or P2X4-G135S-AcGFP.
A) Whole-cell averaged current traces from mock vector control alone (n = 6 cells), P2X4-WT (n = 6 cells) and P2X4-G135S (n = 10 cells) in response to ATP (100 μM). B) Typical Ca2+ influx traces. Ca2+ (3 mM) was added 40 sec prior to the addition of 100 μM ATPγS. Normalized Ca2+ influx from four separated experiments. *p<0.01; **p<0.0001.
DISCUSSION
In this study we have identified a P2RX7-P2RX4 haplotype containing three rare missense mutations segregating with disease in a multi-incident family with high incidence of MS (Fig. 1A). This data not only shows the existence of genetic variants resulting in familial forms of MS, but also highlights a less recognized function of the P2X7 receptor. Our functional studies revealed that the P2X7 205M, P2X7 361S, P2X4 135S haplotype expressed in HEK293 cells causes near total loss of ATP-induced ethidium uptake, which is the standard assay for the P2X7 proinflammatory actions of this receptor (Fig. 2A). This result was opposite to that expected for the loss of P2X7 pore function, which generally protects rather than promotes secretion of proinflammatory cytokines in the CNS (Mariathasan et al. 2006; Qu et al. 2007; Sluyter et al. 2004); and it is in sharp contrast to our recent genetic study of a P2X7 p.R307Q loss of function variant which is associated with a two-fold increased protection against the risk of developing MS (Gu et al. 2015). To resolve this paradox we examined the expression of P2X7 as well as measuring a second function of this receptor, namely a scavenger function in which P2X7 in the absence of ATP can recognize and regulate the phagocytosis of apoptotic cells (Gu et al. 2010; Gu et al. 2011). Unlike the majority of polymorphic variants such as p.R307Q, which are expressed close to wild type levels on the cell surface, the P2RX7-P2RX4 haplotype identified in the index family shows near total absence of P2X7 surface expression on HEK293 cells (Fig. 2B). Cells transfected with P2X7 205M constructs failed to bind the monoclonal L4-antibody, which is specific for the extracellular domain of P2X7 (Buell et al. 1998). This result explains the loss of P2X7-mediated ethidium uptake by the mutant haplotype and suggests an impaired trafficking of P2X7 to the cell surface, although accelerated turnover of surface receptors, cannot be excluded. Failure of the P2X7 mutant haplotype to express on the cell surface resulted in a major loss of phagocytic ability in HEK293 cells, with the uptake of fluorescent beads being significantly reduced compared to wild type transfected cells. Together these functional data show the near complete abolition of both pore function and scavenger function is a result of the failure of P2X7 receptors to express on the cell surface. The role of P2X4 p.G135S on P2X7 function appears minor although there is some evidence of interaction between P2X4 and P2X7 receptors on the surface of immune cells. P2X4 and P2X7 subunits can be co-immunoprecipitated both from transfected and native cells while protein-protein interactions between P2X4 and the carboxyl tail of P2X7 have been shown to in FRET experiments (Guo et al. 2007; Perez-Flores et al. 2015). Our data is in line with a previous study (Guo et al. 2007) and show that co-transfection with P2X4 135S and P2X7 205M constructs gave even greater inhibition of pore function compared to P2X7 205M alone. However, this functional interaction of mutant P2X4 and P2X7 did not significantly further reduce the surface expression of P2X7 receptors.
Functional analysis of P2X4 showed that the 135S mutation identified in MS patients resulted in increased ATP-induced inward currents. Although the P2X4 contribution to inflammation appears minor in comparison to P2X7 (Burnstock 2016; de Rivero Vaccari et al. 2014), disruption of the P2X7-P2X4 interaction has been shown to hinder the physiological response to ATP in immune cells (Perez-Flores et al. 2015), and appears to potentially exacerbate the P2X7 phenotype (Fig. 2A). Further studies are necessary to fully characterize the role and interaction of P2X7 and P2X4 mutations in the onset of MS. However, our study is the first to show impaired phagocytosis as a mechanism of disease for MS, and suggest impaired function of scavenger receptors as one of the critical component in MS pathophysiology.
The pathogenicity of the P2RX7-P2RX4 haplotype identified in a multi-incident family (Fig. 1A) is supported by a significant co-segregation with MS (LOD = 3.07), disrupted biological functions (Fig. 2 & 3), and previously reported associations between common variants and MS risk (Gu et al. 2015). Functional analysis indicates that P2X7 p.N361S is unlikely to be pathogenic; however, genotyping in additional multi-incident families is necessary to elucidate whether the disease haplotype, P2X7 p.T205M or p.P2X4 p.G135S is the major genetic contributor to the high prevalence of MS observed in this family. It should also be noted that the identification of two unaffected mutation carriers suggests that additional genetic or environmental factors may be required in addition to this highly susceptible genetic background to trigger the onset of disease.
In summary, our data shows that the P2RX7-P2RX4 three mutation haplotype identified in a multi-incident MS family greatly impairs the expression of P2X7 on the cell surface, resulting in almost complete abolition of the scavenger function of this archaic purinergic receptor. This data is in sharp contrast to previously characterized polymorphisms in these genes, and suggests the disruption of transmembrane cation channels and impairment of phagocytosis by P2RX7 and P2RX4 mutations as a major contributor to the onset of MS.
Supplementary Material
Acknowledgments
We are grateful to all individuals who generously participated in this study. We thank Kevin Atkins for data collection and extraction. This research was undertaken thanks to funding from the Canada Research Chair (950-228408) and Canada Excellence Research Chair (214444) programs, Canadian Institutes of Health Research (MOP-137051), Vancouver Coastal Health Research Institute, the Milan & Maureen Ilich Foundation (11-32095000), and the Vancouver Foundation (ADV14-1597). This work was also supported by ARC Future Fellowship (BG, FT120100581), NHMRC Project Grant (1048082, 1061419) and the Victorian Government’s Operational Infrastructure Support Grant to the Florey Institute.
Grant numbers: 950-228408, 214444, MOP-137051, 11-32095000, ADV14-1597, FT120100581, 1048082, 1061419.
References
- Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5’-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Buell G, Chessell IP, Michel AD, Collo G, Salazzo M, Herren S, Gretener D, Grahames C, Kaur R, Kosco-Vilbois MH, Humphrey PP. Blockade of human P2X7 receptor function with a monoclonal antibody. Blood. 1998;92:3521–3528. [PubMed] [Google Scholar]
- Burnstock G. P2X ion channel receptors and inflammation. Purinergic Signal. 2016;12:59–67. doi: 10.1007/s11302-015-9493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, Le Rhun E, Mutez E, et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rivero Vaccari JP, Dietrich WD, Keane RW. Activation and regulation of cellular inflammasomes: gaps in our knowledge for central nervous system injury. J Cereb Blood Flow Metab. 2014;34:369–375. doi: 10.1038/jcbfm.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Aggregation Consortium. Lek M, Karczewski K, Minikel E, Samocha K. Analysis of protein-coding genetic variation in 60, 706 humans. BioRxiv. 2015 doi: 10.1038/nature19057. http://dx.doi.org/10.1101/030338. [DOI] [PMC free article] [PubMed]
- Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forwell AL, Bernales CQ, Ross JP, Yee IM, Encarnacion M, Lee JD, Sadovnick AD, Traboulsee AL, Vilarino-Guell C. Analysis of CH25H in multiple sclerosis and neuromyelitis optica. J Neuroimmunol. 2016;291:70–72. doi: 10.1016/j.jneuroim.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu BJ, Field J, Dutertre S, Ou A, Kilpatrick TJ, Lechner-Scott J, Scott R, Lea R, Taylor BV, Stankovich J, Butzkueven H, Gresle M, et al. A rare P2X7 variant Arg307Gln with absent pore formation function protects against neuroinflammation in multiple sclerosis. Hum Mol Genet. 2015;24:5644–5654. doi: 10.1093/hmg/ddv278. [DOI] [PubMed] [Google Scholar]
- Gu BJ, Rathsam C, Stokes L, McGeachie AB, Wiley JS. Extracellular ATP dissociates nonmuscle myosin from P2X7 complex: this dissociation regulates P2X7 pore formation. Am J Physiol Cell Physiol. 2009;297:C430–439. doi: 10.1152/ajpcell.00079.2009. [DOI] [PubMed] [Google Scholar]
- Gu BJ, Saunders BM, Jursik C, Wiley JS. The P2X7-nonmuscle myosin membrane complex regulates phagocytosis of non-opsonized particles and bacteria by a pathway attenuated by extracellular. ATP Blood. 2010;115:1621–1631. doi: 10.1182/blood-2009-11-251744. [DOI] [PubMed] [Google Scholar]
- Gu BJ, Saunders BM, Petrou S, Wiley JS. P2X(7) is a scavenger receptor for apoptotic cells in the absence of its ligand, extracellular. ATP J Immunol. 2011;187:2365–2375. doi: 10.4049/jimmunol.1101178. [DOI] [PubMed] [Google Scholar]
- Gu BJ, Sun C, Fuller S, Skarratt KK, Petrou S, Wiley JS. A quantitative method for measuring innate phagocytosis by human monocytes using real-time flow cytometry. Cytometry A. 2014;85:313–321. doi: 10.1002/cyto.a.22400. [DOI] [PubMed] [Google Scholar]
- Guo C, Masin M, Qureshi OS, Murrell-Lagnado RD. Evidence for functional P2X4/P2X7 heteromeric receptors. Mol Pharmacol. 2007;72:1447–1456. doi: 10.1124/mol.107.035980. [DOI] [PubMed] [Google Scholar]
- Junger WG. Immune cell regulation by autocrine purinergic signalling. Nat Rev Immunol. 2011;11:201–212. doi: 10.1038/nri2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442:527–532. doi: 10.1038/nature04886. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace MD, Gu BJ, Eamegdool SS, Weible MW, Wiley JS, Allen DG, Chan-Ling T. P2X7 receptors mediate innate phagocytosis by human neural precursor cells and neuroblasts. Stem cells. 2015;33:526–541. doi: 10.1002/stem.1864. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, McFarland HF, Paty DW, Polman CH, Reingold SC, Sandberg-Wollheim M, Sibley W, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- Ott J. Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A. 1989;86:4175–4178. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Flores G, Levesque SA, Pacheco J, Vaca L, Lacroix S, Perez-Cornejo P, Arreola J. The P2X7/P2X4 interaction shapes the purinergic response in murine macrophages. Biochem Biophys Res Commun. 2015;467:484–490. doi: 10.1016/j.bbrc.2015.10.025. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, Lublin FD, Metz LM, McFarland HF, O’Connor PW, Sandberg-Wollheim M, Thompson AJ, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- Poser CM, Paty DW, Scheinberg L, McDonald WI, Davis FA, Ebers GC, Johnson KP, Sibley WA, Silberberg DH, Tourtellotte WW. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol. 1983;13:227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. Journal of Immunology. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- Sadovnick AD, Risch NJ, Ebers GC. Canadian collaborative project on genetic susceptibility to MS, phase 2: rationale and method. Canadian Collaborative Study Group. Can J Neurol Sci. 1998;25:216–221. doi: 10.1017/s0317167100034041. [DOI] [PubMed] [Google Scholar]
- Sluyter R, Dalitz JG, Wiley JS. P2X7 receptor polymorphism impairs extracellular adenosine 5′-triphosphate-induced interleukin-18 release from human monocytes. Genes & Immunity. 2004;5:588–591. doi: 10.1038/sj.gene.6364127. [DOI] [PubMed] [Google Scholar]
- Stokes L, Scurrah K, Ellis JA, Cromer BA, Skarratt KK, Gu BJ, Harrap SB, Wiley JS. A loss-of-function polymorphism in the human P2X4 receptor is associated with increased pulse pressure. Hypertension. 2011;58:1086–1092. doi: 10.1161/HYPERTENSIONAHA.111.176180. [DOI] [PubMed] [Google Scholar]
- Traboulsee AL, Bernales CQ, Ross JP, Lee JD, Sadovnick AD, Vilarino-Guell C. Genetic variants in IL2RA and IL7R affect multiple sclerosis disease risk and progression. Neurogenetics. 2014;15:165–169. doi: 10.1007/s10048-014-0403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh J, Vilarino-Guell C, Donald A, Shah B, Yu I, Szu-Tu C, Aasly JO, Wu RM, Hentati F, Rajput AH, Rajput A, Farrer MJ. STX6 rs1411478 is not associated with increased risk of Parkinson’s disease. Parkinsonism Relat Disord. 2013;19:563–565. doi: 10.1016/j.parkreldis.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sadovnick AD, Traboulsee AL, Ross JP, Bernales CQ, Encarnacion M, Yee IM, de Lemos M, Greenwood T, Lee JD, Wright G, Ross CJ, et al. Nuclear receptor NR1H3 in familial multiple sclerosis. Neuron. 2016;90:948–954. doi: 10.1016/j.neuron.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley JS, Sluyter R, Gu BJ, Stokes L, Fuller SJ. The human P2X7 receptor and its role in innate immunity. Tissue Antigens. 2011;78:321–332. doi: 10.1111/j.1399-0039.2011.01780.x. [DOI] [PubMed] [Google Scholar]
- Yiangou Y, Facer P, Durrenberger P, Chessell IP, Naylor A, Bountra C, Banati RR, Anand P. COX-2, CB2 and P2X7-immunoreactivities are increased in activated microglial cells/macrophages of multiple sclerosis and amyotrophic lateral sclerosis spinal cord. BMC Neurol. 2006;6:12. doi: 10.1186/1471-2377-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.