Abstract
Proteins with knotted configurations are restricted in conformational space relative to unknotted proteins. Little is known if knotted proteins have sufficient dynamics to communicate between spatially separated substrate-binding sites. In bacteria, TrmD is a methyl transferase that uses a knotted protein fold to catalyze methyl transfer from S-adenosyl methionine (AdoMet) to G37-tRNA. The product m1G37-tRNA is essential for life as a determinant to maintain protein synthesis reading-frame. Using an integrated approach of structure, kinetic, and computational analysis, we show here that the structurally constrained TrmD knot is required for its catalytic activity. Unexpectedly, the TrmD knot has complex internal movements that respond to AdoMet binding and signaling. Most of the signaling propagates the free energy of AdoMet binding to stabilize tRNA binding and to assemble the active site. This work demonstrates new principles of knots as an organized structure that captures the free energies of substrate binding to facilitate catalysis.
Keywords: TrmD, RNA methylation, m1G37-tRNA, trefoil-knot protein fold
The catalytic function and biological specificity of proteins is dependent on intra-molecular signaling that transmits free energies of substrate binding to the active site. Such signaling leads to the precise assembly of the active site and influences the rate of catalysis. Substrate signaling is regulated by intrinsic protein motions to coordinate different binding events at spatially separated sites into an integrated mechanism1,2. Even large RNA-protein complexes exhibit complex dynamics of substrate-signaling3,4. Intriguingly, a small number of natural proteins (~1.5% in Protein Data Bank, PDB) possess the property of threading through a knotted backbone in highly constrained topology5,6. While topological constraints confer global stability to proteins7, they also suppress internal motions. Some natural protein knots are deep, presenting challenges to exploit dynamics within restricted conformational space. Indeed, knotted proteins have slower and less diffusive folding kinetics relative to unknotted proteins8–10. Whether knotted proteins have sufficient dynamics for substrate signaling and how the magnitudes of signaling are compared to unknotted proteins is unknown. Addressing this question is important for understanding the structure and function of knotted proteins.
TrmD is a bacterial enzyme with a trefoil-knot in the active site, involving three crossings of the protein backbone through a loop. TrmD catalyzes methyl transfer from AdoMet to the N1 of G37 on the 3' side of the tRNA anticodon11,12. The product m1G37-tRNA is essential for life to maintain translational reading-frame13–15. Elimination of TrmD increases protein synthesis frameshifts and causes cell death16. The TrmD knot is closely related to the trefoil-knot in SpoU methyl transferases17, which catalyze 2'-O-methylation to RNA ribose for wide-ranging activities18–23. In crystal structures of SpoU enzymes in complex with AdoMet, AdoHcy (S-adenosyl homocysteine), or sinefungin (SFN), the ligand in the deep crevice of the trefoil-knot11,12,20,22,23 has a bent conformation23, which constrains the adenosine (Ado)- and the methionine (Met)-equivalent moieties to face each other. In contrast, most methyl transferases bind AdoMet in the open space of a dinucleotide fold24, where the Ado and Met moieties are extended from each other25 (Supplementary Fig. 1).
TrmD is a leading antimicrobial drug target26, due to its essentiality for bacterial growth, its broad conservation across bacterial species, and its deep-rooted distinction from the human and archaeal counterpart Trm5, which has the dinucleotide fold27–30. In virtually all aspects of the methyl transfer reaction29,31–36, TrmD is distinct from Trm5. To succeed in drug targeting of TrmD, an understanding of how it differs from Trm5 in substrate signaling is necessary. This is now possible, based on our recent tRNA-bound crystal structure of TrmD in complex with SFN31. TrmD is an obligated homodimer that places each active site at the dimer interface, involving the AdoMet in chain A interacting with the tRNA in chain B, while the AdoMet in chain B is inactivated. While the catalytically active AdoMet and tRNA are in two separate domains in two separate chains, no model exists for how these two substrates communicate with each other and how they keep the other AdoMet inactive.
To answer these questions, we developed a multidisciplinary approach. We report here that the TrmD knot is structurally constrained relative to the Trm5 active site, but that this knot is necessary to fold AdoMet to the bent conformation for methyl transfer. The TrmD knot has complex intrinsic motions that regulate AdoMet signaling, with the strongest upon Ado binding to the knot. Another communication travels across the dimer interface to suppress the other AdoMet from action. Mutations in the knot impair propagation of motions and eliminate the regulated asymmetry of the two chains, permitting the two knots to act independently of each other. This work has both biological significance and chemical novelty. It reveals unexpected internal dynamics of the TrmD knot and identifies new principles of the knot for selective drug targeting.
RESULTS
The structurally constrained TrmD knot
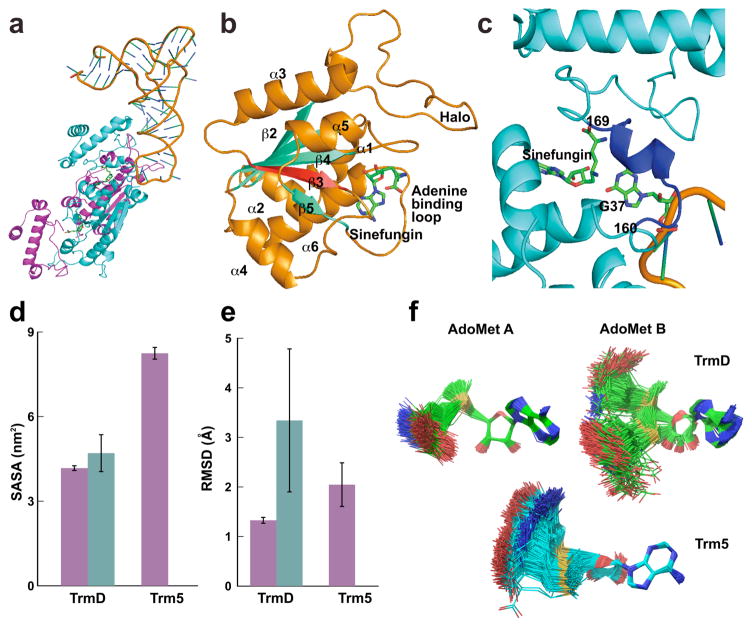
Each chain of the TrmD dimer has three distinct domains: an N-terminal domain (NTD) for binding AdoMet, a C-terminal domain (CTD) for binding tRNA, and a flexible linker in between. The dimer binds two SFNs but only one tRNA in a stoichiometry consistent with kinetic analysis36 (Fig. 1a). Each SFN is accommodated at the deep cleft of a trefoil-knot in the same position as AdoMet in a binary complex11. The trefoil-knot is made up of three β-strands at the central β-sheet, starting with β3, followed by a loop that turns in the back of β3 and emerges into β4. The end of β4 is followed by another loop that turns into β5, which makes a circular insertion into the knot by crossing over β3 and coming out of the knot with a loop that binds the adenine of SFN (Fig. 1b). The single tRNA is bound to the CTD of chain B, whose inter-domain linker stabilizes G37. Methyl transfer occurs by aligning AdoMet A with G37 on linker B for nucleophilic attack. This arrangement supports the half-of-the-sites model36, where only one knot is active at a given time. A unique feature of the tRNA-bound ternary complex31, relative to the AdoMet-bound binary complex11, is the ordering of just the G37-bound linker B (Fig. 1c).
Figure 1.
The ternary crystal structure and dynamics of the TrmD–tRNA–SFN complex. (a) The TrmD dimer binds one tRNA and two SFNs. Amino acid residues 1 to 160 are in the NTD, 169 to 246 in the CTD, and 161 to168 in the flexible linker. (b) A ribbon diagram of the trefoil-knot in chain A, showing β strands in aqua blue, α helices in orange, and SFN in green. The root-mean-square deviation (RMSD) of superposition of the knot with the knot in an AdoMet-bound binary complex is 0.32 Å for 64 Cα atoms. (c) A ribbon diagram of the inter-domain linker (residues 160–169) of chain B, which becomes organized upon binding of G37-tRNA. The RMSD of superposition of the CTD harboring this linker deviates from the corresponding region in the binary complex by 0.94 Å for 85 Cα atoms outside of the linker. A recombinant HiTrmD expressed in and purified from E. coli was used for structural analysis, while the tRNA was made by T7 transcription based on the Thermotoga maritima tRNAGln/CUG sequence31. (d) Mean Solvent Accessible Surface Area (SASA) of the active site A and active site B of HiTrmD (4.2 vs. 4.8 nm2) and MjTrm5 (Mj: Methanococcus jannaschii) (8.2 nm2). Magenta, active site A; blue aqua, active site B. Error bars are s.d. (n = 3 independent simulations). (e) RMSD of AdoMet A and AdoMet B in the active site of HiTrmD (1.3 and 3.3 Å) and MjTrm5 (2.0 Å). Magenta, AdoMet A; blue aqua, AdoMet B. Error bars are s.d. (n = 3 independent simulations). (f) Superposition of AdoMet conformations throughout a course of molecular dynamics simulations. The convergence of simulations was confirmed through independent and multiple rounds of calculations. Simulations were based on the crystal structure31 of HiTrmD and the crystal structure29 of MjTrm5.
We showed that the trefoil-knot indeed constrained the structure of TrmD relative to the unknotted archaeal counterpart28,29 Methanococcus jannaschii Trm5 (MjTrm5). By explicit solvent molecular dynamics simulations, we compared TrmD in the apo form, the AdoMet-bound binary complex, and the tRNA-bound ternary complex, with the ternary structure of MjTrm5. All simulated trajectories were stable over a long time, showing the RMSD (root mean square deviation) of most structures oscillating between 2 and 3 Å (Supplementary Fig. 2). The number of hydrogen (H) bonds between amino acids was stable up to 100 ns in all structures, and up to 400 ns in some structures. In the local structure of TrmD, the trefoil-knot is the least fluctuating relative to other regions (Supplementary Fig. 3), showing the lowest average value of RMSF (root mean square fluctuation) among all Cα atoms both in the apo-enzyme (1.1 Å) and in the tRNA-bound ternary complex (0.75 Å). Thus, the knot itself is intrinsically constrained, regardless of bound substrates. However, compared to the Trm5 active site, the structural mobility of the TrmD knot based on the average RMSF for both chains is smaller by only 0.2 Å.
Additional analysis showed that the average RMSF did not represent the TrmD knot. Instead, separate analysis of the two chains found that the two knots are different in mobility. The catalytically active knot A binds AdoMet in the bent shape and is more constrained relative to the Trm5 active site, as shown by the solvent accessible surface area (SASA) and the RMSD of the AdoMet (Fig. 1d, e). In contrast, the catalytically inactive knot B has a more fluid structure, with a high SASA value and most notably a high RMSD value. Furthermore, the catalytically inactive knot B can bind AdoMet in a range of shapes, from the bent to the extended conformation as in the Trm5 active site (Fig. 1f).
The AdoMet bent conformation necessary for methyl transfer
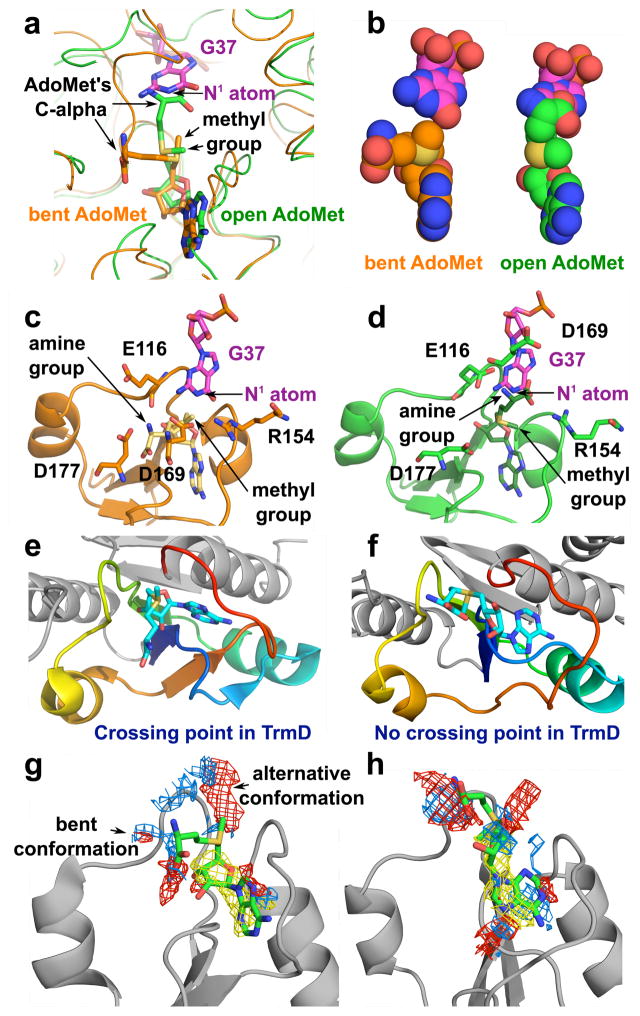
Simulation analysis showed that the bent shape in TrmD allows proper alignment for methyl transfer, whereas the open shape would introduce steric clash. In the bent shape, the methyl group of AdoMet is in an appropriate distance from N1 of G37 (3.5 Å) and the Cα is well separated (6.4 Å), whereas in the open shape, it is further away (4.3 Å) and the Cα is in direct conflict with the target base (0.8 Å) (Fig. 2a, b). Three acidic residues (E116, D169, and D177) form strong H-bonds with the amine of AdoMet in the bent shape, but these interactions (except with E116) are lost when AdoMet is extended (Fig. 2c, d). D169 is the general base31 for proton abstraction from N1 of G37. The distance between the center of mass of D169 and the amine of AdoMet is increased from the bent to the open shape (by 9.9 Å). Thus, the bent shape is the necessary conformation to conduit methyl transfer, while the open shape is sterically impossible.
Figure 2.
Molecular simulations of the bent vs. open shape of AdoMet. (a, b) The shape of AdoMet as shown by sticks and by van der Waals in the TrmD knot for the bent (a) and the open (b) conformation, respectively. (c, d) Positions of TrmD catalytic residues required for methyl transfer from AdoMet in the bent (c) and open (d) shape. (e, f) Accommodation of the bent shape in the knotted TrmD structure31 (PDB 4YVG) (e) and in the unknotted TrmD structure37 (PDB 1OY5) (f). (g) AdoMet in the knotted TrmD structure has the option to adopt the bent conformation or the alternative open conformation31 (PDB 4YVG). (h) AdoMet in the unknotted TrmD structure37 (PDB 1OY5) can only adopt the extended open conformation but not the bent conformation. Simulations in panels (g, h) were performed using SiteMap software.
We found that the Aquifex aeolicus TrmD was crystallized without a knot37. Because the crossing characteristics of the knot are absent from this structure, the cleft needed to bind AdoMet is not formed. Simulation and docking analysis showed that, although the unknotted structure may accommodate AdoMet in a similar location as in the knotted structure (Fig. 2e–f), the bent shape would result in a catalytically inactive complex due to orientation of the methyl group toward the protein. While the active site of the knotted TrmD can allow both the bent and extended conformations of the ligand (Fig. 2g), this is not possible in the active site of the unknotted TrmD, wherein only the extended conformation is allowed and the bent conformation is prohibited by the lack of space and stabilization (Fig. 2h). In Trm5, by contrast, docking analysis showed that, even if it can accommodate a bent shape, the complex would be inactive. The distance between the methyl group and the N1 of G37 in the simulated bent form (8.3 Å) is unfavorable for methyl transfer relative to the distance in the crystal structure29 (2.8 Å) and in the simulated open form (3.3 Å, Supplementary Fig. 1). Also, the bent form in the Trm5 structure would lose interactions with important enzyme residues (e.g., D223) for methyl transfer38. Thus, the bent conformation of AdoMet must be achieved in the TrmD knot and it is energetically unstable in the unknotted Trm5 and cannot be part of the active complex.
The landscape of intra-molecular substrate signaling
We identified mutations in the TrmD knot that altered AdoMet binding and communicated the alteration to reduce tRNA binding and methyl transfer. We used E. coli TrmD (EcTrmD) as a model33–36, which has high sequence identity (>83%) and similarity (93%) to H. influenzae TrmD in crystal structures11,31. A set of 15 residues in EcTrmD broadly conserved in evolution was mutated (Supplementary Note, Supplementary Table 1), 11 of which are in the NTD, 1 in the flexible linker, and 3 in the CTD. We mutated each to Ala and measured the mutational effect on AdoMet binding and assessed whether the loss of free energy of binding was transmitted to the tRNA-site and the active site. Although some of these Ala substitutions were previously tested12, no substrate-signaling information was available. We used pre-steady-state analysis to monitor the chemical step of methyl transfer in rapid equilibrium binding conditions36,39 (Supplementary Tables 2, 3, Supplementary Fig. 4).
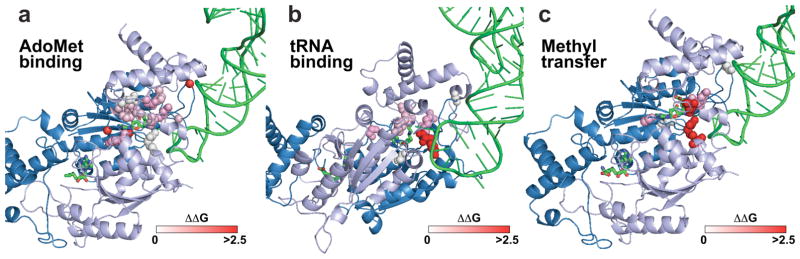
If a mutation decreased AdoMet or tRNA binding or methyl transfer, we calculated each decrease as the loss in free energies required for that specific action. Mapping individual energetic effects onto the ternary complex revealed the landscape of energy coupling (Fig. 3). Many residues that contribute to protein-AdoMet binding energies also contribute to protein-tRNA binding energies, indicating that these residues couple AdoMet binding with tRNA binding. Importantly, residues making the most energetic contribution to tRNA binding are also directly involved in methyl transfer, indicating that once tRNA is stabilized, TrmD is poised for methyl transfer. This landscape points to a unidirectional pathway of energy transmission, utilizing dynamic motions to capture the AdoMet binding energy to drive tRNA binding and to assemble the active site.
Figure 3.
Signaling strengths from individual protein-ligand contacts mapped to the ternary structure of HiTrmD. (a) Surface representation of HiTrmD (blue), showing quantitative effects of individual Ala substitutions (from white to pink to red) on protein-AdoMet binding. (b) Surface representation of HiTrmD, showing quantitative effects of individual Ala substitutions on protein-tRNA binding. (c) Surface representation of HiTrmD showing quantitative effects of individual Ala substitutions on kchem of methyl transfer. The scale at the bottom of each panel calibrates ΔΔG0 = −RT ln[(kchemwt/kchemmutant)], where T = 310 K and R = 1.987 kcal/mol.
Initiation of substrate signaling by Ado binding
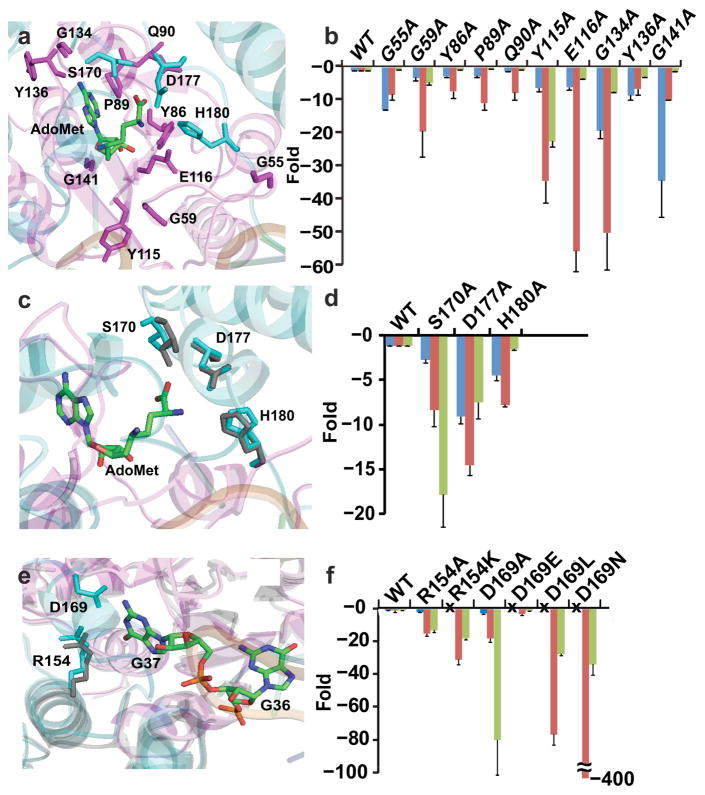
Because AdoMet binding initiates substrate signaling, we asked which of its constituents is responsible for this initiation. The Ado fragment is placed in the trefoil-knot at the end of β5 in a pocket made up of the “cover” loop (S88-G91), the “wall” loop (G113-I118), and the “bottom” loop (S132-G140), with a “halo” loop (P53-M60) on the top. Key residues in each of these loops were mutated and the effect on substrate signaling determined (Fig. 4a, b).
Figure 4.
Mutations leading to fold-changes in kinetic parameters of TrmD methyl transfer. Values from enzyme titration of tRNA were used for free energy analysis. Blue bars show fold-changes in Kd (AdoMet), red bars show fold-changes in Kd (tRNA), and green bars show fold-changes in kchem (methyl transfer). (a, b) Structures and residues tested by Ala substitutions of the Ado pocket (a) and the fold-change relative to the wild-type enzyme for each substitution (b). (c, d) Structures and residues tested by Ala substitutions of the Met pocket (c) and the fold-change relative to the wild-type enzyme for each substitution (d). (e, f) Structures and residues tested by Ala substitutions of the G37 pocket (e) and the fold-change relative to the wild-type enzyme for each substitution (f). The symbol “x” means not tested. Error bars represent s.d. (n = 5 independent measurements).
In the halo loop, G55 contacts the phosphate of G26 in the tRNA elbow region, while G59 contacts the phosphate of A38 in the anticodon loop31. The Ala substitution of each had a profound effect on tRNA binding (8.2- and 19.3-fold). Intriguingly, while neither residue makes contact with Ado, each substitution also had a strong effect on AdoMet binding (12.8- and 3.2-fold). Molecular simulation was used to determine whether dynamic motions mediated communication from the tRNA-contact to the AdoMet-site. We chose G55 for analysis, because it contacts not only the phosphate of G26 but also the base of G27. In 6 simulations of the G55A mutant (Supplementary Fig. 5), three residues in the AdoMet-site (Y86, G113, and L138) had reduced interaction with the ligand relative to the wild-type structure. Additionally, the G55A interaction with the tRNA base G27 was reduced relative to the wild-type structure. These data indicate that disrupting the enzyme-tRNA contact at base G27 reduces dynamic motions to stabilize AdoMet binding. Interestingly, the G55A substitution had no effect on methyl transfer, consistent with molecular simulations showing that two residues important for methyl transfer (E116 and D169) in the mutant maintained contact with the G37 base. In contrast, the G59A substitution reduced methyl transfer by 4.7-fold, indicating that dynamic motions were propagated to the active site.
In the cover loop, Y86, P89, and Q90 overlay on top of the adenine ring, leading the way to the trefoil-knot. The Ala substitution of each had a small effect on AdoMet binding (2.8, 2.6, and 1.2-fold), but a strong effect on tRNA binding (7.3-, 10.9-, and 7.7-fold), indicating substrate signaling from the Ado site to the tRNA site. However, none of the substitutions had a notable effect on methyl transfer, suggesting that substrate signaling was propagated only to the tRNA site. Of interest is P89, which stacks on the adenine ring. Not only did the bulkier P89W have a stronger effect on AdoMet binding (23-fold) than P89A, but the larger effect also penetrated into the active site to reduce methyl transfer (3.4-fold). Thus, signaling from the same contact can have a differential effect; a more severe perturbation can propagate the effect to the active site.
In the wall loop, Y115 and E116 are strictly conserved in a signature SpoU motif that stabilizes the Ado ribose. The Y115A and E116A substitutions, and the more conservative Y115F substitution, all had a noticeable effect on AdoMet binding (6.3-, 6.0-, and 8.0-fold), while also exerting one of the largest effects on tRNA binding (34.3-, 55.5-, and 30.5-fold). Notably, while both Y115A and Y115F originated from the same site and had a similar effect on AdoMet and tRNA binding, only the former reduced methyl transfer (22.3-fold).
The bottom loop includes G134, Y136, and G141, located between β5 and α6, to form the platform that binds the adenine ring. The Ala substitution of all three decreased AdoMet binding (19.0-, 8.4-, and 32.2-fold), with G141A having the largest effect. All three substitutions also had a strong effect on tRNA binding (50-, 6.6-, and 10.0-fold), with G134A having the largest effect. G134A was distinguished with a large effect also on methyl transfer (7.4-fold).
Active site assembly by Met binding
The Met equivalent portion of SFN interacts with the CTD of chain B and is stabilized by S170*, D177*, and H180* (where the asterisks indicate their origins from chain B)31 (Fig. 4c, d). The Ala substitution of each decreased both AdoMet binding (2.6-, 9.0-, and 4.4-fold) and tRNA binding (8.3-, 15.2-, and 7.7-fold). The D177*A substitution had the largest effect on both binding events, while also exerting a strong effect on methyl transfer (7.4-fold). The S170*A substitution caused one of the largest effects on methyl transfer (17.8-fold), while also having a strong effect on the two binding events. The importance of S170* is consistent with its physical position. It is preceded by D169*, the general base for methyl transfer31, and followed by F171*, which responds to Met binding by insertion between NTD and CTD to assemble the active site31. Without Met binding, F171* is disordered and invisible32 and the active site is not formed.
Stabilization of tRNA by G37-binding
In the ternary complex31, G37 of tRNA is flipped out from the anticodon loop and inserted into its binding pocket, where N1 is separated from the ε amino group of SFN (corresponding to the methyl of AdoMet) by 3.5 Å. This distance can accommodate the imino proton (Fig. 4e, f), suggesting that the structure is captured in a state before proton abstraction. The carboxylate of D169* interacts with N1 and N2 of G37. The positively charged side-chain of R154 is in a H-bond distance from O6 of G37, providing stabilization of the developing negative charge31. Although the structure showed no divalent metal ions, we found that a loosely bound Mg2+ stabilized O6 of G37, while also coordinating with D169* to activate the N1 nucleophile35. These observations suggest that the stabilization and proper orientation of G37 is the prerequisite for positioning tRNA for methyl transfer.
R154 is at the end of the trefoil-knot leading into the active site, while D169* is in the organized linker B. The Ala substitution of each substantially reduced tRNA binding (15.2-, 18.2-fold) and had a major effect on methyl transfer (13.2-, 80.9-fold), but neither had an effect on AdoMet binding. This suggests that stabilizing G37 is the driving force to promote tRNA binding, and that once G37 is stabilized; TrmD does not change AdoMet binding but is committed to methyl transfer. The importance of G37 binding for methyl transfer is consistent with previous work34. R154 is strictly conserved in evolution and cannot be replaced by the similarly charged K154K mutation. In contrast, D169* is occasionally replaced with glutamate in evolution, consistent with the observation that the D169*E mutant was fully active. However, the D169*N and D169*L mutants were severely defective, in both tRNA binding and methyl transfer, indicating the importance of the negatively charged side-chain of D169*.
Impaired substrate signaling by Y115A
We used the Y115A mutation as an example to probe the mechanism of substrate signaling. This mutation occurred in the trefoil-knot at a position not directly involved in stabilizing the G37 base, yet the mutation caused a large decrease in both tRNA binding and methyl transfer. We explored the explicit solvent molecular dynamics simulations of the mutant enzyme, together with five other enzymes (Y86A, Q90A, Y115A, E116A, and D169A). The average difference between experimental values of these mutants is ~1 kcal/mol, within the force field error of theoretical calculation of free energies40. We therefore used the MMPBSA simulation approach to calculate ΔG for AdoMet binding to each mutant. This analysis showed high correlation with experimentally determined ΔΔG values (Supplementary Fig. 6, Tables 3, 6), indicating that the simulations captured the structural changes that led to reduced AdoMet binding in each case.
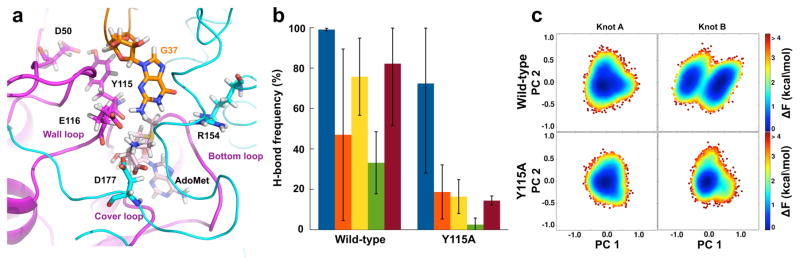
Simulations of the Y115A mutant identified impaired substrate signaling (Fig. 5a, b). In the local protein-tRNA interaction network, the mutation reduced the frequency of D177* to form a H-bond with both AdoMet A and AdoMet B. The reduction is caused by a conformational transition of the CTD, shifting several amino acids along the protein-tRNA interface. Consequently, the enzyme interaction via M197 with G11 of tRNA was lost, while aberrant interactions developed via H200 with U9, and E203 with C10 and U12. Additionally, Y115A increased the distance between R154 and the G37 base, while decreasing the base-E116 interaction to block AdoMet from methyl transfer. Y115A also prevented D50 from interaction with the G36 base of tRNA, thus eliminating the prerequisite for methyl transfer34.
Figure 5.
Effects of the Y115A mutation. (a) Simulation of active site of chain A (magenta), chain B (cyan), and tRNA (orange), showing residues with the most notable changes in structure and dynamics as sticks upon the Y115A substitution. (b) The mean occurrence of H-bonds in the wild-type (left panel) and Y115A structure (right panel). Blue, H-bond between D177* and AdoMet A; orange, H-bond between D177* and AdoMet B; yellow, H-bond between Glu116 and nucleotide G37 in tRNA; green, H-bond between Arg145 and nucleotide G37 in tRNA; burgundy, H-bond between Asp50 and nucleotide G36 in tRNA. (c) Projection of the knots of the wild-type and Y115A mutant structures along the first and second principal component (PC) on the free energy landscape. The wild-type structure is based on HiTrmD31 (PDB 4YVI). Error bars are s.d. calculated from 3 independent trajectories.
In global correlated motions, principal component analysis (PCA) showed that Y115A changed the landscape of the enzyme. Comparison of the free energy landscape on the projection along the first and second principal components between the wild-type and Y115A mutant showed similar correlations in the motion of knot A but different in knot B (Fig. 5c). The knot B of the wild-type enzyme had two global energy minima separated by a low barrier (< 2 kcal/mol), whereas the same region in the mutant possessed only one state, indicating that the second state cannot be reached due to a high-energy barrier. Additionally, analysis of correlation of motions between the two chains of the same structure (Supplementary Table 5) showed that the two knots exhibited different motions in the wild-type enzyme, but showed similar dynamics in the mutant.
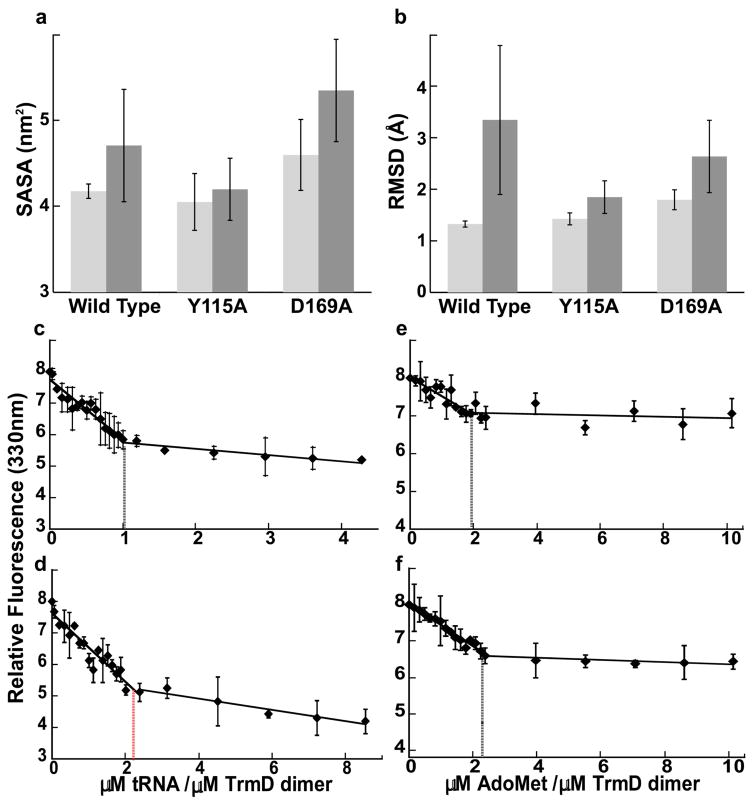
The half-of-the-sites mechanism of TrmD is an asymmetry between chain A and chain B, mediated by substrate signaling across the dimer interface, The Y115A mutation, however, eliminated this asymmetry and equalized the two chains, enabling a similar solvent accessibility for the two active sites and a similar degree of motion between the two AdoMet molecules (Fig. 6a, b). The loss of asymmetry indicates a loss of communication across the dimer interface. Three other mutations Y86A, Q90A, and E116A also eliminated the asymmetry, whereas the D169A mutation retained the asymmetry (Supplementary Fig. 6). Notably, D169A is distinguished by being outside of the knot. These data assert the role of the knot as the signal transducer between the two chains of TrmD.
Figure 6.
The active sites and the ligand binding stoichiometries of the wild-type and Y115A structures of TrmD. (a) The mean value of solvent accessible surface area (SASA) of active site A (light grey) and active site B (dark grey). In active site A, the knot is from chain A and the CTD is from chain B, whereas in active site B, the knot is from chain B and the CTD is from chain A. (b) The mean RMSD of AdoMet A (light grey) and AdoMet B (dark grey). Error bars are s.d. (n = 3 independent simulations). (c, d) Binding stoichiometry of tRNA to TrmD dimers. Determination of binding stoichiometry by monitoring the quenching of TrmD intrinsic tryptophan fluorescence with increasing amounts of tRNA to the wild-type dimer (c) and the Y115A dimer (d). (e, f) Biding stoichiometry of AdoMet to TrmD dimers. Determination of binding stoichiometry by monitoring the quenching of TrmD intrinsic tryptophan fluorescence with increasing amounts of AdoMet to the wild-type dimer (c) and the Y115A dimer (d). Error bars are s.d. (n = 3 independent measurements).
The asymmetry was previously shown to limit the TrmD dimer to bind to only one tRNA31. Using a fluorescence titration assay, we observed a biphasic quenching of the enzyme intrinsic fluorescence upon tRNA binding, with an initial steep phase followed by a second and flatter phase (Fig. 6c–f). The two phases intercepted at a molar ratio of 1:2 for one tRNA per wild-type dimer, but a ratio of 2:2 for the Y115A dimer, consistent with the notion that the two chains of the mutant are no longer coordinated but that each can bind one tRNA. Despite asymmetry or not, the two chains can each bind one AdoMet (Fig. 6a, b). Indeed, both the wild-type enzyme and the Y115 mutant bound two molecules of AdoMet per dimer.
DISCUSSION
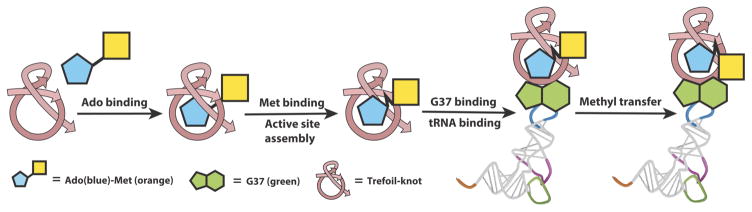
The rarity of their presence and the diversity of their reactions make protein knots both intriguing and challenging to understand. By studying the life-dependent TrmD knot, this study provides conceptual advances on two fundamental principles of protein knots. First, despite the constrained structure, the TrmD knot has active dynamic motions to support substrate signaling upon AdoMet binding. While this knot binds AdoMet to the bottom of the deep crevice, signaling was identified from energy coupling between the knot, the tRNA site, and the active site through mutational analysis. All mutants here retained the dimer structure (Supplementary Fig. 7). The picture that emerged is of a highly complex process (Fig. 7), starting with Ado binding to the knot to initiate signaling, followed by accommodation of Met into the CTD, which enables the assembly of the active site to host G37 of tRNA binding. Once G37 is stably bound, the affinity for the global tRNA is secured and the free energies from such stabilization are directly channeled to the active site to promote methyl transfer. Thus, while tRNA binding can occur independent of AdoMet36, it is the AdoMet binding that positions G37 for methyl transfer. Second, the focal point of signaling is Ado binding to the knot. Key residues (e.g., Y115) mediate signaling by propagation of local conformational changes in the knot, global conformational fluctuations within the knot, and cross-subunit communication between the two knots. Energy transfer in magnitudes are similar to those of the unknotted glutaminyl-tRNA synthetase41,42. Thus, contrary to the conventional notion, a knotted protein is not less competent in substrate signaling. Instead, a knotted protein can use the structurally organized knot to restrict the diffusion of dynamic motions to capture and coordinate binding energies to facilitate catalysis.
Figure 7.
A diagram of substrate signaling in TrmD. This signaling starts with Ado binding to the trefoil-knot, followed by Met binding to assemble the active site, by G37 binding to stabilize tRNA binding, and by methyl transfer at the active site.
A major contribution of this work is the insight into how to target TrmD. Because AdoMet binding to the knot is the origin of signaling, the best inhibitors of TrmD should target the knot and capture the bent conformation in a structurally rigid form. While such inhibitors may have difficulty accessing the knot, binding should become progressively tighter as the enzyme adapts to each inhibitor. This slow-but-tight binding mechanism is attractive, because drug binding changes the enzyme to a state that no longer favors the original ligand. The long residence time of binding reduces concentrations and dosing intervals of each drug and minimizes off-target interactions. While resistance to inhibitors may emerge eventually, likely by enzymes that modify antibiotics, it is less likely to occur to the novel bent shape of TrmD inhibitors.
ONLINE METHODS
Sequence similarity analysis
A multi-sequence alignment based on the sequence of EcTrmD as the query was performed on the National Center for Biotechnology Information (NCBI) webserver, using the Domain Enhanced Lookup Time Accelerated BLAST, DELTA-BLAST algorithm43 with the expect threshold set to 1 x e−6 and the UniProtKB/Swiss-Prot database44. An alignment of 497 sequences was generated using the European Bioinformatics Institutes (EMBL-EBI) Clustal Omega45 MSA (multiple sequence alignment) program webserver with default settings. The resulting MSA was visualized and further optimized by sequence ID, duplicate deletion, and gap optimization (visual and via gap penalty modification) using the Unipro UGENE v. 1.14.2 bioinformatics software46. Conservation percentages from the MSA on homologue sequences were then used to assess the degree of evolutionary conservation of each residue in this study.
Mutagenesis and protein purification
The pQE-30 (Qiagen) plasmid overexpressing EcTrmD with an N-terminal His-tag was utilized for mutagenesis following the QuickChange Mutagenesis protocol (Stratagene). Plasmids were introduced into E. coli TG-1 cells (GE Healthcare) as described47 and mutations were verified by DNA sequence analysis. Wild-type and mutant TrmD enzymes were expressed in E. coli SG13009 (Qiagen) as described27 and purified by affinity binding to a cobalt resin (Talon, Clontech) and by ion-exchange chromatography through MonoQ on the AKTA FPLC system (GE healthcare)36. Protein concentrations were determined by the Bradford Assay (BioRad) with bovine serum albumin as the standard and were corrected for active dimer fraction36. His-tagged TrmD mutants were 86 to 98% pure as determined by SDS-PAGE. Enzymes were verified for nuclease-free by incubation in 10-fold excess over a tRNA at 37 °C for ~3 hrs to confirm the lack of tRNA degradation on a 12% PAGE-urea gel.
E coli tRNALeu synthesis and purification
The substrate for kinetic analysis of EcTrmD was the transcript of E. coli tRNALeu/CAG, which was extensively studied previously32,33,36,39. Transcription was performed using an in-house T7 RNA polymerase33 on a synthetic DNA template made by primer extension of two overlapping oligonucleotides (IDT)48. Heterogeneity at the 3'-end of the transcript was significantly reduced by including two 2'-O-methyl RNA bases at the 5'-end of the anti-sense strand of the DNA template. Transcript was further purified on a 12% PAGE-urea gel.
Rapid chemical quench kinetics
We determined the Kd (AdoMet) and Kd (tRNA) by analysis of the rate constant of methyl transfer kobs as a function of the respective substrate, while maintaining the enzyme and all other components in excess. In these assays, the kobs was independent of the order by which reaction components were added and the Kd of each substrate (Supplementary Fig. 6, Table 2) was similar to values determined from equilibrium analysis34,36. The kchem (kobx, max) represented the intrinsic rate constant of methyl transfer or a conformational rearrangement preceding the transfer. For the wild-type EcTrmD, the kchem from enzyme titration of tRNA (0.089 ± 0.003 s−1, Supplementary Table 2) was similar to the kchem from enzyme titration of AdoMet (0.113 ± 0.003 s−1, Supplementary Fig. 6). Pre-steady-state assays in single turnover conditions were performed on the rapid quench-flow apparatus (Kintek RQF-3) or on the bench, depending on the kinetics of the mutant. The tRNA substrate was heated to 85 °C for 2.5 min followed by addition of magnesium (to 10 mM) and slow cooling to 37 °C over 15 min33. The Kd for tRNA36 and the Kd for AdoMet32 were determined according to the previously published conditions. For example, determination of Kd (AdoMet) was done with increasing concentration of enzyme (1 to 100 μM) at 37 °C in the presence of 4 μM tRNALeu and 0.1 μM 3H-AdoMet (Perkin Elmer, 4200 DPM/pmol). Reaction aliquots were removed at various time points and precipitated in 5% (w/v) trichloroacetic acid on filter pads, which were washed with ethanol, dried, and counted on an LS600 scintillation counter (Beckman). Counts were converted into pmoles using AdoMet specific activity after correction for signal quench on the filter pad.
Fluorescence titration experiments
The stoichiometry of TrmD binding to substrates and products was determined by monitoring the quenching of intrinsic tryptophan fluorescence36. The fluorescence was excited at 295 nm and the emission was monitored from 304 nm to 400 nm at room temperature in a buffer containing 100 mM Tris-HCl, pH 8.0, 100 mM KCl, 0.1 mM EDTA, 6 mM MgCl2, and 4 mM DTT. For Kd measurements, TrmD was at 0.2 μM and AdoMet and AdoHcy were both titrated in the range of 0.1–40 μM. while the tRNALeu transcript was titrated in the range of 0.1–6 μM.
Size exclusion chromatography
Determination of the dimer state of mutant enzymes was performed by size exclusion on the Superdex 75 column (GE Healthcare). Molecular weight of each protein was extrapolated from its retention time, based on the calibration of a series of marker proteins of known molecular weights48. Alternatively, 2 μg of each mutant was loaded on a native protein gel (no SDS or β-mercaptoethanol) along with marker proteins. Bands were visualized by brilliant Coomassie blue stain.
Molecular dynamics
TrmD ternary complex structure31, where the SFN in the ternary crystal structure was replaced with AdoMet by simulations, was used as a starting structure for molecular dynamic analysis. The apo- and binary tRNA-free structures were obtained from the PDB code 1UAK. Mutations of the ternary complex were done on both subunits using PyMOL (http://www.pymol.org/). All of the structures were protonated in pH 8.0 using PDB2PQR server49. All simulations were done in GROMACS 5.0.2 (http://manual.gromacs.org/documentation) with CHARMM36 force field (http://www.ncbi.nlm.nih.gov/pubmed/23832629) in TIP3 water model with 2 fs time steps. Necessary parameters and topologies for the ligand were taken from the force field. The total charges of the systems were neutralized with addition of appropriate amounts of NaCl ions. Energy minimizations were carried out using the steepest descent minimization until the force of less than 0.01 kJ/mol/nm was reached. The systems were then equilibrated at a constant temperature (310 K) and pressure (1 bar) for 100 ps. The temperature and pressure of simulations were maintained using Nose-Hoover algorithm (http://phycomp.technion.ac.il/~phsorkin/thesis/node42.html). Cutoff length of 12 Å was applied to both electrostatic (particle mesh Ewald method – PME) and van der Waals interactions. Hydrogen bonds were calculated with Hbonds plugin (http://www.ks.uiuc.edu/Research/vmd/plugins/hbonds/) from VMD 1.9.1 with bond cutoff at 3.2 Å and angle cutoff at 50°. The rest of the analysis was done using the GROMACS package.
Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) method
Binding energies were calculated for each ligand in each trajectory (wild-type and mutated ternary structures) with snapshots taken every 1 ns. The calculation was done using the g_mmpbsa tool described by Kumari and colleagues50, using polar solvation and SASA energy terms. Dielectric constant of the protein was set to 2 and of water to 78.54. Temperature was 310 K and the calculation was based on solving the linear Poisson-Boltzman equation. Other parameters were kept at default.
Principal component analysis (PCA)
We focused on the large-amplitude motions important for substrate signaling and determined how these motions were affected by the Y115A mutation. In this methodology, the dynamics of protein motions in high dimensional conformational-space observed in simulation were reduced to a few functionally important motions by identification of the best reaction coordinates (eigenvectors) for those motions. Along these coordinates, the largest mean-square atomic fluctuations (the eigenvalue) for all backbone atoms were localized. The most important motions were identified upon descending order of each eigenvalue with its eigenvectors. PCA analysis showed that only the first five eigenvectors (in five dimensions) were sufficient to describe almost all dynamics of a structure (Supplementary Table 4). In the wild-type structure, the CTD exhibited the largest total variance of motions based on the sum of the first five eigenvalues, but this amplitude decreased with successive binding of AdoMet and tRNA (Supplementary Fig. 6b), indicating that ligand binding stabilized motions. Unexpectedly, while the trefoil-knot exhibited the lowest amplitude of motions, which further decreased upon AdoMet binding, the motions actually increased upon tRNA binding. This supports the notion that the knot is an active component in the signaling between AdoMet and tRNA binding. PCA was performed using the following functions from the GROMACS package, where g is covar, g is anaeig and g is analyze. Calculation of the covariance matrix was based on an average structure from each of the trajectories consisting of the protein's backbone atoms. Root Mean Square Inner Product (RMSIP), which measures similarity in the subspace spanned by given eigenvectors (the measured degree of overlap). is defined as:
where pAi and qBj are the ith and jth eigenvectors from a set of eigenvectors A and B. In this work, RMSIP was calculated between different sets of eigenvectors and different number of eigenvectors as well. We calculated the RMSIP for the individual systems (trajectories) by splitting the trajectory in half and comparing the first 5 and 10 eigenvectors from each half of the trajectory. The eigenvalue-normalized overlap s(A,B) derived by B. Hess51 was adopted as a standard measure of convergence52,53. Cosine content was calculated (using g_analyze from GROMACS package) for the first principal component for the trajectory within the first 100 ns.
Free-energy landscape analysis was done using the first two principal components obtained from the PCA. Energy in each protein’s state (sj) is based on the probability of being in this state (P(si)).
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health grants GM081601 and GM114343 (to Y.-M.H.), the European Molecular Biology Organization (EMBO) installation grant 2057 and the National Science Center, Sonata BIS 2012/07/E/NZ1/01900 grant (to J.I.S.), and the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan, JSPS KAKENHI Grant Number 20247008 (to T.I. and S.Y.). We thank I. Masuda, R. Takase, R. Matsubara, S. Maharjan, and K. Donaldson for preparation of figures.
Footnotes
AUTHOR CONTRIBUTIONS
T.C., R.S., and G.L. performed kinetic analysis; A.P.P. and J.I.S. performed computational molecular simulation analysis; T.I. and S.Y. performed structural analysis; E.A.T. performed sequence conservation analysis, and J.I.S. and Y.-M.H. prepared the manuscript. All authors discussed and commented on the manuscript.
COMPETING FINANICAL INTERESTS The authors declare no competing financial interests.
References
- 1.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–82. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 2.Hammes GG, Benkovic SJ, Hammes-Schiffer S. Flexibility, diversity, and cooperativity: pillars of enzyme catalysis. Biochemistry. 2011;50:10422–30. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–9. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ditzler MA, Otyepka M, Sponer J, Walter NG. Molecular dynamics and quantum mechanics of RNA: conformational and chemical change we can believe in. Acc Chem Res. 2010;43:40–7. doi: 10.1021/ar900093g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolinger D, et al. A Stevedore's protein knot. PLoS Comput Biol. 2010;6:e1000731. doi: 10.1371/journal.pcbi.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamroz M, et al. KnotProt: a database of proteins with knots and slipknots. Nucleic Acids Res. 2015;43:D306–14. doi: 10.1093/nar/gku1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sulkowska JI, Sulkowski P, Szymczak P, Cieplak M. Stabilizing effect of knots on proteins. Proc Natl Acad Sci U S A. 2008;105:19714–9. doi: 10.1073/pnas.0805468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO. Structure and folding of a designed knotted protein. Proc Natl Acad Sci U S A. 2010;107:20732–7. doi: 10.1073/pnas.1007602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulkowska JI, Noel JK, Onuchic JN. Energy landscape of knotted protein folding. Proc Natl Acad Sci U S A. 2012;109:17783–8. doi: 10.1073/pnas.1201804109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virnau P, Mallam A, Jackson S. Structures and folding pathways of topologically knotted proteins. J Phys Condens Matter. 2011;23:033101. doi: 10.1088/0953-8984/23/3/033101. [DOI] [PubMed] [Google Scholar]
- 11.Ahn HJ, et al. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. Embo J. 2003;22:2593–603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkins PA, et al. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol. 2003;333:931–49. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Bystrom AS, Bjork GR. The structural gene (trmD) for the tRNA(m1G)methyltransferase is part of a four polypeptide operon in Escherichia coli K-12. Mol Gen Genet. 1982;188:447–54. doi: 10.1007/BF00330047. [DOI] [PubMed] [Google Scholar]
- 14.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat Commun. 2015;6:7226. doi: 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. The UGG Isoacceptor of tRNAPro Is Naturally Prone to Frameshifts. Int J Mol Sci. 2015;16:14866–83. doi: 10.3390/ijms160714866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–9. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 17.Gustafsson C, Reid R, Greene PJ, Santi DV. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–62. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745- deficient mutant. J Bacteriol. 1998;180:359–65. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson BC, Jager G, Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2'-O-methyltransferase activity. Nucleic Acids Res. 1997;25:4093–7. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nureki O, et al. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure (Camb) 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Ochi A, et al. The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process. J Biol Chem. 2013;288:25562–74. doi: 10.1074/jbc.M113.485128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel G, et al. The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure (Camb) 2002;10:1303–15. doi: 10.1016/s0969-2126(02)00852-3. [DOI] [PubMed] [Google Scholar]
- 23.Lim K, et al. Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins. 2003;51:56–67. doi: 10.1002/prot.10323. [DOI] [PubMed] [Google Scholar]
- 24.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–35. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–93. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 26.White TA, Kell DB. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp Funct Genomics. 2004;5:304–27. doi: 10.1002/cfg.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christian T, Evilia C, Williams S, Hou YM. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol. 2004;339:707–19. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 28.Goto-Ito S, et al. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72:1274–89. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 29.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol. 2009;16:1109–15. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 30.Christian T, Gamper H, Hou YM. Conservation of structure and mechanism by Trm5 enzymes. RNA. 2013;19:1192–9. doi: 10.1261/rna.039503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, et al. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc Natl Acad Sci U S A. 2015;112:E4197–205. doi: 10.1073/pnas.1422981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahoud G, et al. Differentiating analogous tRNA methyltransferases by fragments of the methyl donor. RNA. 2011;17:1236–46. doi: 10.1261/rna.2706011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol. 2007;373:623–32. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakaguchi R, et al. Recognition of guanosine by dissimilar tRNA methyltransferases. RNA. 2012;18:1687–701. doi: 10.1261/rna.032029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi R, Lahoud G, Christian T, Gamper H, Hou YM. A divalent metal ion-dependent N(1)-methyl transfer to G37-tRNA. Chem Biol. 2014;21:1351–60. doi: 10.1016/j.chembiol.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christian T, Lahoud G, Liu C, Hou YM. Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J Mol Biol. 2010;400:204–17. doi: 10.1016/j.jmb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, et al. Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold. Proteins. 2003;53:326–8. doi: 10.1002/prot.10479. [DOI] [PubMed] [Google Scholar]
- 38.Christian T, et al. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA. 2010;16:2484–92. doi: 10.1261/rna.2376210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christian T, Evilia C, Hou YM. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry. 2006;45:7463–73. 18. doi: 10.1021/bi0602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirts MR, Pitera JW, Swope WC, Pande VS. Extremely precise free energy calculations of amino acid side chain analogs: Comparison of common molecular mechanics force fields for proteins. The Journal of chemical physics. 2003;119:5740–5761. [Google Scholar]
- 41.Uter NT, Perona JJ. Long-range intramolecular signaling in a tRNA synthetase complex revealed by pre-steady-state kinetics. Proc Natl Acad Sci U S A. 2004;101:14396–401. doi: 10.1073/pnas.0404017101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Hernandez A, Perona JJ. Heat maps for intramolecular communication in an RNP enzyme encoding glutamine. Structure. 2011;19:386–96. doi: 10.1016/j.str.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 43.Boratyn G, et al. Domain enhanced lookup time accelerated BLAST. Biology Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poux S, et al. Expert curation in UniProtKB: a case study on dealing with conflicting and erroneous data. Database. 2014;2014 doi: 10.1093/database/bau016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sievers F, Higgins DG. Current Protocols in Bioinformatics. John Wiley & Sons, Inc; 2002. Clustal Omega. [DOI] [PubMed] [Google Scholar]
- 46.Okonechnikov K, Golosova O, Fursov M tU team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 47.Hou YM, Westhof E, Giege R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc Natl Acad Sci U S A. 1993;90:6776–80. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys) Nat Struct Mol Biol. 2008;15:507–14. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–5. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumari R, Kumar R, Lynn A Open Source Drug Discovery C. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–62. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 51.Hess B. Convergence of sampling in protein simulations. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:031910. doi: 10.1103/PhysRevE.65.031910. [DOI] [PubMed] [Google Scholar]
- 52.Luchko T, Huzil JT, Stepanova M, Tuszynski J. Conformational analysis of the carboxy-terminal tails of human beta-tubulin isotypes. Biophys J. 2008;94:1971–82. doi: 10.1529/biophysj.107.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurylowicz M, Yu CH, Pomes R. Systematic study of anharmonic features in a principal component analysis of gramicidin A. Biophys J. 2010;98:386–95. doi: 10.1016/j.bpj.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.