Abstract
The pathogenic fungus Cryptococcus neoformans delivers virulence factors such as capsule polysaccharide to the cell surface to cause disease in vertebrate hosts. In this study, we screened for mutants sensitive to the secretion inhibitor brefeldin A to identify secretory pathway components that contribute to virulence. We identified an ortholog of the Cdc50 family of the non-catalytic subunit of type IV P-type ATPases (flippases) that establish phospholipid asymmetry in membranes and function in vesicle-mediated trafficking. We found that a cdc50 mutant in C. neoformans was defective for survival in macrophages, attenuated for virulence in mice and impaired in iron acquisition. The mutant also showed increased sensitivity to drugs associated with phospholipid metabolism (cinnamycin and miltefosine), the antifungal drug fluconazole and curcumin, an iron chelator that accumulates in the ER. Cdc50 is expected to function with catalytic subunits of flippases and we previously documented the involvement of the flippase Apt1 in virulence factor delivery. A comparison of phenotypes with mutants defective in genes encoding candidate flippases (designated APT1, 2, 3 and 4) revealed similarities primarily between cdc50 and apt1 suggesting a potential functional interaction. Overall these results highlight the importance of membrane composition and homeostasis for the ability of C. neoformans to cause disease.
Keywords: Flippase, heme, antifungal drug, curcumin, pathogenesis, secretion
Introduction
The pathogenic yeast Cryptococcus neoformans causes a high burden of cryptococcosis in immunocompromised people, particularly those afflicted with HIV/AIDS (Brown et al., 2007; Park et al., 2009). Disease in vertebrate hosts results from the ability of the fungus to grow at 37°C and to deliver key virulence traits to the cell surface. These factors include a polysaccharide capsule, the cell wall pigment melanin, and extracellular enzymes such as urease, phospholipase B and acid phosphatase (Zhu et al., 2004; Djordjevic et al., 2005; Idnurm et al., 2005 Doering, 2009; Zaragoza et al., 2009; Djordjevic, 2010; Kronstad et al., 2011; Almeida et al., 2015). The fungus must also resist killing by phagocytic cells and by oxidative and nitrosative mechanisms (Tucker et al., 2002; Brown et al., 2007). The regulation and mechanisms of trafficking of capsule polysaccharide and extracellular enzymes are being actively pursued (Garcia-Rivera et al., 2004; Djordjevic et al., 2005; Yoneda et al., 2006; Rodrigues et al., 2007; Casadevall et al., 2009; Eisenman et al., 2009; Panepinto et al., 2009; Yoneda et al., 2009; Rodrigues et al., 2012; Lev et al., 2014). For example, we previously showed that vesicle trafficking functions are regulated by the cAMP-signaling pathway that also controls the formation of both the capsule and melanin (Hu et al., 2007a). These functions include Ova1, a predicted phospatidylethanolamine binding protein that negatively influences melanin and capsule formation. We also found that capsule size was reduced in cells treated with inhibitors of Golgi apparatus-mediated transport (e.g., brefeldin A or monensin) or lithium chloride (Hu et al., 2007a).
The involvement of putative type IV P-type ATPases (P4-ATPases or flippases) in the export of virulence factors has also been examined in C. neoformans (Hu et al., 2010; Huang et al., 2016; Rizzo et al., 2014). Flippases translocate phosphatidylserine (PS) and/or phosphatidylethanolamine (PE) from one leaflet of the bilayer to the other and play a role in maintaining the asymmetrical distribution of aminophospholipids in membranes (Alder-Baerens et al., 2006; Lopez-Marques et al., 2011). Phospholipid asymmetry is important for plasma membrane and trans-Golgi network events such as vesicle budding and docking (Muthusamy et al., 2009a; Sebastian et al., 2012). Thus flippases participate in both endocytosis and exocytosis, and are required for efficient Golgi function. In this context, we showed that the flippase Apt1 can complement a drs2 mutant of S. cerevisiae, and contributes to membrane trafficking and sensitivity to oxidative and nitrosative stress as well as drugs targeting ergosterol biosynthesis and secretion (Hu et al., 2010). Interestingly, loss of Apt1 does not influence capsule and melanin formation in vitro but does appear to regulate capsule size during infection. The protein is also required for intracellular growth in macrophages and virulence in a mouse model of cryptococcosis (Hu et al., 2010; Rizzo et al., 2014).
In this study, we fortuitously identified a candidate for a non-catalytic subunit of flippases from the Cdc50 family and characterized its role in virulence. This family of flippase components has been characterized in a number of organisms including S. cerevisiae where there are three members, Cdc50, Lem3 and Crf1 (Sebastian et al., 2012). These proteins are thought to function as chaperones to facilitate the exit of flippases from the endoplasmic reticulum (ER) and to help with their correct localization. A C. neoformans mutant lacking Cdc50 was also recently characterized in the context of sensitivity to the antifungal drug caspofungin (Huang et al., 2016). Our characterization of the C. neoformans cdc50 mutant revealed similarities to phenotypes reported for cdc50 and lem3 mutants in S. cerevisiae. Importantly, we identified a novel role for Cdc50 in iron acquisition that may partly explain the observed virulence defect in mice. We also compared phenotypes for the cdc50 mutant with mutants lacking each of four candidate flippases and discovered similar phenotypes for the cdc50 and apt1 mutants. Overall, these findings suggest a link between functions that maintain phospholipid asymmetry and virulence in C. neoformans.
Results
Insertional mutagenesis to identify functions for virulence factor secretion
Previously, we demonstrated that brefeldin A (BFA), an inhibitor of ADP ribosylation factor (ARF) activity in ER to Golgi trafficking, blocked formation of cell-associated capsule (Hu et al., 2007a). Additionally, we identified a role for ESCRT functions both in formation of the extracellular capsule and use of heme as an iron source (Hu et al., 2013; 2015). To identify additional genes involved in virulence factor delivery and iron uptake, we generated a collection of ~30,000 Agrobacterium-mediated insertion strains in the wild type (WT) background of C. neoformans and screened for mutants with increased sensitivity to BFA. This screen identified 32 mutants that were then tested for defects in production of capsule and melanin, and iron acquisition. Among the candidates, eight had reduced melanin production and 16 had reduced capsule size. Reverse-PCR and sequencing identified one of the candidates as Vps20 (Mvb6), a component of ESCRT complex III (endosomal sorting complex required for transport), and we previously showed that components of this complex contribute to capsule production, growth on heme as the sole iron source and virulence in a mouse inhalation model (Hu et al., 2013; Hu et al., 2015). This finding validated the screen and prompted the characterization of additional mutants. We identified several strains without defects or with subtle changes in the formation of capsule and/or melanin. One of these candidates showed poor growth in BFA and a pronounced growth defect on iron. Reverse-PCR and sequencing revealed that a putative CDC50 homologue (CNAG_06465) was disrupted in the strain by the T-DNA insertion and we focused on this gene because of our previous discovery of a role for the flippase Apt1 in virulence in C. neoformans (Hu et al., 2010; Rizzo et al., 2014). Additionally, we identified CNAG_06465 as a HapX-regulated gene in our earlier analysis of iron regulatory factors (Jung et al., 2010). Interestingly, a screen for BFA sensitive mutants in S. cerevisiae also identified Lem3, another P4 - ATPase subunit in the Cdc50 family (Muren et al., 2001). We also note that CNAG_06465 was recently found to contribute to caspofungin resistance and virulence in C. neoformans (Huang et al., 2016).
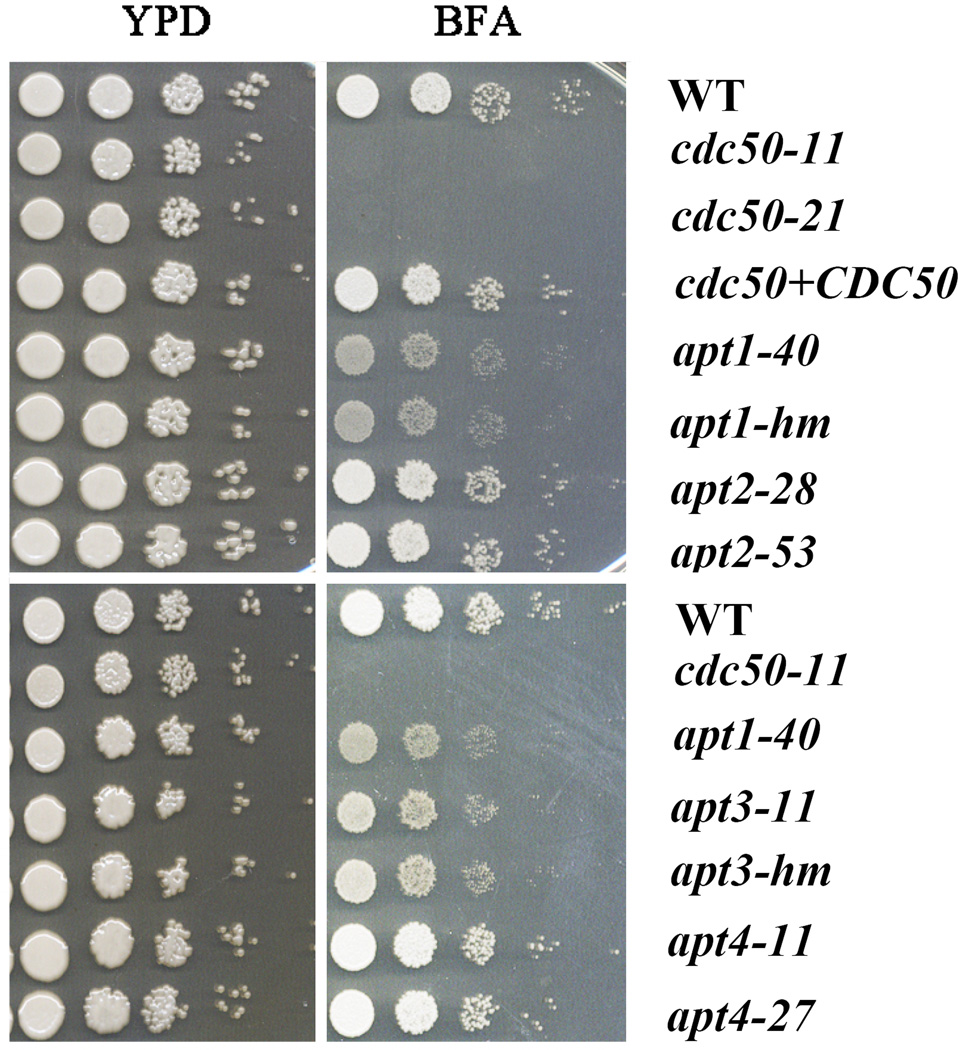
To confirm that BFA sensitivity was due to loss of CDC50, we generated two independent deletion mutants, cdc50-11 and cdc50-21, in the WT background. Deletion of the gene in the mutants was confirmed by colony PCR and Southern hybridization (data not shown). We also complemented the cdc50 mutation by introducing the WT gene into the cdc50-11 mutant strain. Phenotypic characterization confirmed that deletion of CDC50 leads to the increased sensitivity to BFA as well as monensin, an inhibitor that blocks intracellular transport in both the trans-Golgi and post-Golgi compartments (Fig. 1 and data not shown). The cdc50 mutants were also more sensitive to N-ethylmaleimide (NEM), a cysteine alkylating agent that interferes with disulfide bond formation (data not shown). The CDC50 complemented strain restored growth to the WT level on the inhibitors (Fig. 1), indicating that the phenotypes were due to the absence of CDC50.
Figure 1. The cdc50 mutants are sensitive to trafficking inhibitors.
Serial 10-fold dilutions of the indicated strains were spotted on YPD or YPD supplemented with the inhibitor brefeldin A (BFA) (30 µg/ml). The plates were incubated at 30°C for 2 days and photographed. The apt1-mh and apt3-mh strains were from the collection constructed by Hiten Madhani’s group (Liu et al., 2008).
Cdc50 associates with flippases and is important for their transport from the ER and translocase activity (Kato et al., 2002; Chen et al., 2006; Lenoir et al., 2009). In C. neoformans, only one copy of CDC50 (CNAG_06465) was identified by BLASTp searches using the S. cerevisiae protein sequences of Cdc50, Lem3 and Crf1 (YNR048W) in the Cdc50 family (Kato et al., 2002; Hanson et al., 2003; Saito et al., 2004; Lenoir et al., 2009). By BLAST alignment, the predicted 402 amino acid polypeptide of Cdc50 in C. neoformans shares identity of 34%, 36% and 39% to Lem3, Crf1 and Cdc50 in S. cerevisiae, respectively. To place our analysis of Cdc50 in context, we also constructed mutants with defects in four candidate phospholipid flippase-encoding genes. We previously identified four putative flippases in C. neoformans, including Apt1 (CNAG_06469), Apt2 (CNAG_01278), Apt3 (CNAG_00383) and Apt4 (CNAG_05282) (Supplemental Table S1), and characterized the role of Apt1 in the response to stress, fluconazole sensitivity, capsule formation and virulence (Hu et al., 2010; Rizzo et al., 2014). Notably, CDC50 and APT1 are closely located in a 13 kb region on chromosome 13 (Hu et al., 2008a). We deleted APT2, APT3 and APT4 and analyzed two independent mutants for each gene for comparison with the cdc50 mutants and the previously described apt1 mutants (Hu et al., 2010; Rizzo et al., 2014). We also included independently constructed apt1 and apt3 mutant strains from the deletion collection of Liu et al. (Liu et al., 2008). We examined the growth of the mutants of four flippase genes on YPD with BFA and found that the apt1 and apt3 mutants demonstrated increased sensitivity, although to a lesser extent than the cdc50 mutants (Fig. 1). A double mutant lacking CDC50 and APT1 showed sensitivity to BFA that was comparable to the cdc50 mutant and greater than the apt1 mutant (Supplemental Fig. S1). This result is consistent with a role for Cdc50 as an interacting subunit of a complex with Apt1, although other interpretations are possible. Furthermore, Apt1 and Apt3 appear to make redundant contributions because a double mutant (apt1 apt3) was more sensitive to BFA than either single mutant (Supplemental Fig. S1).
Cdc50 is required for survival in macrophages and virulence in mice
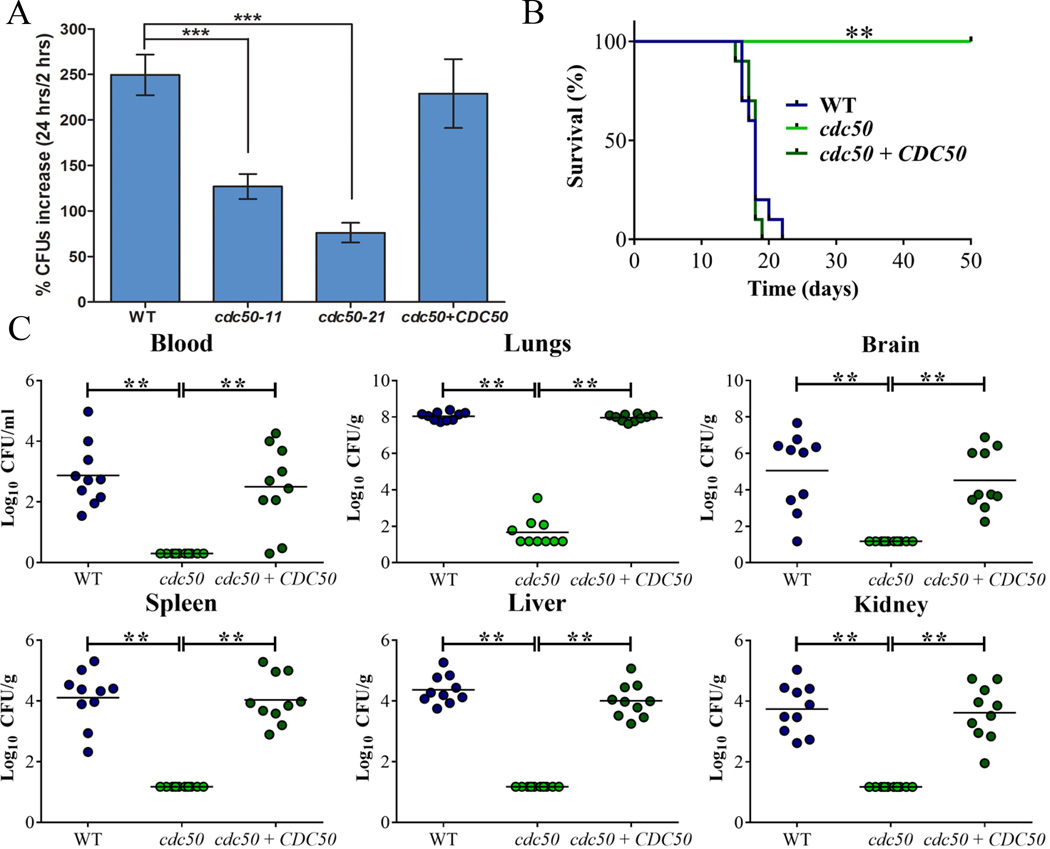
To first establish the relevance of Cdc50 in cryptococcal disease, we examined the survival of the cdc50 mutants during the interaction with a murine macrophage-like cell line and the virulence of the mutant in a mouse model of cryptococcosis. The cells of the WT, cdc50-11, cdc50-21 and the complemented CDC50 strains were incubated with macrophage-like J774A.1 cells. As shown in Fig. 2A, the number of cdc50 mutant cells recovered at 24 hours was significantly lower than for the WT and complemented strains indicating a reduced ability to survive and proliferate in macrophages. The reduced survival of the cdc50 mutants prompted a further assessment of virulence in mice and we therefore challenged 10 mice per strain by intranasal inoculation and monitored disease. In contrast to the WT strain, which caused a lethal disease in all mice by 21 days post-inoculation, the cdc50 mutant showed an avirulent phenotype in this model, and the infected mice survived to the end of the experiment at 50 days post-inoculation (P<0.0001). The complemented strain completely restored virulence to the WT level (Fig. 2B). Further examination of fungal loads in organs harvested from infected mice revealed a much lower fungal burden in the lungs in all 10 mice infected with the cdc50 mutant than in lungs with the WT and complemented strains (Fig. 2C). Similarly, at the end of the assay, almost no cells were retrieved from other organs (blood, kidney, liver, spleen and brain) for mice infected with the mutant, while high numbers of fungal cells were retrieved from these organs for mice infected with the WT and complemented strains (Fig. 2C). Overall, the cdc50 mutant appeared to be unable proliferate in mice and to disseminate to other organs including the brain, although the fungus was not completely cleared from the lungs (Fig. 2C). Together, we conclude that CDC50 is required for survival after phagocytosis and for virulence in mice.
Figure 2. Cdc50 is required for survival in macrophages and for virulence in mice.
A) Cells of the WT strain, two cdc50 mutants, and the CDC50 complemented strain were incubated with macrophage J774A.1 cells. C. neoformans was inoculated at 1 × 105 cells, and the wells were washed after 2 h of incubation to remove extracellular yeast cells. Fungal proliferation was measured by plating on YPD and counting CFUs after 24 h of incubation. The data represent the mean values ± standard error of the mean (S.E.M) of three independent biological experiments done in triplicate. Statistical analysis was performed using an unpaired two-tailed Student’s t test to determine the difference between the WT strain and two cdc50 mutants, separately (***P < 0.0005, significantly different). B) Ten female BALB/c mice were inoculated intranasally with 2 × 105 cells of each of the strains indicated, and the survival of the mice was monitored daily. The cdc50-11 mutant was used for the experiment. Survival differences between groups of mice were evaluated by log rank tests. The P values for the mice infected with the WT and mutant strains were statistically significantly different (P < 0.001). C) Fungal burden was determined in systemic organs (lung, brain, liver, kidney and spleen) and cardiac blood for all mice infected with the strains at the end of the experiment. The Mann-Whitney U test was used for statistical analysis. Differences in the fungal loads between the WT and cdc50 mutants in each organ examined were statistically significant (P < 0.001).
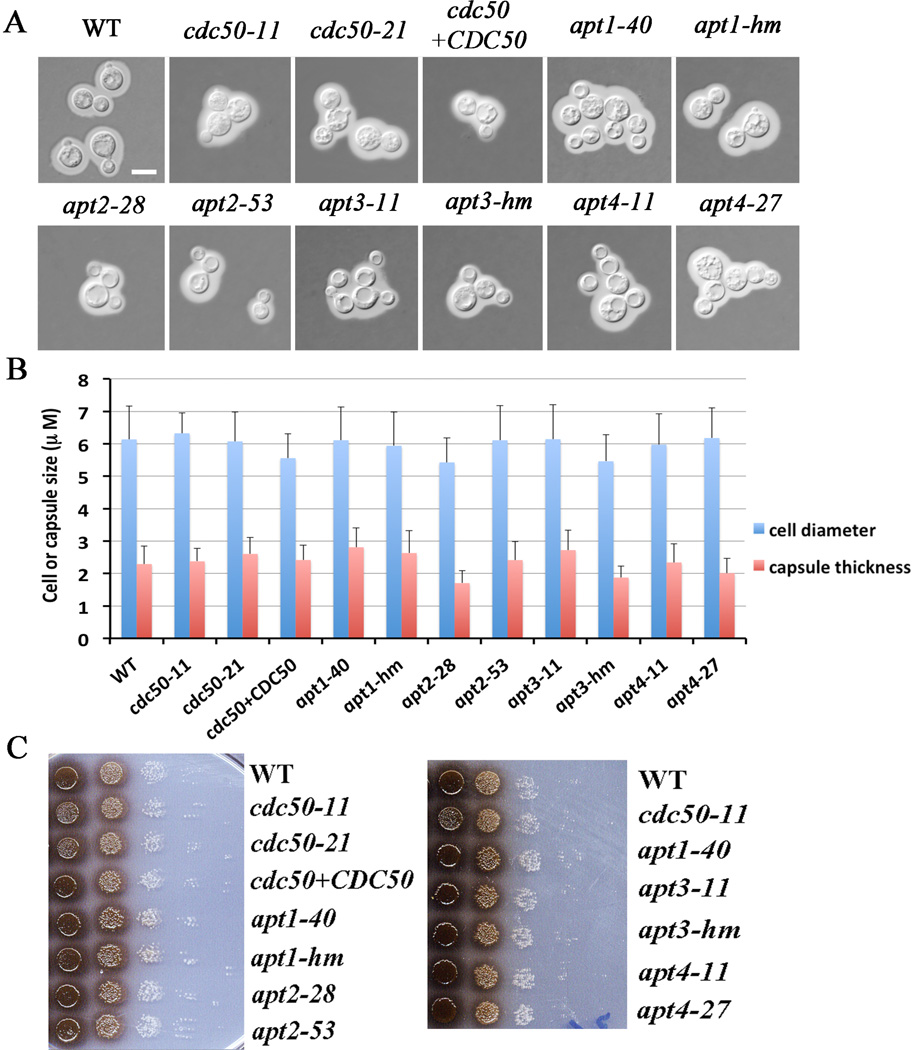
We next analyzed the cdc50 mutant for the major known virulence factors to potentially explain the avirulent phenotype (Idnurm et al., 2005; Kronstad et al., 2011; Kronstad et al., 2012). First, we assessed capsule formation by India ink staining of strains cultured in defined low iron media. We did not observe a significant influence of CDC50 deletion on capsule size in this medium (Fig. 3A, 3B). This is in contrast to a recent report that described a larger capsule but reduced polysaccharide secretion for a CDC50 deletion strain grown in a different medium (DMEM) (Huang et al., 2016), indicating that capsule size may vary depending on the inducing media. We extended the measurement of capsule size to include the apt1, 2, 3 and 4 mutants and found no significant change in capsule size under our conditions (Fig. 3). We also examined the ability of the cdc50 and apt1, 2, 3 and 4 mutants to grow on solid media at 37°C and to produce melanin. We did not observe significant growth defects at 37°C for any of the strains and we only observed a subtle reduction of melanin production for the cdc50 mutants at 37°C (Fig. 3C and data not shown). Taken together, these observations revealed that deletion of CDC50 did not cause major defects in the known major virulence factors sufficient to account for the differences in macrophage survival and virulence.
Figure 3. Loss of Cdc50 does not influence capsule formation in low-iron medium and has a small effect on melanin production.
A) Cells were grown in defined low-iron medium at 30°C for 48 h and capsule formation was assessed by India ink staining for the indicated strains. Bar = 5 µm. B) One hundred cells of each strain from the assays in A were measured to determine the cell diameter and capsule radius. Each bar represents the average of the 100 measurements with standard deviations. Statistical significance was analyzed using the Student’s t-test (P < 0.05) and no differences relative to the WT capsule size were found. C) Melanin production was tested after growth at 30°C for 2 days by spotting serial 10-fold dilutions of the indicated strains onto L-DOPA plates.
Loss of Cdc50 influences protein trafficking and sensitivity to inhibitors that interact with phospholipids
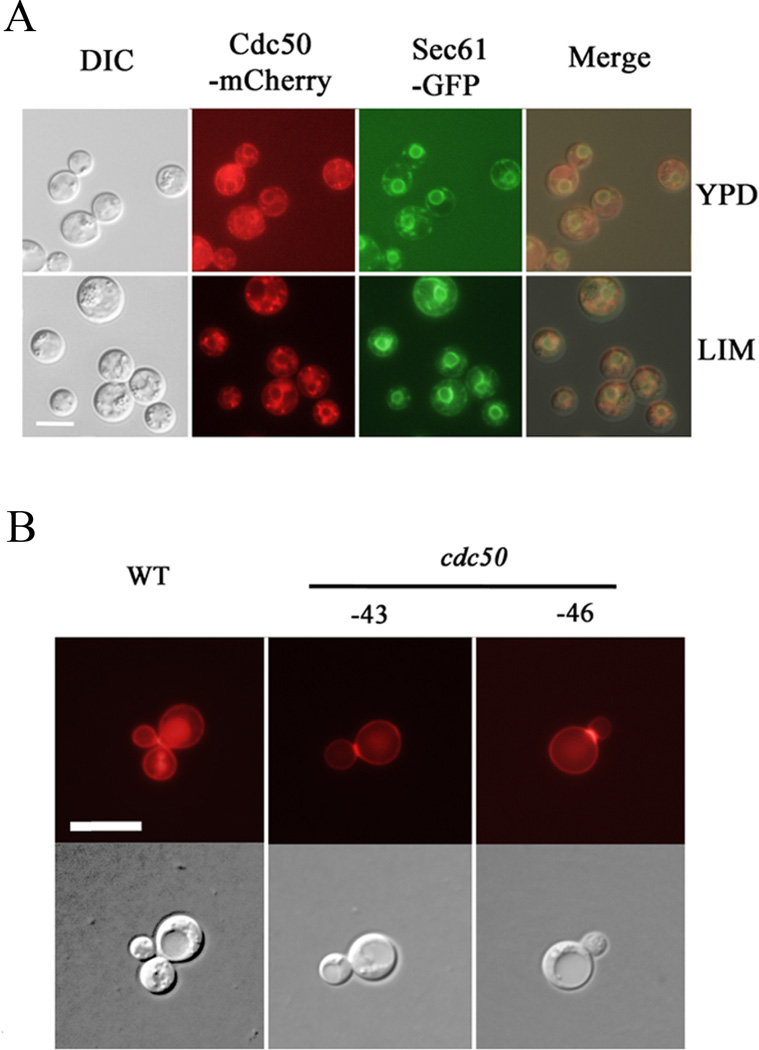
To further investigate the contribution of Cdc50 to virulence, we first confirmed the properties of Cdc50 expected from the role of the orthologous proteins in S. cerevisiae (Cdc50, Lem3 and Crf1) in maintaining lipid asymmetry and supporting vesicle transport pathways (Graham, 2004; Muthusamy et al., 2009a; Sebastian et al., 2012). In S. cerevisiae, both Cdc50 and Lem3 interact with flippases to regulate the translocation of phospholipids and maintenance of the membrane asymmetry. Yeast Cdc50 is localized in trans-Golgi network (TGN) complex and endosomes, while Lem3 is localized in the ER and plasma membrane (Saito et al., 2004; Chen et al., 2006; Azouaoui et al., 2016). We tagged the C. neoformans Cdc50 protein at the C-terminal with mCherry and assessed subcellular location in cells grown in either the rich medium (YPD) or low iron medium (LIM). In both media, fluorescence was visible in cytoplasmic structures that partially co-localized with Sec61-GFP, an ER-targeted protein (Fig. 4A). The Cdc50-mCherry protein was also discernable at the plasma membrane and in punctate structures in the cytoplasm that we hypothesize may be endosome-like structures. The strain with the Cdc50-mCherry fusion (at the CDC50 locus) was phenotypically identical to the WT and complemented strains (Supplemental Fig. S2).
Figure 4. Cdc50 is localized to the ER, and is required for proper localization of acid phosphatase.
A) Co-localization of Cdc50-mCherry and the ER-targeted Sec61-GFP in both YPD and the defined low iron media. In addition to the ER localization (co-localized with Sec61-GFP), the mCherry signal is discernable at the cell periphery. The Cdc50-mCherry protein is also detected in punctuate vesicular structures which may be endosomes. Bar = 5 µM. B) DsRed-tagged Aph1 (acid phosphatase) is targeted to the cell periphery, bud-necks and vacuoles in the WT strain, but mainly enriched in cell periphery and bud-necks in the cdc50 mutants. Significant reduced fluorescence is present in vacuoles. Bar = 10 µm.
In C. neoformans, the acid phosphatase Aph1 contributes to virulence, and DsRed-tagged Aph1 is transported to the cell periphery and vacuoles via endosome-like structures and is enriched in bud necks in the low phosphate condition (Lev et al., 2014). In S. cerevisiae, it has been shown that the flippase Drs2 plays a role in the trans-Golgi network to produce exocytic vesicles for the delivery of the enzymes to the cell surface (Gall et al., 2002). We therefore used Aph1 localization to assess the impact of loss of Cdc50. We found that Aph1-DSRed was located at the cell periphery and in large (vacuoles) and small (endosome-like structures) spherical cytoplasmic structures after WT cells were incubated in minimal medium with low glucose for 3–6 hours, as described by Lev et al. (2014) (Fig. 4B). The signal was also discernable in the budding apices and bud necks (Fig. 4B). In contrast, Aph1-DSRed was also enriched in emerging bud apices and bud necks, but with a significant reduced distribution in both vacuoles and endosomes in the cdc50 mutant (Fig. 4B). A weak signal was also discernable in the fungal cell periphery. We conclude that deletion of CDC50 reduced accumulation of Aph1-DsRed in vacuoles and endosomes, consistent with a role for Cdc50 in protein trafficking in C. neoformans.
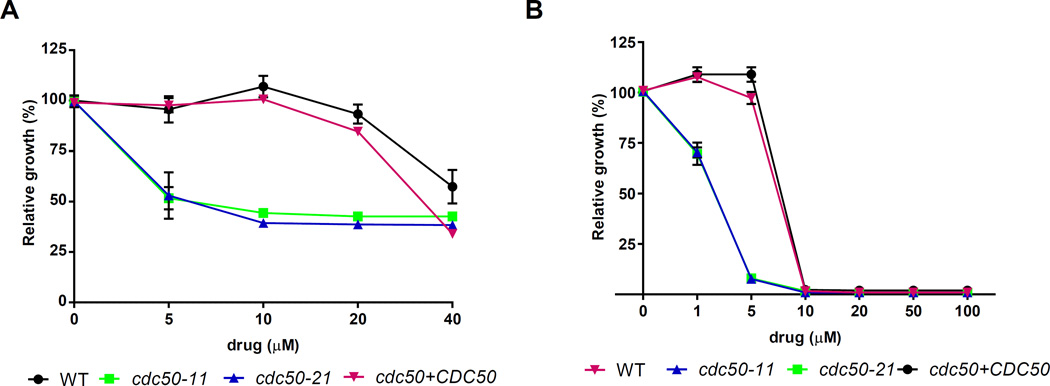
We next tested whether loss of Cdc50 resulted in changes in phospholipid asymmetry by examining PS-specific Annexin V binding and sensitivity to drugs that target phospholipids in the plasma membrane. Flippases translocate phosphatidylserine (PS), phosphatidylcholine (PC) and/or phosphatidyethanolamine (PE) from one leaflet of the membrane bilayer to the other to maintain membrane asymmetry. Both PE and PS are located preferentially on the cytoplasmic leaflet of plasma membrane and a loss of asymmetry results in exposure of PS and PE on the outer leaflet (Muthusamy et al., 2009a; Sebastian et al., 2012). With regard to PS, we observed a slight increase of Annexin V binding in the cdc50 mutants compared with that in the WT and the CDC50 complemented strains (data not shown). This is consistent with the recent report by Huang et al. (2016) that a cdc50 mutant had increased accumulation of Annexin V binding and is more sensitive to PS-specific drug papuamide B. We also assessed sensitivity to cinnamycin, a cyclic antifungal peptide that targets PE exposed on the outer leaflet of the plasma membrane. Cinnamycin is thought to be toxic because it affects transbilayer lipid movement. Interestingly, we observed that deletion of CDC50 caused increased sensitivity to cinnamycin, suggesting greater PE exposure on the outer leaflet (Fig. 5A). This result is different than the situation in S. cerevisiae where Cdc50 is thought to have an influence specifically on PS asymmetry (Chen et al., 1999; Saito et al., 2004; Lenoir et al., 2009). We previously observed that deletion of the putative flippase, Apt1, leads to the increased sensitivity to cinnamycin (Hu et al., 2010). We also examined the influence of miltefosine on the growth of WT, cdc50 mutant and CDC50 complemented strains. Miltefosine is an antitumor drug that also is used to treat leishmaniasis, and it has been previously reported to inhibit the growth of C. neoformans (Widmer et al., 2006). The drug is an alkylphospholipid that is internalized by P4-ATPase activity in Leishmania (Hansen et al., 2003; Widmer et al., 2006; Ravu et al., 2013; Garcia-Sanchez et al., 2014). Surprisingly, deletion of CDC50 in C. neoformans caused increased sensitivity to miltefosine (Fig. 5B), perhaps because of changes in flippase activity. Taken together, our analysis revealed that Cdc50 in C. neoformans plays a conserved role in protein trafficking and maintenance of membrane asymmetry by interfering with the distribution of phospholipids on the membrane, as suggested by altered drug sensitivity.
Figure 5. Cdc50 influences sensitivity to cinnamycin and miltefosine.
Deletion of CDC50 causes increased sensitivity to the PE-binding antifungal peptide cinnamycin (Ro09-0198) (A) and the PC-binding miltefosine (B). The sensitivities of the strains to the drugs are shown at a range of concentrations. After 72 h of incubation at 30°C, optical densities (OD600) were measured and the values of cultures incubated without the peptide were normalized to 100% growth. Three replicates of the experiment were performed.
Cdc50 is required for robust growth on heme and inorganic iron sources
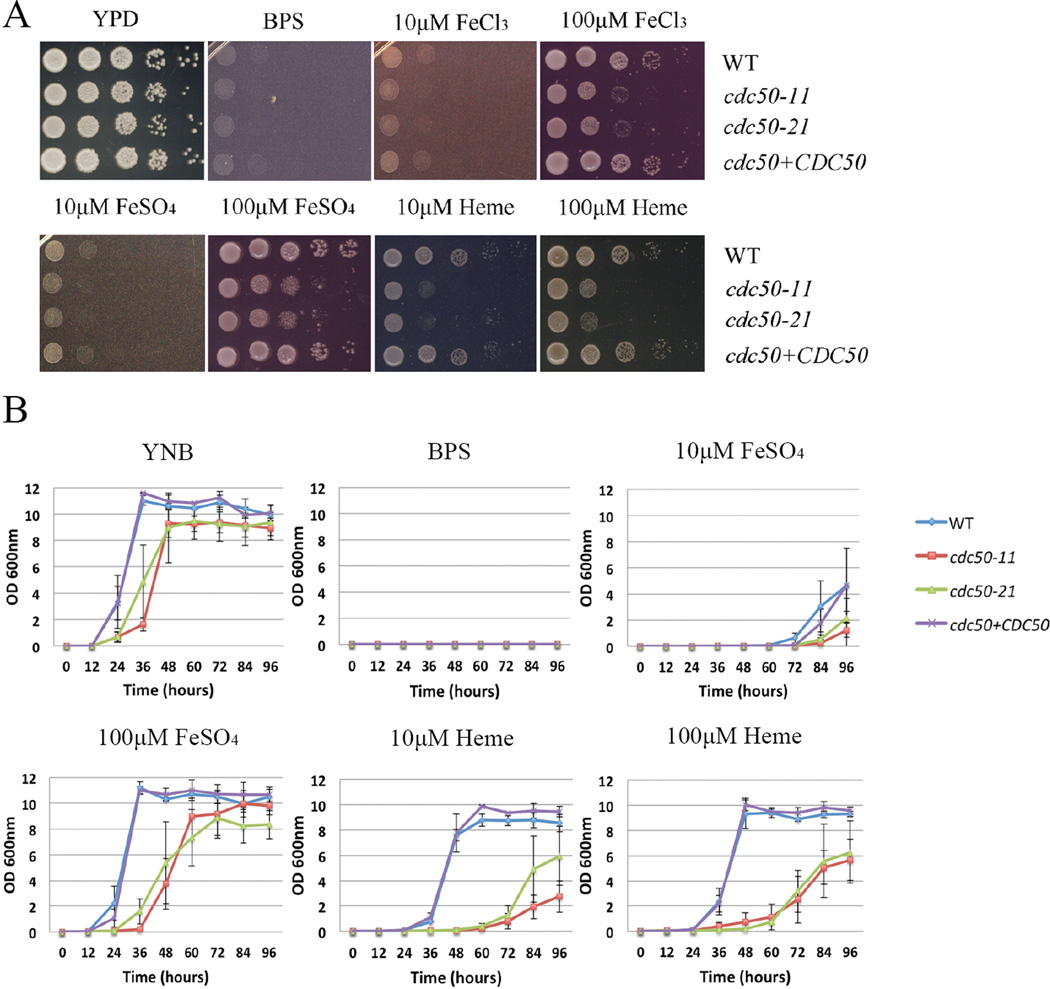
Our initial observation of a growth defect on iron for the cdc50 mutant suggested that Cdc50 might function in proper trafficking of iron acquisition proteins to the cell surface and/or internalization of iron or iron-containing molecules. We examined the ability of the cdc50 deletion mutants to acquire iron from heme and from inorganic sources by first growing strains in yeast nitrogen base–low-iron medium (YNB-LIM) to exhaust intracellular iron stores. Growth was then tested in spot assays on YNB-LIM at neutral pH (pH 7.0), without or with FeCl3, FeSO4 or heme. As predicted, the iron-starved WT, cdc50 mutant and the complemented strains grew robustly on iron-replete YPD medium, but failed to grow on iron-depleted medium (YNB-LIM) (Fig. 6A). All of the iron-starved strains grew poorly on YNB-LIM with addition of 10 µM FeCl3 or 10 µM FeSO4. The WT strain also grew on YNB-LIM with the addition of 100 µM FeCl3, 100 µM FeSO4, and 10 µM or 100µM heme. However, the cdc50 deletion mutants exhibited reduced growth on YNB-LIM supplemented with 100 µM FeCl3, 100 µM FeSO4, or heme at 10 or 100 µM at pH 7.0. Re-introduction of CDC50 to the cdc50 mutant strain restored the growth of the mutant (Fig. 6A). Growth assays were also performed in liquid YNB-LIM medium and, although the cdc50 mutants generally grew more poorly than the WT strain, there was a clear growth defect on heme as the iron source (Fig. 6B).
Figure 6. Cdc50 is required for robust growth on heme or inorganic iron sources.
(A) Tenfold serial dilutions of each strain (labeled on the right) were spotted on the indicated media after iron starvation and the plates were incubated at 30°C for 2 days before being photographed. (B) Iron-starved cells of the indicated strains were inoculated in liquid YNB medium plus 150 µM BPS without and with supplementation with iron sources. The cultures were incubated at 30°C, and OD600 was measured.
We also tested the growth of the apt1, 2, 3 and 4 mutants on YNB-LIM with or without iron sources (heme, FeCl3, or FeSO4). The apt1 mutants had reduced growth on YNB-LIM supplemented with 100 µM FeSO4 (Supplemental Fig. S3), suggesting that Apt1 contributes to iron utilization (although the phenotype was less pronounced than for the cdc50 mutant). Interestingly, loss of Apt1 did not reduce growth on FeCl3 suggesting that Apt1 makes a specific contribution to the use of ferrous iron. The apt2, apt3 and apt4 mutant strains grew at the level of the WT strain on YNB-LIM with all iron sources (Supplemental Fig. S3). Overall, the results of the growth assays on different iron sources support the hypothesis that Cdc50 and Apt1 are required for efficient iron acquisition from heme and/or inorganic sources.
Loss of Cdc50 caused increased sensitivity to the iron chelator curcumin
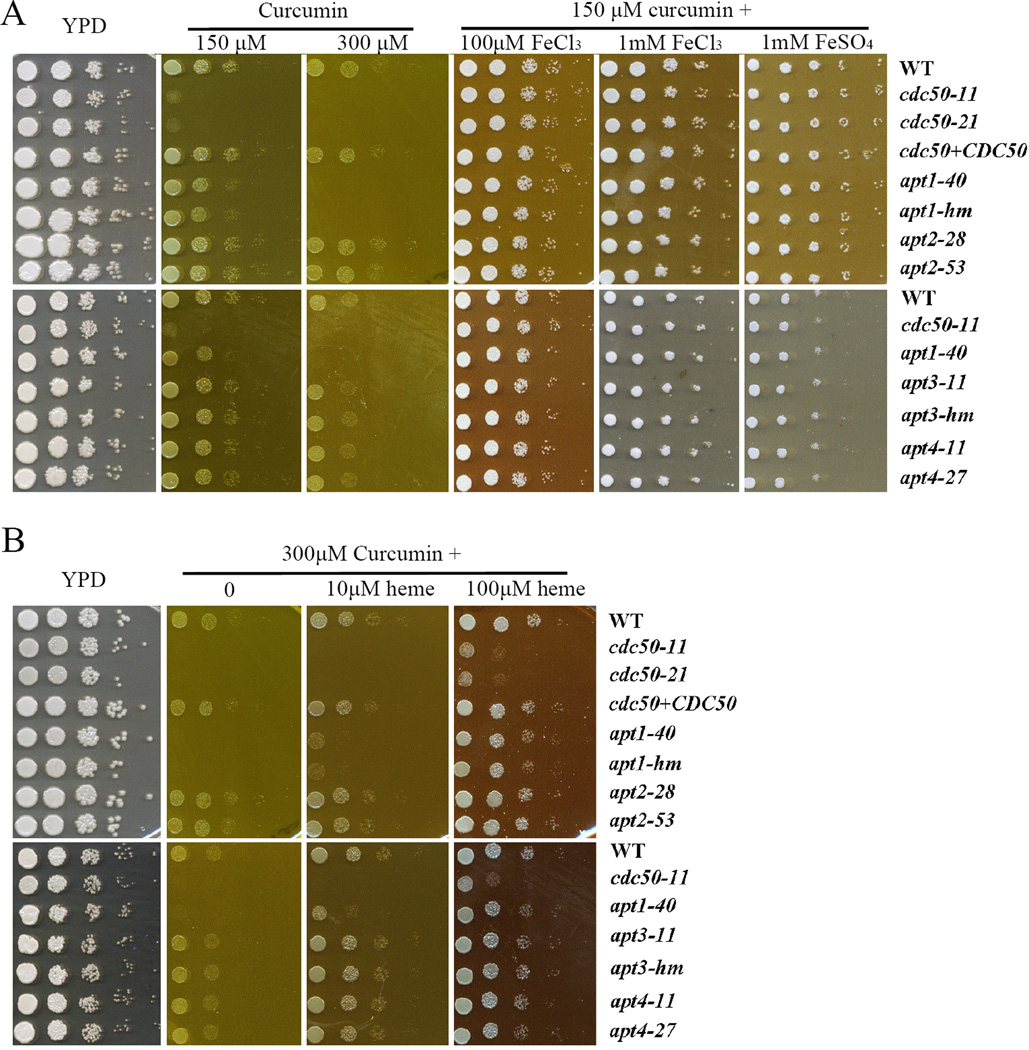
We investigated the iron acquisition defect of the cdc50 mutant in more detail with the drug curcumin, a component of turmeric from the root of the Curcuma longa plant that is used extensively in traditional Indian medicine (Hatcher et al., 2008). Curcumin is a chelator of Fe(III) that antagonizes the growth of S. cerevisiae and Candida albicans (Minear et al., 2011; Neelofar et al., 2011; Azad et al., 2014; Kumar et al., 2014; Lee et al., 2014; Carmello et al., 2015). Interestingly, a screen of the S. cerevisiae deletion collection for sensitivity to curcumin identified mutants lacking iron uptake functions and the Lem3 subunit of flippases (Azad et al., 2014). It is known that curcumin accumulates in the ER in S. cerevisiae and we also confirmed this for C. neoformans (Supplemental Fig. S4) (Minear et al., 2011). We then performed spot assays of the WT and mutant strains on YPD supplemented with the different concentrations of curcumin. The WT and complemented strains grew well on YPD supplemented with either 150 µM or 300 µM curcumin, although the growth on the higher concentrations was generally weaker than on lower concentrations (Fig. 7A). As expected, deletion of CDC50 increased sensitivity to curcumin at either 150 µM or 300 µM (Fig. 7A, Supplemental Fig. S5, data not shown). Supplementation with inorganic iron sources (FeCl3 and FeSO4) at the relatively high concentrations of 100 µM or 1 mM to the YPD plates with curcumin (150 µM or 300 µM) completely restored the growth of mutants to the WT level suggesting that low affinity uptake may bypass the requirement for Cdc50 (Fig. 7A). In contrast, addition of heme at either 10 or 100 µM to the YPD plate with curcumin did not completely restore the growth of the cdc50 mutants (Fig. 7B). This finding suggests a role for Cdc50 in heme uptake and/or utilization. We also noted that the inorganic iron sources and heme allowed the WT strain to overcome curcumin inhibition, and this is consistent with the iron chelation activity of curcumin (Fig. 7). The influence of metals is specific for iron because addition of zinc or copper ions to the YPD plates with or without curcumin did not influence the growth of the strains (Supplemental Fig. S5, and data not shown). Interestingly, assays with the apt1, 2, 3 and 4 deletion mutants demonstrated that only deletion of APT1 caused increased sensitivity to curcumin, although not to the same extent as the cdc50 mutation (Fig. 7, Supplemental Fig. S5). Addition of the inorganic iron sources and heme restored the growth of apt1 strains on curcumin. An apt1 apt3 mutant showed the same sensitivity as the apt1 mutant on curcumin (Supplemental Fig. S1). Taken together the sensitivity to curcumin provides additional evidence that Cdc50 and Apt1 (to a lesser extent) are involved in iron acquisition in C. neoformans.
Figure 7. Loss of Cdc50 or Apt1 leads to increased sensitivity to the iron-chelating drug, curcumin.
A) Tenfold serial dilutions of each strain, including the WT, cdc50 mutants, the CDC50 complemented strains and the flippase mutants, without iron starvation were spotted on the YPD plates without or with curcumin at a concentration of either 150 µM or 300 µM supplemented with 0, 100 µM or 1 mM of the indicated inorganic iron sources. B). Tenfold serial dilutions of each strain without iron starvation were spotted on the YPD plates without or with 300 µM curcumin supplemented with 0, 10, or 100 µM heme. The plates were incubated at 30°C for 2 days before being photographed.
We extended the characterization of curcumin sensitivity by assessing the growth of well-characterized iron acquisition mutants and other strains on solid YPD medium supplemented with 150 µM of curcumin (Supplemental Fig. S6). The strains included the mutants with known iron-related phenotypes due to defects in ESCRT complex genes (vps27, vps23, vps22, snf7, vps20, bro1, vps4) (Hu et al., 2013, Hu et al., 2015), regulators of iron utilization (cir1, sre1, rim101, hapX) (Jung et al., 2006; Chang et al., 2007; Jung et al., 2010; O’Meara et al., 2010), components of the high affinity iron uptake system (cft1 and cfo1) (Jung et al., 2008; Jung et al., 2009), and a putative hemophore (cig1) (Cadieux et al., 2013). Other strains carried mutations in components of the cAMP-PKA signaling pathway (pka1 and pkr1) (D’Souza et al., 2001b; Hu et al., 2007a), functions involved in elaboration of capsule and melanin (cap59, cap60, lac1) (Chang et al., 1998; Garcia-Rivera et al., 2004; Zhu et al., 2004), and a protein kinase involved in carbon metabolism and melanin production at 37°C (snf1) (Hu et al., 2008b). The spot assays revealed that defects in iron regulation and acquisition caused increased sensitivity to curcumin. For example, the mutants in ESCRT-I, II, and III components (vps27, vps23, vps22, snf7, vps20), but not ESCRT-0 (bro1, vps4), are involved in heme utilization and iron uptake from inorganic sources (Hu et al., 2015). Accordingly, the ESCRT-I, II, and III mutants (vps27, vps23, vps22, snf7, vps20) were more sensitive to curcumin than the WT strain. Similarly, mutations of genes involved in the regulation of iron utilization and iron acquisition, including cir1, sre1, rim101, hapX, cft1 and cfo1 resulted in increased sensitivity to curcumin. Interestingly, deletion of one capsule-associated gene, CAP59, but not the other one, CAP60, caused increased sensitivity. Moreover, loss of the regulatory subunit (Pkr1), but not the catalytic subunit (Pka1) of cAMP-dependent protein kinase A (PKA) led to the hypersensitivity to curcumin. Taken together, the spot assays support the conclusions that curcumin inhibits the growth of C. neoformans due to iron chelation, and that Cdc50 and Apt1 contribute to iron and heme acquisition.
Cdc50 deletion causes increased sensitivity to fluconazole
We previously showed that loss of Apt1 in C. neoformans results in increased sensitivity to azole drugs (Hu et al., 2010). Additionally, we found that azole sensitivity is associated with iron and heme acquisition (Jung et al., 2009). We extended this analysis by examining the sensitivity of the cdc50 mutants to the azole antifungal drug fluconazole that targets lanosterol 14-α-demethylase in the ergosterol biosynthesis pathway. The cdc50 mutants exhibited a pronounced growth defect on the plates supplemented with fluconazole, and re-introduction of CDC50 to the mutant strain restored the growth on the drug (Supplemental Fig. S7A). We also found that the apt3 mutants displayed increased sensitivity to fluconazole (Supplemental Fig. S7B) and that the cdc50 mutants had the most pronounced sensitivity compared with the apt1 and apt3 mutants (Supplemental Fig. S7B). These results link flippase activity mediated by Cdc50, Apt1 and Apt3 to azole sensitivity, perhaps via a connection to sterol biosynthesis and trafficking as demonstrated in S. cerevisiae (Muthusamy et al., 2009b). We note that heme plays an important role in ergosterol biosynthesis because some of the enzymes in the pathway are heme-dependent including lanosterol 14-α-demethylase (Crisp et al., 2003; Jung et al., 2008; Kim et al., 2012). Previously we showed that heme or a siderophore can restore growth on fluconazole for the C. neoformans cfo1 and cft1 mutants defective in high affinity iron uptake (Jung et al., 2009; Kim et al., 2012; Saikia et al., 2014; Hu et al., 2015). This may be due to up-regulation of uptake functions in the mutants due to iron starvation or to direct rescue of heme-containing enzymes for ergosterol biosynthesis. In our current study, we found that heme did not rescue the growth of the cdc50 mutants on fluconazole (Supplemental Fig. S7A). Therefore, it is possible that loss of Cdc50 influences heme and iron acquisition and/or ergosterol biosynthesis by a mechanism distinct from that of the high affinity iron uptake system. We also found that the increased sensitivity of the mutants to fluconazole was not exacerbated upon addition of curcumin (Supplemental Fig. S7B).
Loss of Cdc50 influences growth at acidic and alkaline pH, and impairs membrane integrity
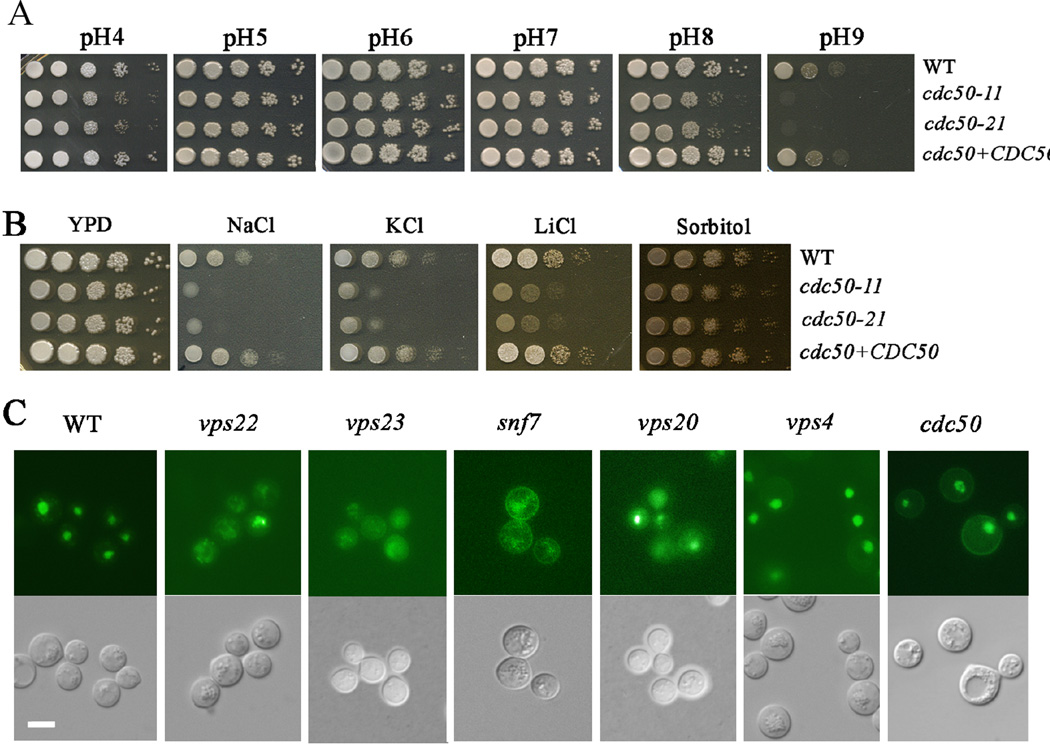
Iron acquisition in fungi is influenced by environmental pH and the pH-responsive transcription factor Rim101 is associated with phospholipid asymmetry in yeast and regulates iron uptake functions in C. neoformans (Xu et al., 2004; Blanchin-Roland et al., 2005; Hayashi et al., 2005; Boysen et al., 2006; Ikeda et al., 2008; Wolf et al., 2010; Ost et al., 2015). We therefore hypothesized that changes in phospholipid asymmetry due to deletion of CDC50 would influence the pH response in C. neoformans. To test this idea, we performed spot assays on YPD plates at a range of pHs from 4 to 9 with the WT, cdc50 mutant and CDC50 complemented strains (Fig. 8A). The WT strain grew well at each pH, although less robustly at alkaline pH (pH 8 and pH 9). In contrast, the cdc50 mutants displayed reduced growth at both acidic (pH4) and alkaline pH (pH8 and pH9). The apt1 mutant also showed reduced growth but only at pH 9.0 (Supplemental Fig. S8). The pH response is regulated by Rim101 in C. neoformans and other fungi, and activation of Rim101 protein is dependent on ESCRT functions (Xu et al., 2004; Blanchin-Roland et al., 2005; Hayashi et al., 2005; Boysen et al., 2006; Wolf et al., 2010; Hu et al., 2013; Ost et al., 2015). The ESCRT complex, Rim101 pathway and its regulatory subunit Rim20, are required for robust growth at alkaline pH (both pH8 and pH9) in C. neoformans and these functions are also involved in response to salt stress but not osmotic stress (O’Meara et al., 2010; Hu et al., 2015; Ost et al., 2015). Moreover, Rim101 is also involved in maintaining and responding to the membrane asymmetry in S. cerevisiae (Ikeda et al., 2008). These observations prompted an examination of additional phenotypes of the cdc50 mutants in light of a possible connection with ESCRT and Rim101 functions.
Figure 8. Loss of CDC50 influences growth at acidic and alkaline pH, and sensitivity to salt stress.
(A) Tenfold serial dilutions of each strain were spotted onto buffered YPD, and the plates were incubated at 30°C for 2 days before being photographed. (B) Tenfold serial dilutions of the strains were spotted onto solid YPD without or with 1.2 M NaCl, 1.5 M KCl, 1.5 M sorbitol, or 100 mM LiCl. The plates were incubated at 30°C for the following times: sorbitol, 3 days; KCl, 5 days; NaCl, 5 days; LiCl, 10 days. (C) Loss of CDC50 did not influence the localization of RIM101-GFP, but deletion of the genes encoding ESCRT complex proteins or Rim101 regulatory components caused mis-localization. Bar = 10 µm.
We first examined the growth of the WT, cdc50 mutant and CDC50 complemented strains on YPD plates supplemented with 1.2M KCl, 1.2M NaCl, 0.2M LiCl or 1.5M sorbitol. The WT strain grew well on all of the media but deletion of CDC50 caused impaired growth on the LiCl, NaCl and KCl plates similar to the phenotypes of the rim101, snf7, vps22, vps23, and vps20 mutants (the ESCRT complex) (O’Meara et al., 2010; Hu et al., 2015; Ost et al., 2015). Cdc50 therefore shows a similar influence on the response to pH and salt stress as the ESCRT-Rim101 pathway. However, deletion of CDC50 resulted in reduced growth on sorbitol, indicating that Cdc50 is involved in osmotic stress response, in addition to its role in response to pH and salt stress (Fig. 8B). The apt1, 2, 3 and 4 mutants did not show marked growth phenotypes under salt stress (Supplemental Fig. S8). The regulation of the pH response by Rim101 is also influenced by the localization of the protein. For example, nuclear Rim101-GFP is mis-localized to cytoplasm in alkaline pH, rim mutants and Vps23 (ESCRT-II) mutant in C. neoformans (Ost et al., 2015). We therefore assessed the Rim101-GFP localization in the cdc50 mutant in comparison with the ESCRT mutants by deleting CDC50, SNF7, VPS23, VPS20 or VPS22 in the Rim101-GFP strain. We found that Rim101-GFP is mis-localized to cytoplasm in the ESCRT (vps23, snf7, vps22, vps20) mutants, but not in the cdc50, bro1 and vps4 mutant strains (Fig. 8C). No change of Rim101-GFP in bro1 and vps4 is expected, as both Bro1 and Vps4 do not play a significant role in the response to alkaline pH (Hu et al., 2015). Taken together, we conclude that Cdc50 exhibits shared phenotypes with the ESCRT-Rim101 pathway in response to the pH and salt stress, but makes distinct contributions with regard to Rim101 localization and osmotic stress. We did note that the snf7 and cdc50 mutants both showed a Rim101-GFP signal at the cell cortext raising the possibility of a role for Cdc50 in the localization of a fraction of the Rim101 protein.
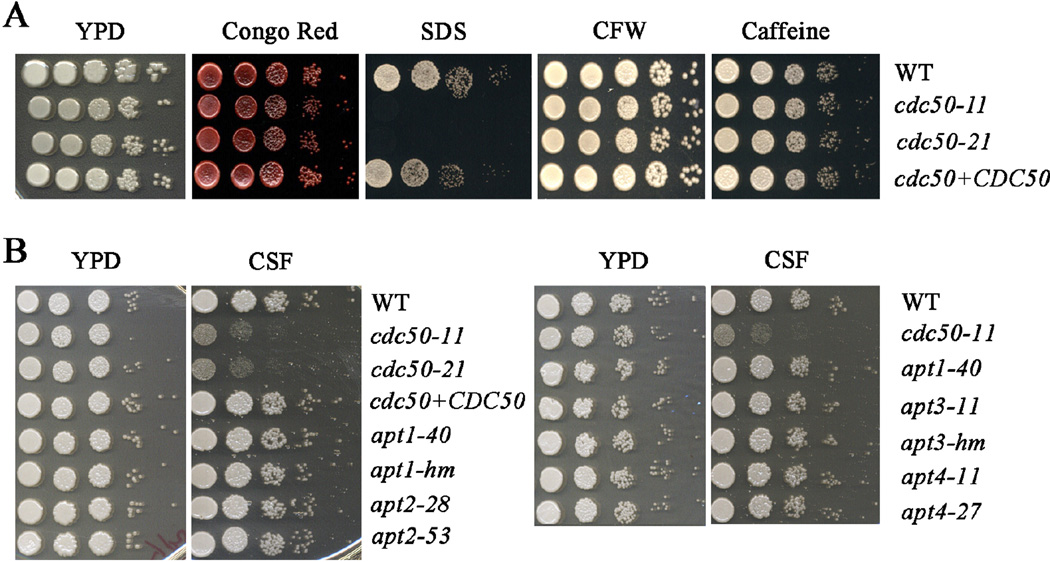
We also examined the influence of deletion of CDC50 on cell wall and membrane integrity by growing strains on YPD plates supplemented with SDS, Congo Red, caffeine, or calcofluor white (Fig. 9A). Deletion of CDC50 causes increased sensitivity to SDS, but not calcofluor white, congo red and caffeine, indicating that Cdc50 is involved in maintaining the cell membrane integrity. Re-introduction of CDC50 to the cdc50 mutant restored the growth to the WT level on SDS (Fig. 9A). Huang et al. (2016) reported that loss of CDC50 caused the hypersensitivity to caspofungin, a drug targeting the fungal cell wall, although C. neoformans is generally insensitive to this drug (Gerik et al., 2005). In light of this observation, we examined the sensitivity of the apt1, 2, 3 and 4 mutants relative to the cdc50 mutant on caspofungin and found that only deletion of cdc50 caused increased sensitivity (Fig. 9B). Moreover, the apt1, 2, 3 and 4 mutants all grew at the WT level on the agents that challenged cell wall and membrane integrity including SDS, Congo Red, caffeine and calcofluor white (data not shown). Overall, these results are consistent with a key role for Cdc50 in membrane integrity and caspofungin sensitivity as reported by Huang et al. (2016). The distinct phenotypes of the apt mutants may be due to separate functions or redundancy.
Figure 9. Deletion of CDC50 influences membrane but not cell wall integrity.
A) Tenfold serial dilutions of the strains were spotted onto the YPD with or without chemicals to challenge cell wall and membrane integrity including Congo Red, sodium dodecyl sulphate (SDS), Calfluor white (CFW) and caffeine. The plates were incubated at 30°C for 2 days before being photographed. B) Tenfold serial dilutions of the strains including the mutants of each candidate flippase gene were spotted onto the YPD with or without the cell wall targeted drug, caspofungin (CSF). The plates were incubated at 30°C for 2 days before being photographed.
Discussion
Flippases function to maintain phospholipid asymmetry in membranes and they participate in vesicle-mediated protein transport in the Golgi and endosomal systems (Muthusamy et al., 2009a; Sebastian et al., 2012; Ravu et al., 2013). There are three members of the Cdc50 family in S. cerevisiae (Cdc50, Lem3 and Crf1) that serve as non-catalytic subunits with flippases (Drs2, Dnf1, Dnf2, Dnf3, Neo1) to establish asymmetric phospholipid distributions in the plasma membrane, the trans-Golgi network (TGN) and the endosomal system (Muthusamy et al., 2009a; Sebastian et al., 2012; Ravu et al., 2013). We identified and characterized the single member of the Cdc50 family in C. neoformans in a screen for BFA sensitivity, and Huang et al. (2016) characterized the same protein as playing a role in caspofungin resistance. Interestingly, Cdc50 from C. neoformans shares properties with both Cdc50 and Lem3 from S. cerevisiae. For example, Cdc50 in S. cerevisiae localizes in the TGN and endosomes while Lem3 is found in the ER and plasma membrane (Hanson et al., 2003; Misu et al., 2003; Saito et al., 2004; Chen et al., 2006; Lenoir et al., 2009; Ono et al., 2009; Hankins et al., 2015). We localized the C. neoformans protein in the ER and endosome-like structures while Huang et al. (2016) reported the protein in the ER and plasma membrane. These results suggest that the C. neoformans protein may have a broader localization than each of the yeast proteins, although additional work is needed to explore an association with the TGN. Loss of Cdc50 in C. neoformans also influenced sensitivity to a number of drugs including BFA, curcumin and fluconazole (as seen for Lem3 in yeast), acidic pH (as seen for Cdc50 in yeast) and alkaline pH. Unlike the yeast versions of Cdc50 and Lem3, the C. neoformans protein did not influence sensitivity to agents that challenge the cell wall such as Congo red, but did share the property of poor growth on SDS thus indicating a problem with membrane integrity.
Loss of Cdc50 in C. neoformans also increased sensitivity to drugs that reflect changes in the asymmetrical distribution or movement of phospholipids in membranes such as papuaminde A (phosphatidylserine) (Huang et al., 2016), cinnamycin (phosphatidlyethanolamine) and miltefosine (phosphatidylcholine). Miltefosine is particularly interesting because this alkylphosphocholine drug is used to treat protozoal and fungal diseases (Hanson et al., 2003; Widmer et al., 2006; Ravu et al., 2013; Garcia-Sanchez et al., 2014). It is curious that cdc50 or lem3 mutants in S. cerevisiae and Leishmania infantum displayed resistance to miltefosine while loss of Cdc50 in C. neoformans led to increased sensitivity. It is possible that miltefosine acts by a different mechanism in C. neoformans or that a compensatory increase in another flippase activity caused increased accumulation of miltefosine in the cdc50 mutant of this fungus. In general, Cdc50 in C. neoformans may also exhibit a distinct response to certain drugs indirectly due to influences on the trafficking of different sets of transporters that control accumulation.
We previously characterized a flippase, Apt1, in C. neoformans and showed that APT1 complemented a drs2 mutation in S. cerevisiae (Hu et al., 2010). In the current study, we compared our apt1 mutant with the cdc50 mutants and included additional C. neoformans mutants with defects in other predicted flippase genes (APT2, APT3 and APT4). We found that cdc50 and apt1 mutants share a number of phenotypes suggesting that they function together to contribute flippase activity. For example, our published analysis revealed that an apt1 mutant is attenuated for survival in macrophages and virulence in mice, and is susceptible to cinnamycin, fluconazole and alkaline pH, as well as nitrosative and oxidative stress (Hu et al., 2010; Rizzo et al., 2014). A comparison with the cdc50 mutant revealed commonalities for all of these phenotypes except the response to nitrosative and oxidative stress. Additional comparisons indicated shared susceptibility to BFA, curcumin and iron/heme starvation for the apt1 and cdc50 mutants, and a cdc50 apt1 mutant had the greater sensitivity to BFA and curcumin seen with the cdc50 mutant (Supplemental Fig. S2). The cdc50 mutants showed additional sensitivities to salt stress and acidic pH. Our data do not preclude the possibility that Cdc50 interacts with the other candidate flippases to influence some of these phenotypes in C. neoformans. In this regard, our analysis of the other apt mutants only revealed a slight sensitivity to BFA for the apt3 mutant and it is possible that major phenotypes are masked by redundancy between Apt2, 3, and 4. Surprisingly, none of apt1, 2, 3 and 4 mutants showed decreased resistance to caspofungin, again perhaps because of redundancy or other functions of Cdc50. Certainly, redundancy has been observed for the flippases in yeast (Graham, 2004), and our analysis of an apt1 apt3 double mutant indicates that both flippases contribute to BFA sensitivity (Supplemental Fig. S2). Taken together, these results provide insights into the complexities of flippases in fungi and suggest that a single Cdc50 in C. neoformans may contribute functions that are shared between Cdc50 and Lem3 in yeast.
Iron acquisition is a critical aspect of C. neoformans virulence and iron sensing is important for regulating the polysaccharide capsule that is the major virulence factor (Jung et al., 2006; Kronstad et al., 2013). The impaired growth of the cdc50 and apt1 mutants on media with heme or inorganic irons as the sole iron source, and the hypersensitivity of the mutants to the specific iron-chelating drug curcumin, suggest that Cdc50 and Apt1 contribute to iron utilization. Cdc50 appears to play a more important and broader role than Apt1 because the phenotypes of cdc50 mutants were more extensive (FeCl3, FeSO4 and heme). The growth defect for the apt1 mutant was observed only on FeSO4. In contrast, heme rescue of curcumin sensitivity was more apparent for the WT strain and the apt1 mutant than for the cdc50 mutant, and the latter strain retained a growth defect in the presence of curcumin and heme. Given possible roles in high and low affinity uptake, heme uptake, internal trafficking of iron-containing molecules (e.g. to the vacuole, mitochondria and ER) and iron processing, it appears that Cdc50 has a broader role, perhaps through interaction with other flippases. Importantly, the lack of rescue of the curcumin inhibition by heme suggests a specific contribution of Cdc50 to heme uptake. Interestingly, Apt1 contributed to the use of ferrous iron and may therefore influence a specific uptake system for reduced iron. An intriguing possibility is that Apt1 directly transports iron given that a P1B4-type ATPase, PfeT, effluxes ferrous iron in Bacillus subtilis (Guan et al., 2015).
Connections between phospholipid composition and iron uptake have been established previously in S. cerevisiae. For example, a screen of the ~4,700 deletion strains identified 17 genes that contributed to hypersensitivity to curcumin (Minear et al., 2011). Of relevance to our study, these genes encoded high affinity iron uptake functions (Fet3, Ftr1), the iron regulator Aft1, copper uptake and regulatory proteins, and proteins for copper and iron homeostasis. Additional identified genes and functions included ERV14 (vesicle secretion), ERG3 (ergosterol sysnthesis), VMA3, 13 and 27 (vacuolar acidification) and LEM3. Overall, this study established that curcumin inhibits growth through an iron chelation mechanism and linked Lem3 to iron acquisition and homeostasis. An additional screen of the yeast deletion collection for mutants with altered trafficking of the transporter Arn1 that mediates uptake of the iron-chelating siderophore ferrichrome also uncovered a connection with phospholipid synthesis and distribution (Guo et al., 2010). Arn1 is found in the plasma membrane in conditions of low ferrichrome and cycles between this location and intracellular vesicles in high ferrichrome (as part of the mechanism of siderophore-iron transport). Guo et al. (2010) found that loss of Drs2 resulted in mislocalization of Arn1-GFP to the plasma membrane in the absence of ferrichrome.
In general, the studies in yeast established a strong connection between flippase activity and iron acquisition that appears to be conserved in C. neoformans. That is, increased sensitivity to curcumin was observed in mutants with defects in genes encoding functions for iron acquisition and utilization in C. neoformans. The genes encode previously characterized regulators of iron utilization (Cir1 and HapX) (Jung et al., 2006; Jung et al., 2010), iron transporters (Cft1 and Cfo1) (Jung et al., 2008; Jung et al., 2009), and components of the ESCRT complexes (Vps27, Vps23, Vps22, Vps20, Snf7) (Hu et al., 2013, Hu et al., 2015). The phenotypes of these mutants support the idea that increased sensitivity to curcumin is due to reduced intracellular iron levels as a result of chelation. Notably, other mutants lacking the pH-response regulator Rim101 and the potential hemophore Cig1 (O’Meara et al., 2010; Cadieux et al., 2013) that specifically influence heme acquisition did not show growth inhibition by curcumin, perhaps because these strains are not iron-limited under the conditions tested. Interestingly, curcumin sensitivity was also observed for mutants lacking Cap59 or Pkr1, two proteins involved in capsule formation (D’Souza et al., 2001a; Garcia-Rivera et al., 2004). Cap59 plays a role in trafficking of capsular materials and deletion of the gene causes acapsular phenotype, while Pkr1 is the regulatory subunit of protein kinase A (PKA) that regulates capsule size (D’Souza et al., 2001b). Interestingly, mutants in CAP59 and PKR1 do not show defects in growth on inorganic iron or heme and this suggests that curcumin sensitivity may be due to other factors such as enhanced accumulation of the drug. PKA is known to influence iron utilization through interactions with the ESCRT pathway in C. neoformans (Hu et al., 2015), and to regulate iron uptake functions in yeast and other fungi (Robertson et al., 2000; Choi et al., 2015).
We demonstrated that curcumin accumulates in the ER of C. neoformans cells, similar to S. cerevisiae (Minear et al., 2011). ER targeting may be relevant to the observed increased sensitivity of the cdc50 mutant to both curcumin and fluconazole, an inhibitor of ergosterol biosynthesis in the ER. Additionally, some of the enzymes involved in ergosterol biosynthesis are heme-dependent including the fluconazole target lanosterol 14-α-demethylase (Erg11). We also noted in this context that the sre1 mutant lacking the sterol-responsive regulator of the response to hypoxia also has increased sensitivity to curcumin (Chang et al., 2007). Taken together, these observations focus attention on a key role for Cdc50 in ER functions related to ergosterol. We hypothesize that the contribution of Cdc50 to flippase activity in establishing and maintaining phospholipid asymmetry is important to the balance between ergosterol and phospholipids. This balance has been demonstrated in S. cerevisiae and Drosophila melanogaster through the characterization of oxysterol binding proteins (Misu et al., 2003; Muthusamy et al., 2009b; Ma et al., 2012). These result focus attention on the potential to target the balance with antifungal drugs and synergistic activity between curcumin and fluconazole has already been described in C. albicans (Sharma et al., 2010; Garcia-Gomes et al., 2012). Although we did not observe a combined influence of both drugs in C. neoformans, additional studies with azole drugs, inhibitors of trafficking, and iron chelators are warranted.
Huang et al. (2016) recently characterized Cdc50 as part of an investigation to understand the lack of sensitivity of C. neoformans to the cell-wall targeting drug caspofungin. In light of these results, we tested caspofungin sensitivity for the putative apt1, 2, 3 and 4 flippase mutants and failed to observe a difference compared with WT, and in contrast with the sensitivity of the cdc50 mutant. It may be necessary to test double or triple deletion mutants to more fully assess the involvement of flippases in caspofungin sensitivity. Similarly, none of the single flippase deletion mutant (apt1, 2, 3 and 4) had increased sensitivity to SDS or any inhibitors of cell wall integrity (Hu et al., 2010, data not shown), suggesting redundancy for the catalytic subunits of the flippases in the maintenance of membrane integrity. The cdc50 deletion mutant also showed both shared and distinct phenotypes compared with the rim101 and the ESCRT complex mutants in response to the acidic and alkaline pH conditions (O’Meara et al., 2010; Hu et al., 2013; Hu et al., 2015). Shared phenotypes to those of ESCRT complex and RIM101 included retarded growth at alkaline pH and in the presence of salt stresses (in specific, NaCl and LiCl). However, in contrast with the ESCRT-Rim101 regulated pH response, the cdc50 mutant strain also demonstrated reduced growth at the acidic pH (pH4), indicating a distinct role. Furthermore, the ESCRT complex contributes to the pH response via a mechanism involving the processing and localization of Rim101 (Hu et al., 2015; Ost et al., 2015), but Cdc50 does not have a major influence on Rim101 localization. It is possible that the influence of Cdc50 is due to changes in plasma membrane integrity or the localization of proteins that mediate the pH response.
Both our study and that of Huang et al. (2016) found that cdc50 mutants are completely avirulent in a mouse model of cryptococcosis. We previously found that mutants lacking Apt1 also displayed attenuated virulence and defective survival in macrophages. As mentioned above, the cdc50 and apt1 mutants share a number of phenotypes, although deletion of CDC50 generally caused more pronounced phenotypes including a more substantial influence on virulence (Hu et al., 2010). Of note, Cdc50 does not appear to have a major influence of common virulence factors such as capsule and melanin formation, and growth at 37°C. Huang et al. (2016) did observe an enlarged capsule for the cdc50 mutant in contrast to our observation that there was little difference from WT; the discrepancy is likely due to different growth conditions for our assays. They also noted that their cdc50 mutant secreted less capsular polysaccharide (Huang et al., 2016). We previously found that apt1 mutants also have a capsule size like WT for cells in culture. However, a more detailed examination showed that apt1 mutants had reduced synthesis of capsule polysaccharide particularly in the host environment (Rizzo et al., 2014). Similar detailed studies of the capsule in the cdc50 mutant should also be performed in the future. However, it seems that influences on capsule and other virulence factors do not adequately account for the cdc50 virulence defect. Instead, it appears that multiple contributions account for the virulence defect including altered membrane asymmetry, improper trafficking of proteins such as the acid phosphatase Aph1 to the cell surface, defects in iron acquisition, and increased sensitivity to stress including extremes of pH. Given our observations that mutants lacking the high affinity iron transporters Cft1 or Cfo1, or the global iron regulator Cir1, displayed attenuate virulence in mice, we favor the possibility that changes in phospholipid asymmetry defects in the acquisition of iron (and perhaps other nutrients) make a major contribution to the poor proliferation of cdc50 mutants in mice (Jung et al., 2006; Jung et al., 2008; Jung et al., 2009; Kronstad et al., 2011; Kronstad et al., 2012).
Experimental Procedures
Strains, plasmids, and media
The serotype A strain H99 (MATα) of C. neoformans var. grubii and mutant derivatives were maintained on YPD medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar). The nourseothricin, neomycin, and hygromycin resistance cassettes were from plasmids pCH233, pJAF1, and pJAF15 (obtained from Dr. J. Heitman), respectively. YPD medium plates containing neomycin (200 µg/ml) were used to select the cdc50, apt1, 2, 3, and 4 deletion transformants. Defined low-iron medium (LIM) and yeast nitrogen base (YNB with amino acids; pH 7.0) plus 150 µM bathophenanthroline disulfonate (BPS) (YNB_LIM) were used as iron-limiting media. YPD and/or YNB media supplemented as indicated were used for phenotypic characterization. The minimal medium with low glucose for Aph1 induction was as follows: 0.1% glucose, 10 mM MgSO4, 0.5% KCl, 13 mM glycine, 3 µM thiamine, and 10 µM CuSO4. All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless indicated otherwise. The strains employed in this study are listed in Supplemental Table S2.
Agrobacterium tumefaciens-mediated transformation of C. neoformans
Transformation was performed as previously described (Walton et al., 2006; Idnurm et al., 2007; Hu et al., 2013). Plasmid pPZP-Neo1 (a gift of Dr. Alex Idnurm), which carries a neomycin resistance cassette, was used to transform the wild type (WT) strain. Briefly, Agrobacterium tumefasciens AGL1 cells were grown overnight with shaking at room temperature in Luria-Bertani medium with kanamycin. Cells were washed and re-suspended in liquid induction medium with 200 µM acetosyringone (AS) at an optical density at 600 nm (OD600) of 0.15 and incubated for 6 h (OD600 of 0.6). C. neoformans WT cells were grown overnight in YPD medium, washed in induction medium, and re-suspended at 106 or 107/ml. Subsequently, 200 µl of each of the C. neoformans and A. tumefaciens cultures were mixed and spotted on induction agar medium (with AS). The plates were incubated for 2 to 3 days before the mixtures were resuspended in liquid YPD medium. Cells were then plated onto YPD medium with neomycin (200 µg/ml) and cefotaxime (100 µg/ml), and incubated at 30°C for 3 to 4 days.
Mutant screening and inverse PCR
A collection of ~30,000 transformants was screened for growth defects on YPD with 20 µg/ml of brefeldin A (BFA). Briefly, overnight cultures of each strain were grown in YPD medium in a 96-well plate format and 5 µl of each culture was transferred to a well containing 200 µl of YPD with BFA. Growth was determined by measurement of OD600 on a Tecan plate reader after incubation for 3 days at 30°C. Strains with defective growth on YPD with BFA were further confirmed with spot assays on YPD with 20 µg/ml of BFA. Inverse PCR was used to determine the disruption sites in candidate mutants using genomic DNA and the methods of (Zhang et al., 2000) and (Hu et al., 2007b; Hu et al., 2013) for DNA digestion, ligation, and PCR amplification. PCR products were sequenced, and insertion sites were determined by BLAST with the genome sequence database (www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/).
Identification of flippase genes, construction of deletion mutants and tagged strains, and complementation of the cdc50 deletion mutant
The candidate genes encoding P4-ATPases (aminophospholipid translocases (APTs) or flippases) were previously identified by a BLASTP search of the H99 genome database (www.broadinstitute.org) using the sequences of the flippases Drs2, Dnf1, Dnf2, Dnf3 and Neo1 from S. cerevisiae (Hu et al., 2010). This analysis identified candidate genes designated APT1 – 4 (Supplemental Table S1). Deletion of APT1 and complementation of the apt1 mutation have been described previously (Hu et al., 2010; Rizzo et al., 2014) and two independent mutants were obtained for APT2, 3 and 4. Additionally, deletion mutants for APT1 and APT3 were obtained from the Fungal Genetics Stock Center (http://www.fgsc.net/crypto/crypto.htm) collection constructed by the Madhani group; these are designated apt1-mh and apt3-mh (Liu et al., 2007). All deletion mutants were constructed by homologous recombination using gene-specific knockout cassettes, which were amplified by three-step overlapping PCR (Davidson et al., 2000) with the primers listed in Supplemental Table S3. The resistance markers for nourseothricin (NAT), neomycin (NEO) and hygromycin (HYG) were amplified by PCR using primers 2 and 5 and the plasmids pCH233, pJAF1 and pJAF15, respectively, as the templates. In general, the gene-specific knockout primers 1 and 2 and 4 and 6 were used to amplify the flanking sequences of their respective genes; and primers 1 and 6 were then used to amplify the gene-specific deletion construct containing the resistance marker. All constructs for deletions were introduced into the WT strain by biolistic transformation, as described previously (Davidson et al., 2000). Two independent cdc50 deletion mutants (cdc50-11 and cdc50-21) were analyzed in the experiments. To complement the cdc50 deletion mutant, an overlap PCR was performed on WT genomic DNA and plasmid pJAF15, respectively, to include the CDC50 gene (Cdc50-Rec-P1F and Cdc50-Rec-P1R), a hygromycin resistance cassette (Cdc50-Rec-P2F and Cdc50-Rec-P2R) and a 3’ flank region of CDC50 gene (Cdc50-Rec-P3F and Cdc50-Rec-P3R), and primers Cdc50-Rec-P1F and Cdc50-Rec-P3R were then used to amplify the complementation construct containing the resistance marker (Supplemental Table S3). The cdc50-11 strain was transformed with the CDC50 complementation construct by biolistic transformation with selection on hygromycin (200 µg/ml). Reintroduction of CDC50 was confirmed by colony PCR and genomic hybridization.
A modified overlapping PCR strategy was used to generate the constructs for the Sec61-GFP and Cdc50-mCherry strains. Briefly, the left and right arms for the Sec61-GFP fusion construct were amplified from WT genomic DNA using the primer set Sec61-GFP-P1F and Sec61-GFP-P1R, and the primer set Sec61-GFP-P3F and Sec61-GFP-P3R, respectively. The GFP gene and the hygromycin (HYG) resistance gene were amplified from the plasmid pGH023 using primers Sec61-GFP-P2F and Sec61-GFP-P2R. Overlap PCR was performed using primers Sec61-GFP-P1F and Sec61-GFP-P3R to yield a 5kb construct. The left and right arms for the Cdc50-mCherry fusion construct were amplified from WT genomic DNA using the primer set Cdc50-mCherry-P1F and Cdc50-mCherry-P1R, and the primer set Cdc50-mCherry-P3F and Cdc50-mCherry-P3R, respectively. The mCherry gene and the neomycin (Neo) resistance gene were amplified from the plasmid pGH025 using primers Cdc50-mCherry-P2F and Cdc50-mCherry-P2R. Overlap PCR was performed using primers Cdc50-mCherry-P1F and Cdc50-mCherry-P3R to yield a 5kb construct. The Sec61-GFP and Cdc50-mCherry constructs were then used to transform the WT strain by biolistic transformation. Transformants were screened for resistance to hygromycin and G418, and the proper location and orientation of the gene fusions at the SEC61 or CDC50 loci were determined by PCR. Primer sequences are listed in Supplemental Table S3.
Capsule formation and melanin production
Capsule formation was examined by differential interference contrast microscopy after incubation for 24 h to 48 h at 30°C in defined LIM and staining with India ink. Melanin production was examined on L-3,4-dihydroxyphenylalanine (L-DOPA) plates containing 0.1% glucose.
Stress and drug response assays
To examine the response of C. neoformans WT, cdc50 and CDC50 complemented strains to various stress conditions, exponentially growing cultures were washed, re-suspended in H2O and adjusted to a concentration of 2 × 104 cells /ml. The cell suspensions were diluted 10-fold serially, and 5 µl of each dilution was spotted onto YPD and/or YNB plates supplemented with different compounds. Plates were incubated for 2–10 days at 30°C or 37°C, and photographed. The responses of strains to oxidative, salt, osmotic stress and to agents that challenge cell wall integrity were examined. The specific assays were performed on YPD and/or YNB plates supplemented with or without 1.2 M KCl, 1.2 M NaCl, 100 mM LiCl, 0.1% SDS, 0.5 mg/ml Congo red. Sensitivity to trafficking inhibitors brefeldin A and monensin were examined by spotting the cell dilutions on YPD containing 30 µg/ml of BFA, 625 µg/ml of monensin. The anti-fungal drug fluconazole (5 or 10 µg/ml) was also tested.
Analysis of iron-related phenotypes
To assess iron-dependent growth on solid media, 10-fold serial dilutions of cells were spotted on agar plates with or without supplemented iron sources. Plates were incubated at 30°C for 2 days (or as indicated) before being photographed. Growth of the strains was also assessed in liquid media. Cells for growth assays in liquid media were pre-grown overnight at 30°C with shaking in YPD. The cells were then washed twice with low iron water, inoculated into YNB-LIM at 4 × 106 cells/mL and grown at 30°C for two days to starve the cells for iron. After starvation, the cells were harvested, washed and inoculated in YNB-LIM with or without supplemented iron sources to a final concentration of 5 × 104 cells/mL. Cultures were incubated at 30°C, and growth was monitored by measuring the optical density at 600 nm using a DU530 Life Science UV/Visible spectrophotometer (Beckman Instruments). To access the response of the WT, cdc50 and CDC50 complemented strains to curcumin, 10-fold serial dilutions of cells (without iron pre-starvation) were spotted on agar plates with 150, 300 or 300 µM of curcumin (Sigma-Aldrich, USA), with or without supplemented inorganic iron sources (100 µM or 1 mM of FeSO4 or FeCl3), or heme (10, 100 µM), or copper (1mM of CuSO4), or zinc (1mM of ZnSO4). Plates were incubated for 2–7 days at 30°C and photographed.
Macrophage survival assays
The effect of CDC50 deletion on fungal survival during incubation with macrophages was assessed as previously described (Caza et al., 2016). Briefly, the murine macrophage-like cell line J774A.1 was maintained at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum, 1% nonessential amino acids, 100 µg/ml penicillin-streptomycin, and 4 mM L-glutamine (Invitrogen). The cell line was used between passages 5 and 10. Cells of the WT, two cdc50 mutants, and the CDC50 complemented mutant were opsonized with monoclonal antibody 18B7 against capsule (1 µg/ml) (a generous gift from Arturo Casadevall), and macrophages were stimulated with 150 ng/ml phorbol myristate acetate (PMA) for 2 hours prior to co-incubation at a multiplicity of infection (MOI) of 1:1. Macrophages were inoculated at 1 × 105 cells and washed after 2 h of inoculation to remove unattached, extracellular fungal cells. After 24 h of incubation, sterile, ice-cold distilled H2O was applied to each well to lyse the macrophages (confirmed microscopically). Fungal growth was measured by plating cells on YPD and determining CFUs. The assay was performed in triplicate for each strain, and the experiment was repeated three times with consistent results. Student’s t test was used to determine the statistical significance of differences in fungal survival.
Assessment of virulence in a murine model
Female BALB/c mice, 4 to 6 weeks old, were obtained from Charles River Laboratories (Pointe-Claire, Québec, Canada) and used in an inhalation model of cryptococcosis. A cell suspension of 2 × 105 cells in a 50-µl volume was used for intranasal instillation and ten mice were inoculated per strain. The status of the mice was monitored once per day post-inoculation. Mice reaching the humane end-point were euthanized by CO2 anoxia. The protocol for the virulence assay (protocol A13-0093) was approved by the University of British Columbia Committee on Animal Care. For determination of the fungal load in organs, infected mice were euthanized by CO2 inhalation and organs were excised, weighed and homogenized in 1 ml of PBS using a MixerMill (Retsch, Cole-Parmer, Montreal, Canada). Serial dilutions of the homogenates were plated on YPD plates containing 50 µg/ml chloramphenicol and colony-forming units were counted after an incubation for 48 h at 30°C. Differences in virulence were statistically assessed by log rank tests for survival and by using the two-tailed non-parametric Mann Whitney U-test from the GraphPad Prism 7 program (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and the National Institutes of Health (RO1AI053721). We thank Julianne Djordjevic for providing the Aph1-DSRed construct, J. Andrew Alspaugh for the Rim101-GFP strain and Arturo Casadevall for antibody against capsule polysaccharide.
Footnotes
This study was presented at the 13th European Conference on Fungal Genetics, Paris, France (3–6 April, 2016)
References
- Alder-Baerens N, Lisman Q, Luong L, Pomorski T, Holthuis JC. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol Biol Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F, Wolf JM, Casadevall A. Virulence-Associated Enzymes of Cryptococcus neoformans . Eukaryot Cell. 2015;14:1173–1185. doi: 10.1128/EC.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad GK, Singh V, Thakare MJ, Baranwal S, Tomar RS. Mitogen-activated protein kinase Hog1 is activated in response to curcumin exposure in the budding yeast Saccharomyces cerevisiae . BMC Microbiology. 2014;14:317. doi: 10.1186/s12866-014-0317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouaoui H, Montigny C, Jacquot A, Barry R, Champeil P, Lenoir G. Coordinated Overexpression in Yeast of a P4-ATPase and Its Associated Cdc50 Subunit: The Case of the Drs2p/Cdc50p Lipid Flippase Complex. Methods Mol Biol. 2016;1377:37–55. doi: 10.1007/978-1-4939-3179-8_6. [DOI] [PubMed] [Google Scholar]
- Blanchin-Roland S, Da Costa G, Gaillardin C. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology. 2005;151:3627–3637. doi: 10.1099/mic.0.28196-0. [DOI] [PubMed] [Google Scholar]
- Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Campbell LT, Lodge JK. Cryptococcus neoformans, a fungus under stress. Curr Op Microbiol. 2007;10:320–325. doi: 10.1016/j.mib.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, et al. The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans . J Inf Dis. 2013;207:1339–1347. doi: 10.1093/infdis/jit029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmello JC, Pavarina AC, Oliveira R, Johansson B. Genotoxic effect of photodynamic therapy mediated by curcumin on Candida albicans . FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov018. fov018. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Nosanchuk JD, Williamson P, Rodrigues ML. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009;17:158–162. doi: 10.1016/j.tim.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Hu G, Price M, Perfect JR, Kronstad JW. The zinc finger protein Mig1 regulates mitochondrial function and azole drug susceptibility in the pathogenic fungus Cryptococcus neoformans . mSphere. 2016;1 doi: 10.1128/mSphere.00080-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans . Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans . Infect Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. Journal Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR. Roles for the Drs2p–Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Choi J, Jung WH, Kronstad JW. The cAMP/protein kinase A signaling pathway in pathogenic basidiomycete fungi: Connections with iron homeostasis. J Microbiol. 2015;53:579–587. doi: 10.1007/s12275-015-5247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp RJ, Pollington A, Galea C, Jaron S, Yamaguchi-Iwai Y, Kaplan J. Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J Biol Chem. 2003;278:45499–45506. doi: 10.1074/jbc.M307229200. [DOI] [PubMed] [Google Scholar]
- D’Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans . Mol Cell Biol. 2001a;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001b;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans . Fungal Gen Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- Djordjevic JT. Role of phospholipases in fungal fitness, pathogenicity, and drug development - lessons from Cryptococcus neoformans . Frontiers Microbiol. 2010;1:125. doi: 10.3389/fmicb.2010.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic JT, Del Poeta M, Sorrell TC, Turner KM, Wright LC. Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem J. 2005;389:803–812. doi: 10.1042/BJ20050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans . Annu Rev Microbiol. 2009;63:223–247. doi: 10.1146/annurev.micro.62.081307.162753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. Vesicle-associated melanization in Cryptococcus neoformans . Microbiol. 2009;155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. Drs2p–dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Gomes AS, Curvelo JA, Soares RM, Ferreira-Pereira A. Curcumin acts synergistically with fluconazole to sensitize a clinical isolate of Candida albicans showing a MDR phenotype. Med Mycol. 2012;50:26–32. doi: 10.3109/13693786.2011.578156. [DOI] [PubMed] [Google Scholar]
- Garcia-Rivera J, Chang YC, Kwon-Chung KJ, Casadevall A. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot Cell. 2004;3:385–392. doi: 10.1128/EC.3.2.385-392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Sanchez-Canete MP, Gamarro F, Castanys S. Functional role of evolutionarily highly conserved residues, N-glycosylation level and domains of the Leishmania miltefosine transporter-Cdc50 subunit. Biochem J. 2014;459:83–94. doi: 10.1042/BJ20131318. [DOI] [PubMed] [Google Scholar]
- Gerik KJ, Donlin MJ, Soto CE, Banks AM, Banks IR, Maligie MA, et al. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans . Mol Microbiol. 2005;58:393–408. doi: 10.1111/j.1365-2958.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- Graham TR. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Arguello JM, Helmann JD. PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol. 2015;98:787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Au WC, Shakoury-Elizeh M, Protchenko O, Basrai M, Prinz WA, Philpott CC. Phosphatidylserine is involved in the ferrichrome-induced plasma membrane trafficking of Arn1 in Saccharomyces cerevisiae . J. Biol Chem. 2010;285:39564–39573. doi: 10.1074/jbc.M110.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell. 2015;26:4674–4685. doi: 10.1091/mbc.E15-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PK, Malone L, Birchmore JL, Nichols JW. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem. 2003;278:36041–36050. doi: 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Fukuzawa T, Sorimachi H, Maeda T. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol Cell Biol. 2005;25:9478–9490. doi: 10.1128/MCB.25.21.9478-9490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Bakkeren E, Do E, Jung WH, Kronstad JW. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans . Mol Microbiol. 2015;96:973–992. doi: 10.1111/mmi.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun. 2013;81:292–302. doi: 10.1128/IAI.01037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008b;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kamp A, Linning R, Naik S, Bakkeren G. Complementation of Ustilago maydis MAPK mutants by a wheat leaf rust, Puccinia triticina homolog: potential for functional analyses of rust genes. Mol Plant-Microbe Interact. 2007b;20:637–647. doi: 10.1094/MPMI-20-6-0637. [DOI] [PubMed] [Google Scholar]
- Hu G, Kronstad JW. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans . Eukaryot Cell. 2010;9:74–83. doi: 10.1128/EC.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Liu I, Sham A, Stajich JE, Dietrich FS, Kronstad JW. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans . Genome Biol. 2008a;9:R41. doi: 10.1186/gb-2008-9-2-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Steen BR, Lian T, Sham AP, Tam N, Tangen KL, Kronstad JW. Transcriptional regulation by protein kinase A in Cryptococcus neoformans . PLoS Path. 2007a;3:e42. doi: 10.1371/journal.ppat.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Liao G, Baker GM, Wang Y, Lau R, Paderu P, et al. Lipid flippase subunit Cdc50 mediates drug resistance and virulence in Cryptococcus neoformans . mBio. 2016;7:e00478–e004516. doi: 10.1128/mBio.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans . Nat Rev Microbiol. 2005;3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Giles SS, Perfect JR, Heitman J. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans . Eukaryot Cell. 2007;6:60–72. doi: 10.1128/EC.00214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Kihara A, Denpoh A, Igarashi Y. The Rim101 pathway is involved in Rsb1 expression induced by altered lipid asymmetry. Mol Biol Cell. 2008;19:1922–1931. doi: 10.1091/mbc.E07-08-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W, Kronstad JW. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans . Eukaryot Cell. 2009;8:1511–1520. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans . PLoS Path. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron source preference and regulation of iron uptake in Cryptococcus neoformans . PLoS Path. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans . PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, et al. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae . J Biol Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- Kim J, Cho YJ, Do E, Choi J, Hu G, Cadieux B, et al. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet Biol. 2012;49:955–966. doi: 10.1016/j.fgb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, et al. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell. 2012;11:109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Attarian R, Cadieux B, Choi J, D’Souza CA, Griffiths EJ, et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, Jung WH. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans . Trends Microbiol. 2013;21:457–465. doi: 10.1016/j.tim.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dhamgaye S, Maurya IK, Singh A, Sharma M, Prasad R. Curcumin targets cell wall integrity via calcineurin-mediated signaling in Candida albicans. Antimicrob Agents Chemother. 2014;58:167–175. doi: 10.1128/AAC.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Lee DG. An antifungal mechanism of curcumin lies in membrane-targeted action within Candida albicans . IUBMB life. 2014;66:780–785. doi: 10.1002/iub.1326. [DOI] [PubMed] [Google Scholar]
- Lenoir G, Williamson P, Puts CF, Holthuis JC. Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem. 2009;284:17956–17967. doi: 10.1074/jbc.M109.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Crossett B, Cha SY, Desmarini D, Li C, Chayakulkeeree M, et al. Identification of Aph1, a phosphate-regulated, secreted, and vacuolar acid phosphatase in Cryptococcus neoformans . mBio. 2014;5:e01649–01614. doi: 10.1128/mBio.01649-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans . Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Marques RL, Holthuis JC, Pomorski TG. Pumping lipids with P4-ATPases. Biol Chem. 2011;392:67–76. doi: 10.1515/BC.2011.015. [DOI] [PubMed] [Google Scholar]
- Ma Z, Liu Z, Huang X. Membrane phospholipid asymmetry counters the adverse effects of sterol overloading in the Golgi membrane of Drosophila. Genetics. 2012;190:1299–1308. doi: 10.1534/genetics.111.137687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minear S, O’Donnell AF, Ballew A, Giaever G, Nislow C, Stearns T, Cyert MS. Curcumin inhibits growth of Saccharomyces cerevisiae through iron chelation. Eukaryot Cell. 2011;10:1574–1581. doi: 10.1128/EC.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu K, Fujimura-Kamada K, Ueda T, Nakano A, Katoh H, Tanaka K. Cdc50p, a conserved endosomal membrane protein, controls polarized growth in Saccharomyces cerevisiae . Mol Biol Cell. 2003;14:730–747. doi: 10.1091/mbc.E02-06-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muren E, Oyen M, Barmark G, Ronne H. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast. 2001;18:163–172. doi: 10.1002/1097-0061(20010130)18:2<163::AID-YEA659>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Muthusamy BP, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009a;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy BP, Raychaudhuri S, Natarajan P, Abe F, Liu K, Prinz WA, Graham TR. Control of protein and sterol trafficking by antagonistic activities of a type IV P-type ATPase and oxysterol binding protein homologue. Mol Biol Cell. 2009b;20:2920–2931. doi: 10.1091/mbc.E08-10-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelofar K, Shreaz S, Rimple B, Muralidhar S, Nikhat M, Khan LA. Curcumin as a promising anticandidal of clinical interest. Can J Microbiol. 2011;57:204–210. doi: 10.1139/W10-117. [DOI] [PubMed] [Google Scholar]
- O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Path. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fukuda R, Ohta A. Involvement of LEM3/ROS3 in the uptake of phosphatidylcholine with short acyl chains in Saccharomyces cerevisiae . Biosci Biotech Biochem. 2009;73:750–752. doi: 10.1271/bbb.80675. [DOI] [PubMed] [Google Scholar]
- Ost KS, O’Meara TR, Huda N, Esher SK, Alspaugh JA. The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator. PLoS Genet. 2015;11:e1005159. doi: 10.1371/journal.pgen.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Komperda K, Frases S, Park YD, Djordjevic JT, Casadevall A, Williamson PR. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans . Mol Microbiol. 2009;71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Ravu RR, Chen YL, Jacob MR, Pan X, Agarwal AK, Khan SI, et al. Synthesis and antifungal activities of miltefosine analogs. Bioorg Med Chem Lett. 2013;23:4828–4831. doi: 10.1016/j.bmcl.2013.06.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J, Oliveira DL, Joffe LS, Hu G, Gazos-Lopes F, Fonseca FL, et al. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans . Eukaryotic cell. 2014;13:715–726. doi: 10.1128/EC.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LS, Causton HC, Young RA, Fink GR. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc Natl Acad Sci USA. 2000;97:5984–5988. doi: 10.1073/pnas.100113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Djordjevic JT. Unravelling secretion in Cryptococcus neoformans: more than one way to skin a cat. Mycopath. 2012;173:407–418. doi: 10.1007/s11046-011-9468-9. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Oliveira D, Hu G, Kronstad J. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans . Inf Immun. 2014;82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae . Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim Biophys Acta. 2012;1821:1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Manoharlal R, Negi AS, Prasad R. Synergistic anticandidal activity of pure polyphenol curcumin I in combination with azoles and polyenes generates reactive oxygen species leading to apoptosis. FEMS Yeast Res. 2010;10:570–578. doi: 10.1111/j.1567-1364.2010.00637.x. [DOI] [PubMed] [Google Scholar]
- Tucker SC, Casadevall A. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc Natl Acad Sci USA. 2002;99:3165–3170. doi: 10.1073/pnas.052702799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell. 2006;17:3768–3780. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer F, Wright LC, Obando D, Handke R, Ganendren R, Ellis DH, Sorrell TC. Hexadecylphosphocholine (miltefosine) has broad-spectrum fungicidal activity and is efficacious in a mouse model of cryptococcosis. Antimicrob Agents Chemother. 2006;50:414–421. doi: 10.1128/AAC.50.2.414-421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JM, Johnson DJ, Chmielewski D, Davis DA. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot cell. 2010;9:1203–1215. doi: 10.1128/EC.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans . Mol Biol Cell. 2004;15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. An unusual organelle in Cryptococcus neoformans links luminal pH and capsule biosynthesis. Fungal Genet Biol. 2009;46:682–687. doi: 10.1016/j.fgb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. The capsule of the fungal pathogen Cryptococcus neoformans . Adv App Microbiol. 2009;68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gurr SJ. Walking into the unknown: a ‘step down’ PCR-based technique leading to the direct sequence analysis of flanking genomic DNA. Gene. 2000;253:145–150. doi: 10.1016/s0378-1119(00)00289-4. [DOI] [PubMed] [Google Scholar]
- Zhu X, Williamson PR. Role of laccase in the biology and virulence of Cryptococcus neoformans . FEMS Yeast Res. 2004;5:1–10. doi: 10.1016/j.femsyr.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.