Abstract
A harmine-derived beta-carboline, CM16, inhibits cancer cells growth through its effects on protein synthesis, as described in “A harmine-derived beta-carboline displays anti-cancer effects in vitro by targeting protein synthesis” (Carvalho et al., 2017)[1]. This data article provides accompanying data on CM16 cytostatic evaluation in cancer cells as well as data related to its effects on transcription and translation. After confirming the cytostatic effect of CM16, we investigated its ability to arrest the cell cycle in the glioma Hs683 and SKMEL-28 melanoma cell lines but no modification was evidenced. According to the global protein synthesis inhibition induced by CM16 [1], transcription phase, a step prior to mRNA translation, evaluated by labelled nucleotide incorporation assay was not shown to be affected under CM16 treatment in the two cell lines. By contrast, mRNA translation and particularly the initiation step were shown to be targeted by CM16 in [1]. To further decipher those effects, we established herein a list of main actors in the protein synthesis process according to literature survey for comparative analysis of cell lines displaying different sensitivity levels to CM16. Finally, one of these proteins, PERK, a kinase regulating eIF2-α phosphorylation and thereby activity, was evaluated under treatment with CM16 in a cell-free system.
Keywords: beta-carboline, protein synthesis, cancer cells
1. Specifications table
| Subject area | Biology |
| More specific subject area | Protein synthesis inhibition of cancer cells in vitro |
| Mechanism of action of potential anticancer drug | |
| Type of data | Graphs and table |
| How data was acquired | Flow cytometer, microplate reader, search on databases |
| Data format | Analyzed graphs and raw data retrieval (table) |
| Experimental factors | As in the description of the data and materials and methods |
| Experimental features | As in the description of the data and materials and methods |
| Data source location | Lab. de Cancérologie et Toxicologie Experimentale, Université Libre de Bruxelles, Brussels, Belgium. |
| Life Technologies, Madison, USA | |
| Data accessibility | Data is with this article |
2. Value of the data
-
•
This data offers an extended comprehension of CM16 mechanism of action as a protein synthesis inhibitor in cancer cells.
-
•
Assays performed to evaluate transcription and translation initiation provide valuable data and may be used as tools in other cell-based investigations of potential protein synthesis inhibitors.
-
•
The data presented shows that different methods add to and enrich the investigation of the mechanism of action of proteins synthesis inhibitors in cancer cells. Therefore, these approaches might be useful in similar studies.
3. Data
Firstly, data on CM16-induced cytostatic effects is presented. As shown in [1] CM16 displays cytostatic effects at its IC50 in glioma Hs683, melanoma SKMEL-28 and breast adenocarcinoma MDA-MB-231 cells. Thus, CM16 effect on the cell cycle of both glioma Hs683 (Fig. 1A) cells and SKMEL-28 (Fig. 1B) are presented. After data showing CM16 inhibiting translation [1], further investigation on the effects of CM16 on newly synthesized mRNA (transcription) were carried out and generated the data here shown (Fig. 2A-B). CM16 effects on PERK activity, is shown in Fig. 3. The data on Table 1 refers to the genes related to translation that were analyzed for their transcriptomic expression in the cell lines most and least sensitive to CM16 effects, according to the NCI 60-cell-line growth inhibitory evaluation [1].
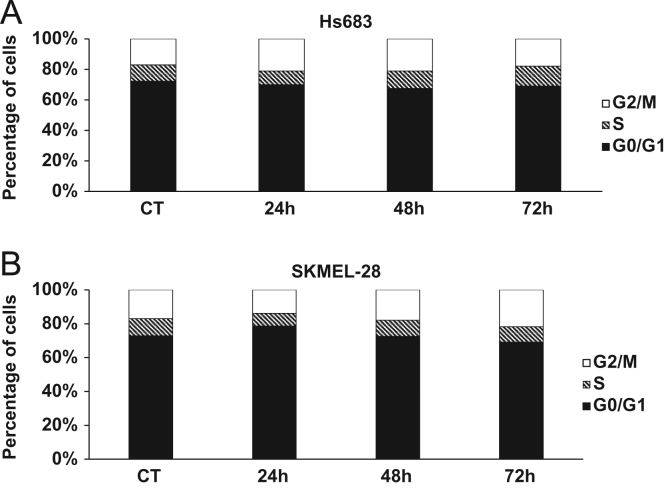
Fig. 1.
Cell cycle effects of CM16 on A: Hs683 at 0.1 µM; and B: SKMEL-28 at 0.5 µM. Data are expressed as the mean percentage of cells in each phase of the cell cycle of four replicates. As proliferation inhibition were observed on the three cancer models under study at their GI50 and the lack of evidence of any effects of CM16 on the cell cycle of Hs683 and SKMEL-28, we did not perform the cell cycle analysis on MDAMB-231.
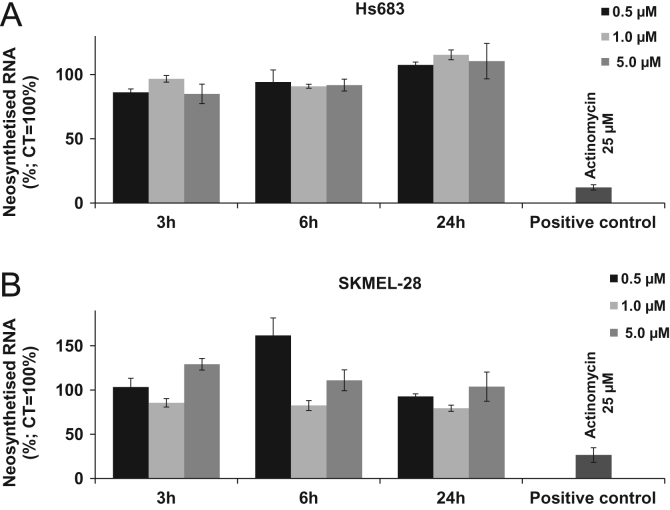
Fig. 2.
Effects of CM16 on newly synthesized mRNA in A: Hs683 and B: SKMEL-28 cell lines. Results are expressed as the mean neosynthesized RNA amounts normalized to the control (100%) ± S.E.M. of six replicates. No significant effects were observed for up to 24 h in the presence of 5.0 µM CM16 in those two cell lines, thus we did not further assayed the breast cancer cell line MDA-MB-231.
Fig. 3.
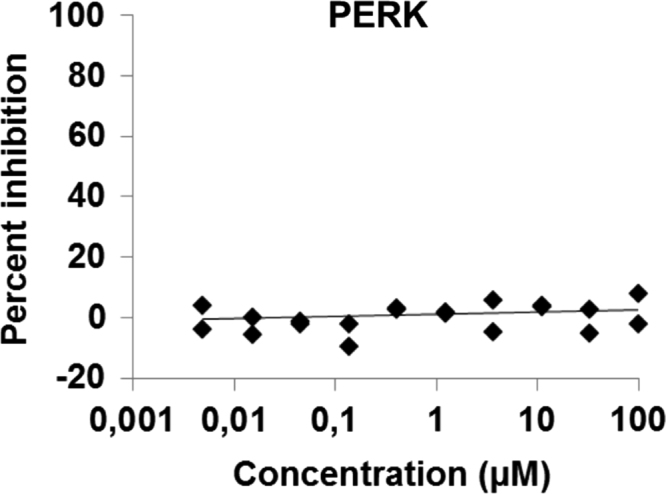
PERK kinase activity in vitro in the presence or absence of CM16.
Table 1.
List of genes analyzed for the transcript intensity from the NCI cell line panel.
| Protein | Protein code (UniProt) | Gene (HGNC Symbol) | Gene code (Entrez Gene) |
|---|---|---|---|
| Eukaryotic translation initiation factor 2 subunit 1 | P05198 | EIF2S1 | 1965 |
| Eukaryotic translation initiation factor 2 subunit 2 | P20042 | EIF2S2 | 8894 |
| Eukaryotic translation initiation factor 2 subunit 3 | P41091 | EIF2S3 | 1968 |
| Translation initiation factor eIF-2B subunit alpha | Q14232 | EIF2B1 | 1967 |
| Translation initiation factor eIF-2B subunit beta | P49770 | EIF2B2 | 8892 |
| Translation initiation factor eIF-2B subunit gamma | Q9NR50 | EIF2B3 | 8891 |
| Translation initiation factor eIF-2B subunit delta | Q9UI10 | EIF2B4 | 8890 |
| Translation initiation factor eIF-2B subunit epsilon | Q13144 | EIF2B5 | 8893 |
| Eukaryotic translation initiation factor 4E | P06730 | EIF4E | 1977 |
| Eukaryotic translation initiation factor 4E-binding protein 1 | Q13541 | EIF4EBP1 | 1978 |
| Eukaryotic translation initiation factor 4 gamma 1 | Q04637 | EIF4G1 | 1981 |
| Eukaryotic translation initiation factor 4 gamma 2 | P78344 | EIF4G2 | 1982 |
| Eukaryotic translation initiation factor 4 gamma 3 | O43432 | EIF4G3 | 8672 |
| MAP kinase-interacting serine/threonine-protein kinase 1 | Q9BUB5 | MKNK1 | 8569 |
| MAP kinase-interacting serine/threonine-protein kinase 2 | Q9HBH9 | MKNK2 | 2872 |
| Eukaryotic initiation factor 4A-I | P60842 | EIF4A1 | 1973 |
| Eukaryotic initiation factor 4A-II | Q14240 | EIF4A2 | 1974 |
| Eukaryotic initiation factor 4A-III | P38919 | EIF4A3 | 9775 |
| Programmed cell death protein 4 | Q53EL6 | PDCD4 | 27250 |
| Eukaryotic translation initiation factor 5A-1 | P63241 | EIF5A | 1984 |
| Eukaryotic translation initiation factor 5A-2 | Q9GZV4 | EIF5A2 | 56648 |
| Eukaryotic translation initiation factor 5B | O60841 | EIF5B | 9669 |
| Eukaryotic translation initiation factor 6 | P56537 | EIF6 | 3692 |
| Eukaryotic translation initiation factor 1 | P41567 | EIF1 | 10209 |
| Eukaryotic translation initiation factor 1A, X-chromosomal | P47813 | EIF1AX | 1964 |
| Eukaryotic translation initiation factor 1A, Y-chromosomal | O14602 | EIF1AY | 9086 |
| Probable RNA-binding protein EIF1AD | Q8N9N8 | EIF1AD | 84285 |
| Eukaryotic translation initiation factor 3 subunit A | Q14152 | EIF3A | 8661 |
| Eukaryotic translation initiation factor 3 subunit B | P55884 | EIF3B | 8662 |
| Eukaryotic translation initiation factor 3 subunit H | O15372 | EIF3H | 8667 |
| Eukaryotic translation initiation factor 3 subunit I | Q13347 | EIF3I | 8668 |
| Eukaryotic translation initiation factor 3 subunit M | Q7L2H7 | EIF3M | 10480 |
| Eukaryotic translation initiation factor 3 subunit E | P60228 | EIF3E | 3646 |
| Eukaryotic translation initiation factor 3 subunit F | O00303 | EIF3F | 8665 |
| Eukaryotic translation initiation factor 2-alpha kinase 3 | Q9NZJ5 | EIF2AK3 | 9451 |
| Eukaryotic translation initiation factor 2-alpha kinase 4 | Q9P2K8 | EIF2AK4 | 440275 |
| Interferon-induced, double-stranded RNA-activated protein kinase | P19525 | EIF2AK2 | 5610 |
| Eukaryotic translation initiation factor 2-alpha kinase 1 | Q9BQI3 | EIF2AK1 | 27102 |
| Elongation factor 1-alpha 1 | P68104 | EEF1A1 | 1915 |
| Elongation factor 2 | P13639 | EEF2 | 1938 |
| Serine/threonine-protein kinase mTOR | P42345 | MTOR | 2475 |
| RAC-alpha serine/threonine-protein kinase | P31749 | AKT1 | 207 |
| RAC-beta serine/threonine-protein kinase | P31751 | AKT2 | 208 |
| RAC-gamma serine/threonine-protein kinase | Q9Y243 | AKT3 | 10000 |
| Ribosomal protein S6 kinase beta-1 | P23443 | RPS6KB1 | 6198 |
| Ribosomal protein S6 kinase beta-2 | Q9UBS0 | RPS6KB2 | 6199 |
| Myc proto-oncogene protein | P01106 | MYC | 4609 |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | P42336 | PIK3CA | 5290 |
| Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | P60484 | PTEN | 5728 |
| Hamartin | Q92574 | TSC1 | 7248 |
| Tuberin | P49815 | TSC2 | 7249 |
| Cellular tumor antigen p53 | P04637 | TP53 | 7157 |
| Retinoblastoma-associated protein | P06400 | RB1 | 5925 |
| 3-phosphoinositide-dependent protein kinase 1 | O15530 | PDPK1 | 5170 |
| Mitogen-activated protein kinase 1 | P28482 | MAPK1 | 5594 |
| Vascular endothelial growth factor A | P15692 | VEGFA | 7742 |
| 78 kDa glucose-regulated protein | P11021 | HSPA5 | 3309 |
Data retrieved from: www.proteinatlas.com; www.uniprot.org; www.genenames.org and http://www.ncbi.nlm.nih.gov/gene in September 2015.
4. Experimental design, materials and methods
4.1. Cell lines and compound
The human cancer cell lines, oligodendroglioma Hs683 (ATCC code HTB-138) and melanoma SKMEL-28 (ATCC code HTB-72) were herein used. Cells were cultivated at 37 °C with 5% CO2 in RPMI culture medium supplemented with 10% FBS, 200U penicillin–streptomycin, 0.1 mg/ml gentamicin and 4 mM L-glutamine. CM16 was synthetized as previously described [2] and the experiments were designed with the cell lines described above treated with different concentrations of CM16, based on its IC50.
4.2. Analysis of CM16 effects on cell cycle
Cell cycle analysis was performed with flow cytometry through the measurements of DNA content with propidium iodide. Hs683 and SKMEL-28 were seeded in cell culture flasks and left untreated or treated with CM16 at its respective IC50 in each cell line for 24 h, 48 h and 72 h. The samples were then centrifuged (10 min, 1500 rcf, 4 °C), resuspended in PBS and pellets were resuspended in cold ethanol 70% for fixation. Staining with 0.08 mg/ml propidium iodide solution (0.08 mg/ml PI; 0.2 mg/ml RNAse in PBS) followed after a PBS wash. The samples were incubated at 37 °C for 30 min and stored at 4 °C for a few hours. Analysis was performed with the Cell Lab Quanta (Beckman Coulter, Analis, Suarlée, Belgium). The experiment was performed once in quadruplicate.
4.3. Analysis of CM16 effects on transcription
Neosynthesized RNA was evaluated through incorporation of a nucleoside analog, 5-ethynyl-uridine, using the Click iT-RNA HCS (Invitrogen, Life Technologies, Merelbeke, Belgium). The alkyne-containing nucleosides react with a fluorescent dye containing the azide moiety after their incorporation into cellular RNA. Briefly, Hs683 or SKMEL-28 cells were seeded and after attachment they were either left untreated (negative control) or treated with CM16 or the positive control actinomycin (Life Technologies, Paisley, UK). After the treatment with the analog 5-ethynyl uridine (4 mM) for two h, the cells were fixed, stained (Alexa Fluor 488 and 594) and fluorescence readings (ex/em: 495/520 nm) were carried out in microplate reader (SynergyMX Biotek, Winooski, USA: ex/em: 350/460 nm). Normalization according to cell number was carried out as described in the user manual with Hoescht counterstaining. The experiment was performed once in sextuplicate.
4.4. PERK inhibition
PERK activity was evaluated by the Life Technologies screening service (Lantha Screen, Madison, USA). The in vitro assay used is based on FRET between a terbium-labeled antibody and the phosphorylated product of the active kinase: TR-FRET increases proportionally to their binding and thereby to the quantity of the phosphorylated product. CM16 compound at different concentrations or the control solutions were mixed with the kinase/substrate/ATP mixture into the wells. After 60 min of reaction at room temperature, the detection mix was added and left to equilibrate for an h prior to fluorescence reading.
Acknowledgements
The Ph.D of A.C. is financially supported by the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (Grant 0674-13/3;CAPES; Brazil). C.M. acknowledges the grant from the Télévie (Grant 7.4529.13; FRS-FNRS; Belgium). R.K. is a director of research with the Fonds National de la Recherche Scientifique (FRS-FNRS; Belgium) that supported the present project (#3.4525.11: Anti-cancéreux dérivés de l’harmine from 2010 to 2014). Part of this study is also supported by the grant by the Belgian Brain Tumor Support (BBTS; Belgium). We thank Mohsin Mssassi for his help during his stay in the Laboratoire of Cancérologie et Toxicologie Expérimentale (ULB, Belgium). JP acknowledges grants from the Canadian Institutes of Health Research (MOP-106530 and MOP-115126). A.K. acknowledges the National Cancer Institute (CA186046-01A1) and Welch Foundation (AI-0045).
Footnotes
Transparency data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.dib.2017.05.006.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Carvalho A., Chu J., Meinguet C., Kiss R., Vandenbussche G., Masereel B. A harmine-derived beta-carboline displays anti-cancer effects in vitro by targeting protein synthesis. Eur. J. Pharmacol. 2017 doi: 10.1016/j.ejphar.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meinguet C., Bruyère C., Frédérick R., Mathieu V., Vancraeynest C., Pochet L. 3D-QSAR, design, synthesis and characterization of trisubstituted harmine derivatives with in vitro antiproliferative properties. Eur. J. Med. Chem. 2015;94:45–55. doi: 10.1016/j.ejmech.2015.02.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material