Abstract
The transformation of a normal cell to cancer requires the derail of multiple pathways. Normal signaling in a cell is regulated at multiple stages by the presence of feedback loops, calibration of levels of proteins by their regulated turnover, and posttranscriptional regulation, to name a few. The tumor suppressor protein FBXO31 is a component of the SCF E3 ubiquitin ligase and is required to arrest cells at G1 following genotoxic stresses. Due to its growth-suppression activity, it is underexpressed in many cancers. However, the molecular mechanism underlying the translational regulation of FBXO31 remains unclear. Here we show that the oncogenic microRNAs miR-93 and miR-106a repress FBXO31, resulting in the upregulation of Slug, which is involved in epithelial-mesenchymal transition and cell invasion. FBXO31 targets and ubiquitylates Slug for proteasomal degradation. However, this mechanism is repressed in breast tumors where miR-93 and miR-106a are overexpressed. Our study further unravels an interesting mechanism whereby Slug drives the expression of miR-93 and miR-106a, thus establishing a positive feedback loop to maintain an invasive phenotype. Together, these results establish the presence of interplay between microRNAs and the ubiquitination machinery, which together regulate cancer cell invasion.
Introduction
Breast cancer is one of the leading causes of death worldwide and is the second most common cancer in women. Over 1.3 million cases of invasive breast cancer are diagnosed worldwide, and more than 450,000 women die of breast cancer each year. Although significant progress has been made in understanding the pathology of breast cancer, for further improvement of therapy, it is important to identify new therapeutic targets. Understanding the molecular players involved in the regulation of breast cancer progression and metastasis is the key to developing improved treatment strategies. The ubiquitin-proteasome machinery is involved in many diseases including cancer [1]. Inhibitors and activators of E3 ubiquitin ligases are promising targets for therapy, as they dictate the proteins to be ubiquitylated and the manner of their ubiquitylation. Therefore, an in-depth understanding of this class of genes is important.
FBXO31 is a member of the F-box protein family that plays an important role in cell cycle progression, DNA damage response, tumorigenesis, and neuronal development [2], [3], [4], [5], [6]. It is located on chromosome 16q24.3 and is inactivated in many cancers due to loss of heterozygosity [2]. It is a part of the SCF E3 ubiquitin ligase complex through association with SKP1, Cullin1, and RBX1. It targets multiple cellular substrates such as cyclin D1, MDM2, p38, and Ctd1 and promotes their polyubiquitylation-mediated proteasomal degradation [5], [6], [7], [8]. Previous studies show that it is a senescence-inducing gene and plays a predominant role in preventing BRAF-induced transformation [2], [3], [4]. More recently, it has been shown that it functions as a dedicated DNA damage checkpoint protein by arresting cells at G1 phase of the cell cycle through proteasomal degradation of cyclin D1 and by preventing MDM2-mediated proteasomal degradation of p53 [5], [6].
Cellular proteins are maintained at the basal level through transcriptional, posttranscriptional, or posttranslational regulation. Posttranscriptional regulation has emerged as one of the major players in malignancy. Small noncoding RNAs, microRNAs (miRNAs), have emerged as crucial gene regulators at the posttranscriptional level, and their expression levels are frequently altered in cancer and other diseases [9], [10]. MiRNAs are transcribed as a primary miRNA by RNA polymerase II or III [11]. This primary miRNA is processed by Drosha and Pasha to generate a precursor miRNA, which is further processed by Dicer to form the mature miRNA in the cytoplasm [12]. The mature miRNA then associates with the RISC complex to function as a posttranscriptional or translational regulator. It is estimated that more than 60% of human proteins coding genes are regulated by miRNAs [13].
MiRNAs are involved in the regulation of various biological processes such as cell proliferation, differentiation, tumorigenesis, angiogenesis, and metastasis [14], [15], [16], [17], [18], [19], [20], [21], [22]. Previous studies reported that miRNA clusters such as miR-106a-363 and miR-106b-25 play an important role in breast cancer malignancy [23], [24]. In addition, miR-106b-25 cluster is reported to function as an oncogene in hepatocellular carcinoma, and miR-93, a member of miR-106b-25 cluster, acts as an oncogene in human glioblastoma [20], [25], [26].
Given the predominant role of FBXO31 in growth arrest and DNA damage checkpoint activation, it becomes important to understand its comprehensive regulation. In this study, we identified miR-93 and miR-106a as negative regulators of FBXO31 during normal cellular growth, but they fail to do so under genotoxic stresses. Further, we showed that these miRNAs promote scratch wound healing and cellular invasion through stabilization of Slug, which in turn is proteasomally degraded by FBXO31. Most interestingly, we unraveled that Slug directly drives the expression of miR-93 and miR-106a by binding to their promoter. Our study thus expands the understanding of molecular mechanisms involved in onset and progression of cancer by highlighting the presence of a feedback loop involving miR-93, miR-106a, FBXO31, and Slug for the first time.
Material and Methods
Cell Lines, shRNAs, and Plasmids
The human breast cancer cell lines MCF7, MDA-MB-231, MDA-MB-435, T47D, and NCI/ADR-Res were provided by Prof. Michael R. Green (University of Massachusetts Medical School, USA). MCF7 cells were grown in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) medium, and rest of the cell lines were grown in RPMI media containing 10% FBS at 37°C in an atmosphere of 5% CO2 under humid conditions. pCMV-myc-FBXO31 and F-box domain deletion mutation vector pCMV-myc-FBXO31∆F were kindly provided by Prof. David F. Callen (University of Adelaide, Australia). psG5-Flag-hSlug was kindly provided by Prof. Sophie Tartare-Deckert (Bâtiment Universitaire Archimed, France). His-Ub was bought from Addgene [27]. pCMV-MIR vector and pMIR-REPORT vectors were kindly provided by Dr. Samit Chattopadhyay (National Centre for Cell Science, India).
FBXO31 knockdown cell lines were generated by stable transduction of MCF7 cells with the following lentiviral shRNAs as described previously [5]. Lentiviral shRNA clones were kindly provided by Prof. Michael R. Green, University of Massachusetts Medical School, USA.
Stable Knockdown Cells Generation
HEK-293T cells were seeded 1 day prior to transfection. Lentiviral shRNA plasmid along with lentiviral packaging plasmids (pPAX2 and pMD2.G) was co-transfected using polyethyleneimine. Transfection mixture was incubated at room temperature for 30 minutes and then added dropwise to the cells. Virus soup was collected after 48 hours of transfection and filtered through 0.45-μm syringe filter. MCF7 cells were transduced with filtered virus soup in the presence of 8 μg/ml of polybrene. Infected cells were selected by treating with 1 μg/ml of puromycin.
Cloning of miRNA and 3′UTR Reporter System
Human miR-20b, miR-93, miR-106a, and miR-106b were amplified using genomic DNA as template, and the amplified polymerase chain reaction (PCR) product was inserted into XhoI site of the pCMV-MIR vector by using Infusion cloning kit according to manufacturer's protocol (Takara). The point mutants of miRs were generated by Quik Change site-directed mutagenesis kit. The 3′UTR sequence (1065 bp) of human FBXO31–bearing miRNAs binding sites was amplified and inserted into the SpeI and HindIII sites of the pMIR-REPORT luciferase vector (named as Luc-WT). Seed sequence of miR-93 and miR-106a in the 3′UTR of FBXO31 was mutated by Quik Change site-directed mutagenesis kit. The mutants were named as Luc-Mut1 and Luc-Mut2.
Primer sequences were as follows:
Luc-WT (forward): 5′-ACCACTAGTCCACATCCTTGCCGCCACAT-3′.
Luc-WT (reverse): 5′-CACAAGCTTGCAGTGGAGGCATCAGTCCA-3′.
Luc-Mut1 (forward): 5′-GAATAGAAGCAGCATGCGAGTTGGAAATCCGGCC-3′.
Luc-Mut1 (reverse): 5′-GGCCGGATTTCCAACTCGCATGCTGCTTCTATTC-3′.
Luc-Mut2 (forward): 5′-CAGTCCAGCCACCCCCCAGCGAGTTATGTAGAGAG-3′.
Luc-Mut2 (reverse): 5′-CTCTCTACATAACTCGCTGGGGGGTGGCTGGACTG-3′.
Mut miRNA-93 (forward): 5′-GGGGGCTCCAAATTTCTGTTCGTGCAGG-3′.
Mut miRNA-93 (reverse): 5′-GCACGAACAGAAATTTGGAGCCCCAGGAC-3′.
Mut miRNA-106a (forward): 5′-GGGGGCTCCAAATTTCTGTTCGTGCAGG-3′.
Mut miRNA-106a (reverse): 5′-GCACGAACAGAAATTTGGAGCCCCAGGAC-3′.
Cell Transfection
Cells were seeded 1 day prior to transfection, and polyethylenimine (PEI 25000 from Polysciences, USA) was used as transfection reagent. Transfection mixture was prepared the following day in 150 mM NaCl solution by mixing DNA and polyethylenimine [DNA (μg):polyethylenimine (μg)] in a ratio of 1:2.4. Transfection mixtures were incubated at room temperature for 15 minutes and then added to the media in dropwise manner.
Dual Luciferase Assay
MCF7 cells were seeded in a 24-well plate, and cells were transfected with indicated plasmids using polyethylenimine as described above. The cells were harvested and lysed after 48 hours of transfection. Luciferase activity was measured according to the manufacturer's protocol (Dual-Luciferase Reporter Assay System; Promega, Madison, WI), and Renilla luciferase was used for normalization.
Quantitative Real-Time Reverse Transcriptase PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was used to extract total RNA according to the manufacturer's protocol. The first-strand cDNA was synthesized with random primers or miRNA stem loop specific primers and MultiscribeTM reverse transcriptase (Applied Biosystems). Quantitative real-time PCR was performed in the Eppendorf Master Cycler RealPlex using SYBR Green Kit from Takara. Fold change was calculated as described previously [28].
Primer sequences were as follows:
FBXO31 (forward): 5′-GGATGTACCTGCCACCTCAT-3′.
FBXO31 (reverse): 5′-CCACTGTAGCCGACTTCCTC-3′.
Slug (forward): 5′-ACAGAGCATTTGCAGACAGG-3′.
Slug (reverse): 5′-TGCTACACAGCAGCCAGAT-3′.
E-Cadherin (forward): 5′-AAGGTGACAGAGCCTCTGGAT-3′.
E-Cadherin (reverse): 5′-GATCGGTTACCGTGATCAAAATC-3′.
GAPDH (forward): 5′-TGCCGGTGACTAACCCTGCG-3′.
GAPDH (reverse): 5′-AGGCGCCCAATACGACCAAATCAGA-3′.
miR-93: stem loop: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3′.
miR-93 (forward): 5′-TTAACAAAGTGCTGTTCGTGC-3′.
miR (reverse): 5′-GTGCAGGGTCCGAGGT-3′.
miR-20b (forward): 5′-TTAACAAAGTGCTCATAGTGC-3′.
miR-20b: stem loop: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3′.
miR-106a-(forward): 5′-TACTAAAAGTGCTTACAGTGC-3′.
miR-106a: stem loop: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3′.
miR-106b (forward): 5′-ATAAATAAAGTGCTGACAGTGC-3′.
miR-106b: stem loop: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCTGC-3′.
miR-93 (forward): 5′-TACGCGGCCGCTCGAGGCCTTTTCCCCACTTCTTAA-3′.
miR-93 (reverse): 5′-TGAGTTTCTGCTCGATCCCGGAGTTCAGCTGTCC-3′.
miR-106b (forward): 5′-TACGCGGCCGCTCGAAGTGCCCAAATTGCTGGAGG-3′.
miR-106b (reverse): 5′-TGAGTTTCTGCTCGAACTGAGGTCCAAGACGGGAG-3′.
miR-106a (forward): 5′-TACGCGGCCGCTCGACCTGTTTCAGGAATATTAACTA-3′.
miR-106a (reverse): 5′-TGAGTTTCTGCTCGACACATTATAAGAAGCAGCTCA-3′.
miR-20b (forward): 5′-TACGCGGCCGCTCGACTGGCACAGGCTGCCTAATA-3′.
miR-20b (reverse): 5′-TGAGTTTCTGCTCGAAGCAATGTCTTTGAATATTCTCT-3′.
18S rRNA (forward): 5′-GTAACCCGTTGAACCCCATT-3′.
18S rRNA (reverse): 5′-CCATCCAATCGGTAGTAGCG-3′.
Immunoblotting Analysis
Cells were harvested, washed with ice-cold PBS, and then lysed with lysis buffer (50 mM Tris pH 7.4, 5 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.5 mM Na orthovanadate, and 0.5% Triton X-100) to prepare protein extracts. Protein concentrations were measured by the Bradford method using bovine serum albumin as standard [29]. The proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a PVDF membrane and were probed with specific primary antibody kept overnight at 4°C with gentle rocking. Antibodies against FBXO31, Flag, β-actin, and α-tubulin were procured from Sigma. TP53 antibody was purchased from Santa Cruz, Slug from Abcam, E-cadherin from BD Biosciences, and c-myc from Roche. Snail, Twist, and K48-ubiquitin antibodies were purchased from Cell Signaling Technology. β-Actin and α-tubulin were used as loading control.
Cycloheximide Pulse Chase Assay
MCF7 cells were treated with cycloheximide (40 μg/ml) for indicated time points. Cells were then harvested, washed with ice-cold PBS, and lysed with lysis buffer as described above. Immunoblotting was performed with mentioned antibodies. Band intensity was measured using ImageJ software and was normalized with loading control. Protein at 0-hour time point was taken as 100%, and the percentage of remaining protein was calculated with respect to 0 hour.
Co-Immunoprecipitation Assay
MCF7 cells were co-transfected with Flag-Slug and myc-FBXO31 for 36 hours and then treated with MG132 (5 μM; Calbiochem) for an additional 8 hours. Six hundred micrograms of protein extract was incubated with indicated antibodies for 10 to 14 hours at 4°C with gentle rocking. Then, protein G agarose beads were added to protein-antibody mixture and incubated for an additional 1.5 hours. Unbound proteins and antibody were separated by washing the beads three times, and immunoprecipitates were eluted from beads by boiling with sample buffer for 5 minutes. Eluted immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with indicated antibodies.
Ubiquitination Assay
MCF7 cells were co-transfected with mentioned plasmids. After 36 hours of transfection, cells were treated with 5 μM MG132 for an additional 8 hours. Protein extracts were incubated overnight with Ni-NTA beads at 4°C with gentle rocking. The beads were washed three times using lysis buffer with an intermittent incubation of 5 minutes each. The bound proteins were eluted by boiling with sample buffer as described above. The eluted proteins were separated through SDS-PAGE and immunoblotted with indicated antibodies.
Scratch Wound Healing Assay
Scratch wound healing assay was performed as described previously [30]. Briefly, transfected MCF7 cells were scratched with 200-μl tip to generate the open space. Cells were grown in DMEM supplemented with 5% FBS containing 5 ng/ml of actinomycin D to prevent the cell proliferation. The open space was tracked using live cell imaging microscope (Nikon Time Lapse microscope model TE 2000E). The percentage of open area was calculated using Tscratch software.
Cell Invasion Assay
Invasion experiment was performed as described previously [31]. Briefly, transfected cells (2 × 104/well) were suspended in serum-free DMEM in the upper chamber, and the lower chamber was loaded with complete DMEM. After 12 hours, the invaded cells were stained with 0.5% crystal violet. Images were taken with Nikon photograph camera.
Colony Formation Assay
Colony formation assay was performed as described previously [5]. Briefly, transfected MCF7 cells (5 × 103) were seeded in 35-mm plate and were grown in complete DMEM containing 0.8 mg/ml of G418 for 3 weeks. Then, the cells were fixed with 3.7% formaldehyde solution followed by staining with 0.5% crystal violet. Stained cells were then washed with PBS three times with gentle rocking at room temperature to remove the residual crystal violet solution.
Senescence Associated β-Gal Staining
MCF7 cells were co-transfected with indicated plasmids. Cells were then treated with γ-rays (10 Gy) and were grown at 37°C for 7 days. Senescence associated β-gal staining was performed as previously described [32]. Briefly, cells were fixed with 3.7% formaldehyde for 15 minutes and were incubated at 37°C with staining solution [150 mM NaCl, 2 mM MgCl2, 5 mM K3 Fe(CN)6, 5 mM K2Fe(CN)6, 40 mM citric acid, and 12 mM sodium phosphate [pH 6.0] containing 1 mg/ml of 5-bromo-4-chloro-3-indolyl-D-galactoside]. Digital pictures were taken on an Olympus TH4-200 by 20× objective lens.
RNA Immunoprecipitation (RIP)
RIP was performed to confirm the binding of miR-93 and miR-106a to the 3′UTR of FBXO31. RIP was performed as described previously [33]. Briefly, after 36 hours of transfection, cells were γ-irradiated (10 Gy) and kept for an additional 12 hours. The cells were then UV cross-linked (50 mJ/cm2) followed by fixation with 3.7% formaldehyde and quenching with 2 M glycine. The lysates were then incubated for 2 hours at 4°C with antibody-coated protein G agarose beads. Then, RNA was extracted from bound complex using TRIzol. cDNA was prepared, and PCR was performed to detect miRNA and 3′UTR of FBXO31 using specific primers.
Immunohistochemistry (IHC)
The tissue samples were obtained from SDM College of Medical Sciences, as per established core procedures and Institutional Ethical Board approval. Tissue samples were stained with hematoxylin-eosin to determine the histological type and grade of tumors. Twenty tissue samples from breast cancer patients, including cancerous tissue and adjacent nonmalignant epithelium, were analyzed for FBXO31 and Slug using immunohistochemical staining as described previously [34]. In brief, after deparaffinization and endogenous peroxidase blockage, the sections were heated in 0.01 M citrate buffer solution (pH 6.0) in water bath at 98°C for 20 minutes; then incubated with the rabbit and goat polyclonal antibody to FBXO31 and Slug (Santa Cruz Biotechnology), respectively, at 1:100 dilution overnight at 4°C; and visualized using 3,3′-diaminobenzidine detection kit (Vector labs). For the negative control, anti-rabbit and anti-goat IgG whole molecule (Sigma-Aldrich) was used at 1:1000 dilution. IHC-stained samples were evaluated by two pathologists, and all samples were blinded. Staining intensity of these proteins in neoplastic cells was graded on a scale of 0 (no staining) to 3+ (strong staining). The protein expression was scored based on the percentage of positive cells: score 0 = 0% of stained positive cells; score 1 = weakly stained tissue or 1%-25% of positive cells; score 2 = moderate stained tissue or 26%-50% of positive stained cells; and score 3 = strongly stained tissue or more than 50% of stained cells.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously [35]. Briefly, MCF7 cells treated with and without etoposide (5 μM for 12 hours) were cross-linked with 3.7% formaldehyde for 15 minutes at room temperature with gentle rocking. Cross-linking reaction was quenched by addition of 125 mM glycine for 10 minutes. The cell lysates were sonicated to generate fragmented DNA-protein complex and were then incubated with anti-Slug antibody overnight at 4°C. Preblocked protein A agarose beads were incubated with lysate-antibody complex for 2 hours followed by reverse cross-linking by proteinase K. DNA was eluted by phenol-chloroform method. PCR was performed using microRNA specific primers.
The following primers were used for PCR.
miR-93 region 1 (forward): 5′-GGGGACCATGTTAAATGGGGA-3′.
miR-93 region 1 (reverse): 5′-GCCCTCCAGCAATTTGGGCAC-3′.
miR-93 region 2 (forward): 5′-ACGTGGAGATGAGGCGAGAGG-3′.
miR-93 region 2 (reverse): 5′-TTCCCTCCACGGACGCTGTGC-3′.
miR-93 region 3 (forward): 5′-GCTACCGCACTGTGGGTACTT-3′.
miR-93 region 3 (reverse): 5′-GGAGCTCAGCCAAGAAGACAA-3′.
Statistical Analysis
Each experiment was repeated at least three times. Values shown are standard deviation (SD) except otherwise mentioned, and data were represented as mean ± SD. Statistical analysis was performed by using Sigma Stat 4.0 (Systat Software Inc., San Jose, CA). Student’s t test was performed to analyze the statistical significance. The values *P < .01 and **P < .001 denote significant differences between the groups (n > 3 at the least).
Results
MicroRNAs 93 and 106a Suppress the Translational Level of FBXO31 via Binding to Its 3′UTR
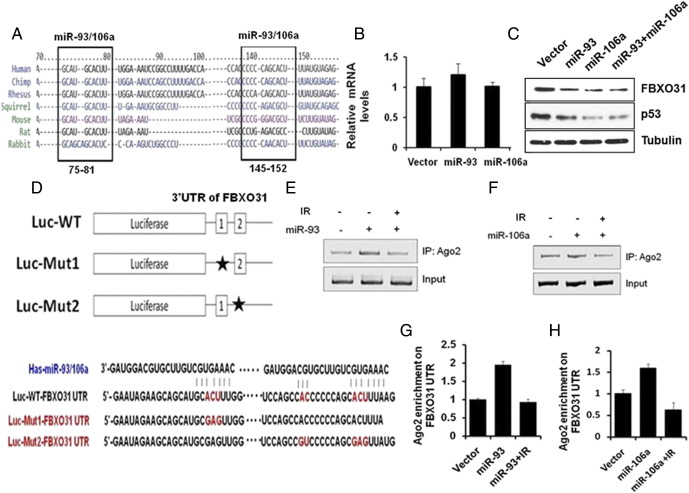
To explore the possible involvement of miRNA-mediated regulation of FBXO31 expression, an in silico analysis was performed by using online computational algorithm as mentioned in Materials and Methods. In silico analysis revealed that the 3′UTR of FBXO31 has putative binding sites for six potential microRNAs (Supplementary Table 1). These microRNAs have two similar putative binding sites on the 3′UTR of FBXO31 which are conserved among mammals: a 7-mer-m8 seed sequence AGCACTTTA (75-81 bp from the start of 3′UTR) and a 8-mer seed sequence ATGCACTTTG (145-152 bp from the start of 3′UTR) (Supplementary Figure 1A).
Supplementary Figure 1.
(A) Seed sequence of miR-93 and miR-106a in the 3′UTR of FBXO31 is conserved among the vertebrates. FBXO31 3′UTR sequences of mammalians were aligned using Clustal Omega. Putative binding sites of microRNAs are marked by square. (B) MiR-93 and miR-106a do not affect transcriptional levels of FBXO31. (C) MiRNAs do not regulate FBXO31 synergistically. MCF7 cells were transfected with mentioned plasmids for 48 hours. Protein extracts were immunoblotted with indicated antibodies. (D) Schematic represents the generation of mutants of FBXO31 3′UTR. Mutants were generated by site directed mutagenesis. (E–H) Both miR-93 and miR-106a bind to 3′UTR of FBXO31. MiR-93 and 106a were transfected in MCF7 cells for 48 hours and were treated with and without IR. RNA-IP was performed using anti-AGO2 antibody as described in the Materials and Methods.
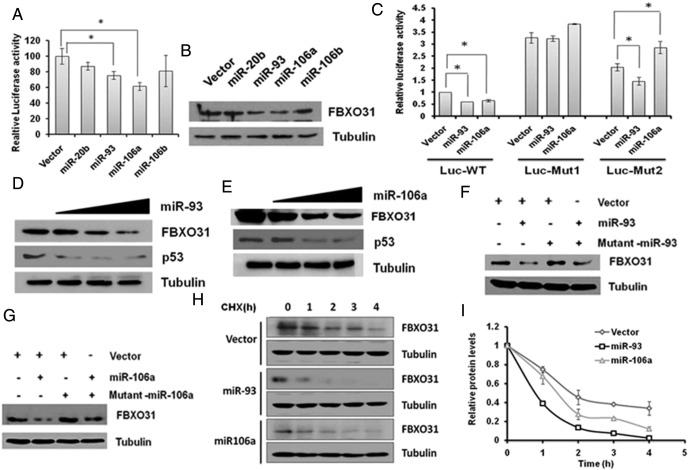
We evaluated the effect of in silico predicted miRNAs on the expression of FBXO31 using a luciferase reporter assay. The activity of luciferase reporter containing the FBXO31 3′UTR was suppressed to different extents in the presence of miRNAs -20b, -93, -106a, and -106b (Figure 1A). Next, immunoblotting data revealed that these miRNAs also suppressed the level of FBXO31 (Figure 1B). Among these miRNAs, miR-93 and miR-106a showed the potent effects on FBXO31 expression without affecting mRNA level (Figure 1B and Supplementary Figure 1B). However, these miRNAs do not regulate FBXO31 synergistically (Supplementary Figure 1C). Based on above results, we selected these two miRNAs to understand the molecular mechanism of FBXO31 regulation.
Figure 1.
MiR-93 and 106a negatively regulate the expression of FBXO31 at the protein level by binding to its 3′UTR. (A) Relative luciferase activity in the presence of miRNAs. Luciferase assay was performed with pCMV-MIR vector containing the 3′UTR of FBXO31. Luciferase reported vector was co-transfected with either empty vector or miRNAs for 48 hours in MCF7 cells. The data were normalized to empty vector, and results are presented as mean ± SD, *P < .001. (B) MiR-93 and miR-106a repressed the translational level of FBXO31. MCF7 cells were transfected with indicated miRNAs and were harvested at 48 hours posttransfection. Whole cell protein extracts were immunoblotted with indicated antibodies. Tubulin was used as loading control. (C) MiR-93 targets first seed sequence, whereas miR-106a needs both the seed sequences. Luciferase activity was measured after 48 hours of co-transfection with indicated plasmids. The data for wild-type 3′UTR of FBXO31 were normalized to empty vector, and the rest of the data were normalized to wild-type 3′UTR of FBXO31. The data are presented as mean ± SD, *P < .001. (D and E) MiR-93 and miR-106a regulate FBXO31 in a dose-dependent manner. Indicated plasmids were transfected (0-, 1-, 2-, and 3-fold) for 48 hours, and then whole cell protein extracts were immunoblotted with indicated antibodies. (F and G) Mutant-miRs interfere in the function of miR-93 and miR-106a. Cells were transfected with miR-93/miR-106a with or without their respective mutant miRNA for 48 hours. Whole cell protein extracts were immunoblotted with indicated antibodies. Tubulin is used as loading control. (H and I) MiR-93 and 106a alter the FBXO31 protein level. Pulse chase cycloheximide assay was performed by overexpression of microRNAs, and different time points were collected after treatment with cycloheximide. (I) Quantitative representation of pulse chase cycloheximide assay.
Next, to confirm the specificity of miR-93– and miR-106a–mediated regulation of FBXO31, we generated mutants by altering the seed sequence of miR-93 and miR-106a in the 3′UTR of FBXO31 luciferase reporter (Supplementary Figure 1D). Results revealed that the luciferase activity was significantly increased upon mutation of either of the seed sequences (Fig. 1C). Analysis of the results demonstrated that miR-106a requires both seed matches, whereas miR-93 preferentially targets the first seed sequence for regulating FBXO31. Further, RNA immunoprecipitation was performed to confirm that these miRNAs bind to the 3′UTR of FBXO31 (Supplementary Figure 1, E–H).
Next, a dose-dependent increase of either miR-93 or miR-106a resulted in the decrease in FBXO31 levels (Figure 1, D and E). This effect was abolished by co-expression of point mutant of these microRNAs (Figure 1, F and G). In addition, cycloheximide pulse chase assay revealed that miR-93 or miR-106a significantly reduced the stability of FBXO31 (Figure 1, H and I).
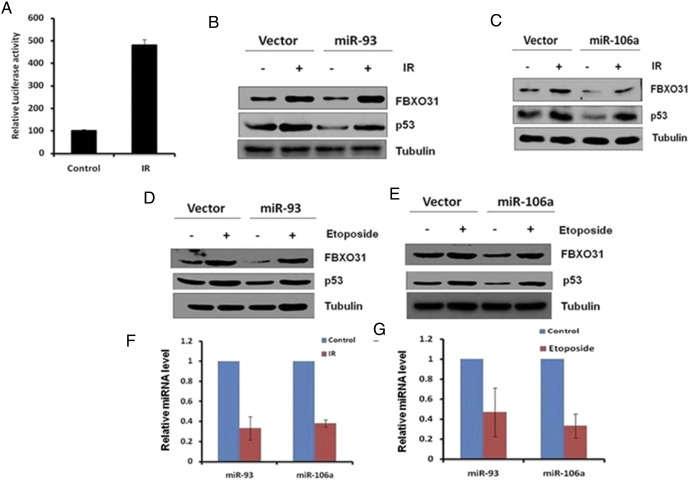
MicroRNAs-93 and 106a Fail to Regulate FBXO31 in Response to Genotoxic Stresses
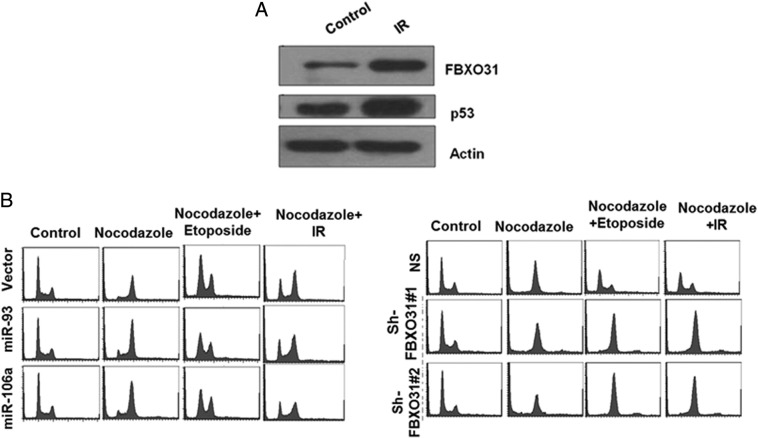
A previous study showed that FBXO31 stabilizes under various genotoxic stresses, suggesting that regulation of FBXO31 may be different under these conditions [5]. Hence, the involvement of miR-93 and miR-106a in the regulation of FBXO31 under genotoxic stress was examined. In agreement with the previous report, luciferase activity of FBXO31 3′UTR and the endogenous FBXO31 levels were significantly increased upon ionizing radiation (IR) (Figure 2A and Supplementary Figure 2A). Interestingly, ectopic expression of miR-93 and miR-106a did not affect the stabilization of FBXO31 in response to IR (Figure 2, B and C) and etoposide treatment (Figure 2, D and E). This effect was further evident from our cell cycle data wherein overexpression of these miRNAs could not prevent the FBXO31-mediated G1 arrest (Supplementary Figure 2B). These results taken together suggest that miRNAs could not suppress FBXO31 expression either due to their inability to bind the FBXO31 3′UTR or due to an alteration of their expression levels. Interestingly, the levels of miR-93 and miR-106a were found to be significantly decreased upon IR (Figure 2F) and etoposide treatment (Figure 2G). Further, RNA immunoprecipitation data demonstrated that these miRNAs are incapable of binding to 3′UTR of FBXO31 under genotoxic stress (Supplementary Figure 1, D–G). Collectively, these results suggest that although miR-93 and miR-106a repress FBXO31 under normal conditions, they were unable to do so under genotoxic stresses due to their reduced expression as well as their reduced binding at the 3′UTR of FBXO31.
Figure 2.
MicroRNAs-93 and 106a fail to regulate FBXO31 in response to genotoxic stresses. (A) Luciferase activity of FBXO31 3′UTR reporter increased upon treatment of IR. MCF7 cells were transfected with FBXO31 3′UTR luciferase construct, and luciferase activity was measured with and without irradiation. Cells were collected at 4 hours postirradiation. (B–E) Ectopically expressed microRNAs failed to prevent FBXO31 stabilization under genotoxic stresses. MCF7 cells were transfected with indicated miRNAs for 48 hours. The transfected cells were exposed to either IR (B and C) or etoposide treatment (D and E). Whole cell protein extracts were immunoblotted with indicated antibodies. (F and G) MicroRNA levels decreased under stress conditions. MCF7 cells were treated with either IR or Etoposide, total RNA was isolated, and cDNA was prepared followed by real-time RT-PCR.
Supplementary Figure 2.
(A) FBXO31 stabilizes upon IR. MCF7 cells were treated with 10-Gy IR. Protein extracts were immunoblotted with indicated antibodies. (B) MicroRNAs do not affect the FBXO31-mediated G1 arrest under genotoxic stresses. Transfected MCF7 cells (left panel) and FBXO31 knockdown cells (right panel) were either untreated or treated with IR (10 Gy) or etoposide (5 μm). After 4 hours of treatment, cells were further treated with and without nocodazole (350 nM) for 20 hours. Cells were then harvested, fixed, and analyzed by flow activated cell sorter.
FBXO31 Suppresses Cell Invasion and Migration by Downregulating the EMT Regulator Slug
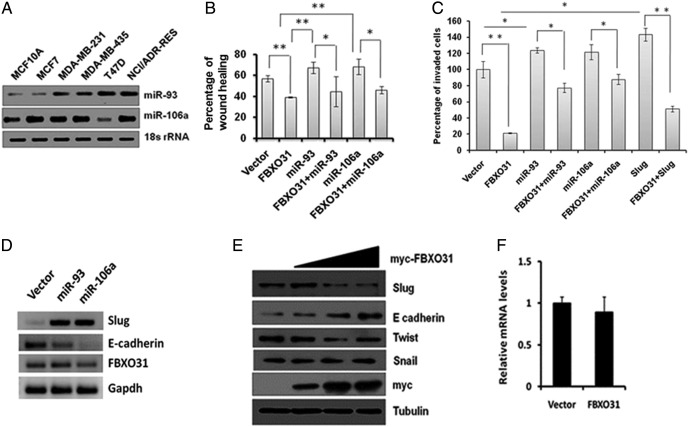
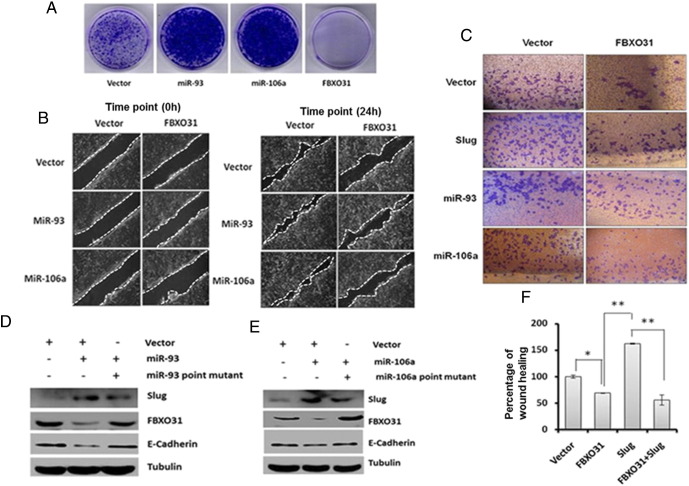
Owing to its growth suppressive effect, expression of FBXO31 is maintained at low level under normal growth conditions. Examination of the levels of miRNAs in different breast cancer cell lines revealed an increase in levels of both miR-93 and miR-106a in most cell lines (Supplementary Figure 3A). We further found that ectopic expression of miR-93 and miR-106a led to a significant increase in the number of colonies formed in the long-term survival assay, which may be partly due to inactivation of FBXO31 (Figure 3A). This is in agreement with the previous reports showing the oncogenic function of miR-93 and miR-106a [20], [25], [26]. Further, most of the oncogenes promote the cellular transformation through activation of EMT. We therefore tested the effect of miR-93 and miR-106a on scratch wound healing and invasion. Results demonstrated that ectopic expression of miR-93 and miR-106a promotes both scratch wound healing and invasion, suggesting that these miRNAs may regulate EMT promoters (Figure 3, B and C, and Supplementary Figure 3, B and C). Slug is among the well-established EMT promoter that enhances wound healing [36]. Therefore, we checked whether these miRNAs have any role in Slug stability and found that mRNA and protein levels of Slug were significantly increased upon ectopic expression of miR-93 and 106a with concomitant suppression of E-cadherin (Supplementary Figure 3D and Figure 3, D and E), indicating that stabilization of Slug is responsible for miR-93– and miR-106a–mediated EMT. Further, ectopic expression of FBXO31 suppressed the miR-93– and miR-106a–mediated scratch wound healing and invasion, suggesting that FBXO31 may affect the expression level of Slug (Figure 3, B and C, and Supplementary Figure 3, B and C).
Supplementary Figure 3.
(A) MiR-93 and miR-106a are highly expressed in breast cancer cell lines. Real-time RT-PCR was performed to measure the relative miRNA levels in breast cancer cell lines as described in Materials and Methods. (B) Quantitative data of scratch wound healing assay. (C) Quantitative data of cell invasion assay. (D) mRNA levels of Slug and E-cadherin upon overexpression of miR-93 and miR-106a. (E) FBXO31 decreases the stability of Slug in a dose-dependent manner. FBXO31 was ectopically expressed in MCF7 cells for 48 hours, and whole cell protein extracts were immunoblotted with indicated antibodies. (F) Overexpression of FBXO31 does not affect the transcriptional level of Slug.
Figure 3.
MiRNAs promote scratch wound healing and cell invasion by regulating Slug. (A) Mir-93 and miR-106a promote the growth of MCF7 cells. Equal numbers of transfected cells were seeded, grown in G418-containing media for 3 weeks, and stained with 0.5% crystal violet for visualization of colonies. (B) MiR-93 and mir-106a promote scratch wound healing. Transfected MCF7 cells were scratched to generate wound, and the cell migration was checked by live cell imaging as described in Materials and Methods. Left and right panels indicate 0 and 24 hours of scratch. (C) MiR-93 and mir-106a promote cell invasion. Equal numbers of transfected MCF7 cells were added in Boyden chamber to check the invasive capacity of the cells. (D and E) Ectopic expression of miR-93 and miR-106a augments stabilization of Slug. Immunoblotting of MCF7 cells expressing either vector or miRNAs or a combination of miRNA and their mutant form for indicated antibodies. (F) FBXO31 suppresses the scratch wound healing activity of Slug. Percentage of scratch wound healing was measured as described in the Materials and Methods. The data are presented as mean ± SD, *P < .01, **P < .001.
Collectively, our results suggest that there could be a cross talk between miRNAs, FBXO31 and Slug. We then checked the effect of FBXO31 on Slug, and the immunoblotting data showed that overexpression of FBXO31 significantly decreased the levels of Slug in a dose-dependent manner and had minimal effect on Twist and Snail (Supplementary Figure 3E). FBXO31-mediated decrease of Slug is further evident by the scratch wound healing and invasion assay wherein ectopic expression of FBXO31 inhibited Slug-mediated invasion and wound healing (Figure 3, C and F, and Supplementary Figure 3, B and C). However, mRNA level of Slug was not affected by FBXO31 (Supplementary Figure 3F). Collectively, these results suggest that miR-93 and miR-106a promote cell survival and accelerate wound healing as well as invasion by stabilization of Slug through suppression of FBXO31.
FBXO31 Promotes the Ubiquitination and Proteasomal Degradation of Slug
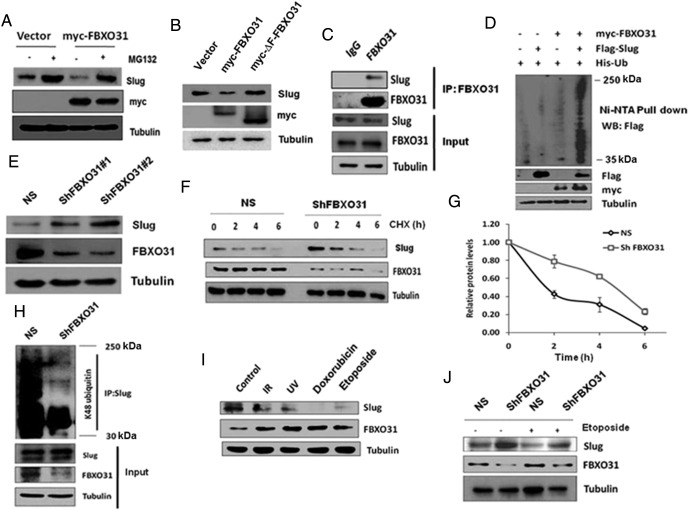
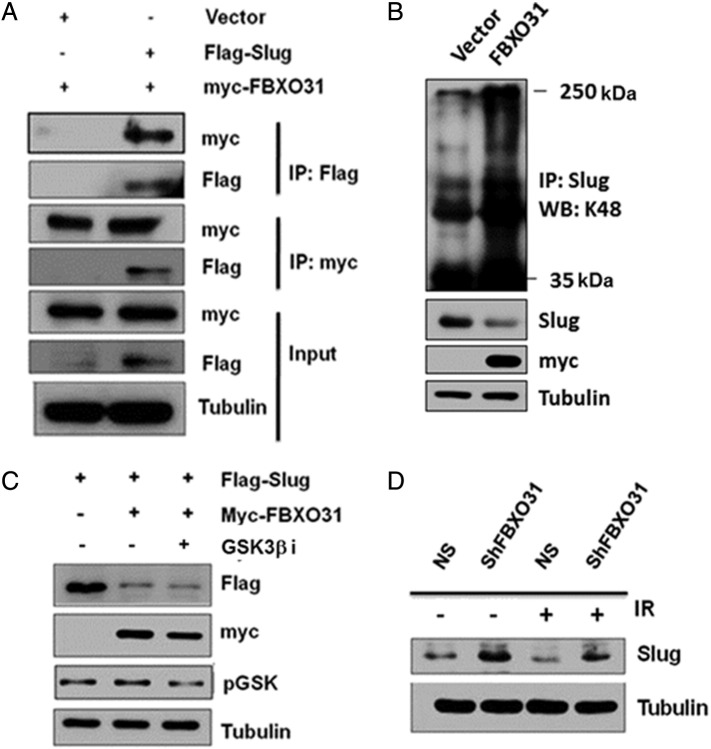
The above results suggest that FBXO31 antagonizes the function of Slug presumably by negatively regulating its levels. Because FBXO31 is part of an E3 ubiquitin ligase, we sought to ask whether it is directly involved in the proteasomal degradation of Slug. FBXO31 failed to cause degradation of Slug in the presence of proteasome inhibitor MG132, suggesting that FBXO31 regulates Slug in a proteasome-dependent manner (Figure 4A). Further, we found that ectopic expression of a mutant form of FBXO31 lacking the F-box motif (ΔF-FBXO31) failed to downregulate Slug, indicating that FBXO31 regulates Slug through the SCF E3 ubiquitin ligase complex (Figure 4B). Furthermore, co-immunoprecipitation experiments indicated that FBXO31 physically interacts with Slug (Figure 4C and Supplementary Figure 4A) and facilitates its polyubiquitination (Figure 4D and Supplementary Figure 4B). Recent reports suggested that GSK3β-mediated phosphorylation of Slug facilitates its turnover [36]. However, inhibition of GSK3β did not stabilize Slug levels in the presence of FBXO31, indicating that FBXO31 does not need GSK3β-mediated phosphorylated form of Slug for its proteasomal degradation (Supplementary Figure 4C).
Figure 4.
FBXO31 degrades Slug at the proteasomal level. (A) FBXO31 degrades Slug through proteasomal pathway. Immunoblot monitoring Slug in MCF7 cells expressing vector or FBXO31 in the absence and presence of MG132. MG132 was added at 40 hours posttransfection, and cells were collected at 48 hours posttransfection. Whole cell protein extracts were immunoblotted with indicated antibodies. (B) FBXO31 degrades Slug through SCF complex. Wild-type (myc-FBXO31) and F-box motif deleted FBXO31 (myc-∆F-FBXO31) were transfected in MCF7 cells for 48 hours. Protein extracts were immunoblotted with indicated antibodies. (C) FBXO31 interacts with Slug at the endogenous level. Whole cell protein extracts were immunoprecipitated with anti-FBXO31 antibody, and immunoprecipitates were immunoblotted for indicated antibodies. (D) FBXO31 promotes the polyubiquitylation of Slug. Ubiquitination assay was performed upon co-transfection of indicated plasmids for 36 hours. Protein extracts were pulled down by Ni-NTA beads. Pulled fractions were immunoblotted with anti-Flag antibody to detect the ubiquitylated Slug. (E) Slug is stabilized in FBXO31 knockdown cells. Stable knockdown cells were generated by lentiviral transduction as described in the Materials and Methods. Whole cell protein extracts were immunoblotted with indicated antibodies. (F) Slug has slower turnover kinetics in FBXO31 knockdown cells. Both NS and FBXO31 knockdown cells were treated with cycloheximide for indicated time periods, and whole cell extracts were immunoblotted with indicated antibodies. (G) Quantitative data of cycloheximide pulse chase assay. The intensity of the slug bands in panel F was normalized to the tubulin loading control and quantified using ImageJ to study the turnover of slug. (H) Lysine 48-linked polyubiquitination of Slug is significantly decreased in FBXO31 knockdown cells. Whole cell protein extracts from NS and FBXO31KD cells were immunoprecipitated with anti-Slug antibody, and immunoprecipitates were immunoblotted with K48-linked ubiquitin antibody. (I) Slug is downregulated under genotoxic stress conditions. MCF7 cells were treated with IR (15 Gy), UV (50 mJ/m2), doxorubicin (10 μM), and etoposide (10 μM). IR and UV treatments were given for 4 hours, whereas doxorubicin and etoposide treatments were given for 12 hours. Whole cell lysates were immunoblotted with indicated antibodies. (J) FBXO31 controls degradation of Slug under genotoxic stresses. NS and FBXO31KD cells were treated with 10 μM etoposide for 12 hours, and protein extracts were immunoblotted with indicated antibodies.
Supplementary Figure 4.
(A) FBXO31 interacts with Slug. Myc-FBXO31 was co-transfected with either vector or Flag-Slug for 48 hours. Then, protein extracts were immunoprecipitated with either anti-myc or anti-Flag antibody. Immunoprecipitates were immunoblotted with indicated antibodies. (B) FBXO31 promotes the K48 linked polyubiquitylation of Slug. MCF7 cells were co-transfected with indicated plasmids for 36 hours. Protein extracts were pulled down with anti-Slug antibody, and immunoprecipitates were immunoblotted with K48-Ub antibody. (C) FBXO31-meidated degradation of Slug is GSK3β independent. MCF7 cells were co-transfected with indicated plasmids and were incubated in the presence or absence of 500 nM of GSK3β inhibitor. (D) FBXO31 degrades Slug upon IR treatment.
Next, we sought to determine whether FBXO31 regulates Slug at the physiological level and found that knockdown of FBXO31 resulted in a significant stabilization of Slug (Figure 4E). In addition, cycloheximide pulse chase study also revealed an enhanced stability of Slug in FBXO31 knockdown cells (Figure 4, F and G). Knockdown of FBXO31 also resulted in decreased levels of polyubiquitylated Slug (Figure 4H). Collectively, our results demonstrated that FBXO31 maintains the cellular level of Slug.
Our initial observations have indicated that miR-93 and miR-106a are suppressed and FBXO31 is stabilized upon genotoxic stresses. Hence, we further sought to understand the expression levels of Slug under genotoxic stresses. Interestingly, immunoblotting data indicated that Slug is decreased under genotoxic stress conditions (Figure 4I). To check whether FBXO31 has any role in Slug regulation under genotoxic stress, FBXO31 knockdown and control cells were exposed to genotoxic agents like etoposide and IR. Results showed that FBXO31 destabilizes Slug under genotoxic stress (Figure 4J and Supplementary Figure 4D).
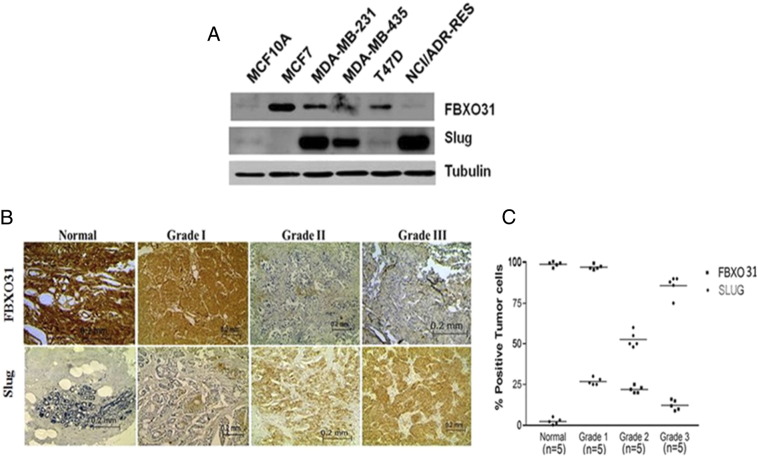
Reciprocal Expression Pattern of FBXO31 and Slug in Breast Cancer Progression
Our in vitro data suggest that FBXO31 negatively regulates Slug, prompting us to check their levels in different breast cancer cell lines as well as patient samples. Results showed the existence of an inverse correlation between Slug and FBXO31 levels in most of the breast cancer cell lines (Figure 5A). Next, to investigate the expression levels of FBXO31 and Slug in breast cancer patient samples, immunohistochemical staining of patient tissue with matched normal breast tissue was performed. Our data indicate that FBXO31 is expressed in all noncancerous tissues (Figure 5, B and C). However, as cancer progressed from grade I to grade III, FBXO31 staining intensity decreased, and high-grade breast cancer tissue samples exhibited negative staining. On the other hand, Slug expression was low/null in normal tissue and was significantly increased in grade I to III in infiltrating ductal carcinoma. The expression pattern of Slug significantly correlated with tumor size, stage of the cancer, and tumor grade as shown in the figure (Figure 5, B and C).
Figure 5.
Reciprocal expression pattern of FBXO31 and Slug in breast cancer progression. (A) Immunoblot monitoring the expression of FBXO31 and Slug in different breast cancer cell lines. (B) Representative immunohistochemical staining with antibodies specific for FBXO31and Slug with tissue sections from normal breast and grade I, grade II, and grade III breast cancer. (C) Quantitative data of IHC staining.
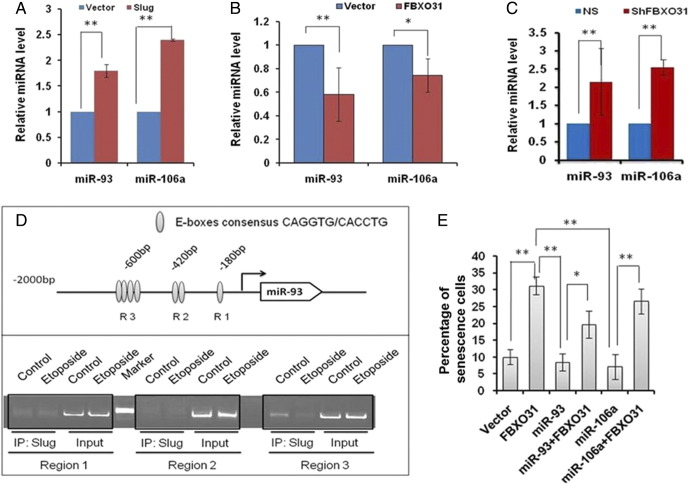
FBXO31 Regulates a Feedback Loop Wherein Slug Drives miR-93 and miR106a Expression
Previous reports have demonstrated that Slug regulates microRNAs at the transcriptional level by binding to the E-box motif in the promoter region [37]. Our results also hint to the existence of a cross talk between miRNAs, FBXO31, and Slug. Therefore, we went on to check if Slug regulates miR-93 and miR-106a as well and found that it regulates these miRNAs at the transcriptional level (Figure 6A). We further checked the effect of FBXO31 on the levels of miRNAs because it degrades Slug. Interestingly, levels of both the miRNAs were decreased upon ectopic expression of FBXO31, and their levels were elevated upon depletion of FBXO31 (Figure 6, B and C). These results taken together indicate that FBXO31 may regulate miRNA levels through Slug. To validate the involvement of Slug in driving miRNA expression, we performed ChIP to determine whether Slug binds at E-box sequences present at the promoter of miR-93 (Figure 6D, upper panel). ChIP results revealed that Slug binds to the third E-box region (R3) of miR-93 under normal condition, which is significantly inhibited upon etoposide treatment (Figure 6D, lower panel).
Figure 6.
FBXO31 regulates miR-93 and miR106a via degradation of Slug. (A) Slug enhances the expression of miR-93 and miR-106a at the transcriptional level. MCF7 cells were transfected with either empty vector or Slug, and transcriptional level of miR-93 and 106a was measured by real-time RT-PCR. (B) MicroRNAs levels were ablated upon overexpression of FBXO31. MCF7 cells were transfected with empty vector/FBXO31 for 48 hours. Total RNA was used to prepare cDNA, and levels of miR-93 and 106a were measured by real-time RT-PCR. (C) Depletion of FBXO31 enhanced the miR-93 and miR-106a levels. (D) The location of E-box consensus binding sites (5′-CAGGTG-3′ or 5′-CACCTG-3′) in the promoter of human miR-93 (upper panel). ChIP assay for recruitment of Slug at the promoter of miR-93 (lower panel). (E) MiR-93 and miR-106a inhibit FBXO31-mediated senescence. Transfected cells were irradiated with IR and were allowed to proliferate for 10 days. Then, cells were stained for β-galactosidase assay. Approximately 500 cells were counted for each case. The data are presented as mean ± SD, *P = .001, **P < .001.
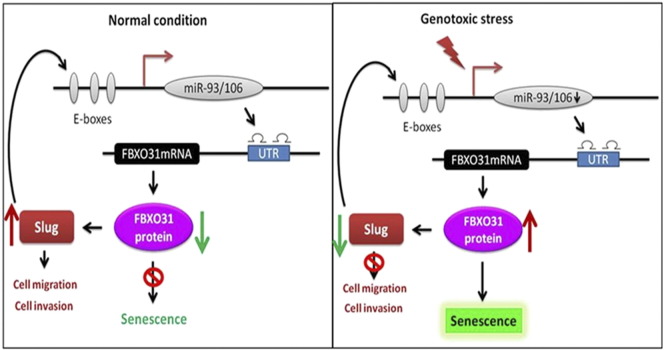
Because FBXO31 is a known inducer of senescence, we investigated the effect of miR-93 and miR-106a on FBXO31-mediated senescence [2], [4], [5]. The senescence-associated β-galactosidase staining showed that overexpression of miR-93 and miR-106a inhibits senescence that is induced by FBXO31 (Figure 6E). Collectively, our study demonstrates that miR-93 and miR-106a act as oncogene by interfering with the tumor suppressive function of FBXO31 and provides the evidence for existence of a feedback mechanism between FBXO31, Slug, miR-93, and miR-106a (Figure 7).
Figure 7.
Proposed model describes feedback loop in FBXO31, Slug, and miRNAs in normal (left) and genotoxic stress (right) conditions.
Discussion
FBXO31 is a member of the F-box family, which functions as a substrate-recognizing unit in SCF (Skp1, Cullin1, Rbx1, and F-box) E3 ubiquitin ligase complex and mostly targets the phosphorylated substrates [5]. Initial studies reported that FBXO31 functions as a tumor suppressor in many cancers such as breast cancer [2], melanoma [5], hepatocellular carcinoma [38], and gastric cancer [39]. It induces senescence by arresting cells at the G1 phase of the cell cycle through proteasomal degradation of either cyclin D1 or MDM2 or both [4], [5], [6]. Having growth suppressive effect and involvement in DNA damage response pathway, posttranslational level of FBXO31 is maintained at a low level under normal growth condition. These findings encouraged us to investigate the regulation of FBXO31.
Our unpublished data demonstrate that FBXO31 is not regulated at the transcriptional level throughout the cell cycle (data not shown). Hence, we examined the possibility of regulation of FBXO31 at the posttranscriptional level by miRNAs. In support of this, our in silico analysis predicted four miRNAs (miR-17, miR-20, miR-93, and miR-106a) to be involved in the regulation of FBXO31 at the translational level. Recently, miR-17 and miR-20 have been reported to regulate FBXO31 in gastric cancer [39]. Our study revealed that miR-93 and miR-106a significantly suppressed the FBXO31 expression. This led us to investigate the regulation of FBXO31 by these miRNAs in detail.
Previous studies reported that miR-93 acts as an oncomiR by negatively regulating the tumor suppressor gene programmed cell death 4 in gastric carcinoma [40]. Oncogenic function of miR-93 is also reported in prostate cancer, ovarian cancer, lung cancer, head and neck squamous cell carcinoma, and non–small cell lung cancer [41], [42], [43], [44], [45]. It is also documented that miR-93 promotes glioma through regulation of AKT and IL8 [46]. Further, miR-93 has been shown to suppress VEGF and IL-8 in neuroblastoma [47]. All these reports establish that miR-93 is an oncomiR and is closely associated with cancer progression. In this study, we are reporting for the first time that oncomiR-93 functions by suppressing FBXO31 in breast cancer. Our results demonstrate that miR-93 is highly expressed in higher grades of breast cancer cell lines, which is in accordance with other cancers. In addition, we found that miR-93 significantly stabilizes Slug through suppression of FBXO31 at the posttranscriptional level, and in turn, stabilization of Slug leads to transcriptional upregulation of miR-93 expression. Thus, our results strongly suggest that miR-93–mediated Slug activation through the suppression of FBXO31 may be associated with breast cancer malignancy.
MiR-106a is also reported to be frequently upregulated in gastric cancer and promotes malignancy through activation of AKT pathway and inhibition of apoptotic pathway through inactivation of FAS [48]. It is also associated with promoting malignancy in NSLC, ovarian cancer, and gastric cancer by regulating PTEN, p130, pRB, and tissue inhibitors of metalloproteinase 2, respectively [49], [50], [51]. Our results demonstrate that miR-106a stabilizes Slug expression through significant suppression of FBXO31 at the posttranscriptional level. This relationship is further observed in different breast cancer cell lines, suggesting that miR-106a plays a crucial role in promoting malignancy.
Our study also demonstrated that FBXO31 promotes lysine 48-linkage specific polyubiquitylation–mediated proteasomal degradation of Slug and thereby prevents cell migration and invasion. Clinical data revealed that expression level of FBXO31 and Slug is inversely correlated in breast cancer. FBXO31 prevents the malignancy by degrading Slug at the proteasomal level and thereby limits the expression of both miRNAs under genotoxic stresses. To summarize, our results demonstrate that there is a feedback mechanism that exists between FBXO31, Slug, miR-93, and miR-106a.Therapeutic intervention aimed at breaking this loop could prove important for the treatment of breast cancer.
The following are the supplementary data related to this article.
List of Predicted miRNAs Targeting FBXO31
Supplementary Materials.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgement
We thank Prof. Michael R. Green, Prof. David F. Callen, Prof. Sophie Tartare-Deckert, and Dr. Samit Chattopadhyay for providing reagents. We also thank Dr. Agnita Kundu for editorial assistance. Part of this work was financially supported by a Department of Biotechnology grant (BT/PR6690/GBD/27/475/2012) to M. K. S. and partly by the National Centre for Cell Science, Department of Biotechnology, Ministry of Science and Technology, Government of India (to M. K. S.). R. K. M., D. P., and N. G. are senior UGC research fellows, and Y. A. is a senior DBT research fellow.
Footnotes
Financial support: Part of this work is supported by grant RGYI-BT/PR6690/GBD/27/475/2012 and National Centre for Cell Science intramural fund.
References
- 1.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Callen DF. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65:11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 3.Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmüller J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One. 2013:e57530. doi: 10.1371/journal.pone.0057530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci U S A. 2015;112:8632–8637. doi: 10.1073/pnas.1510929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Han L., Li B., Yang J., Huen M.S., Cheung A.L. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase 6 (MKK6) J Biol Chem. 2014;289:21508–21518. doi: 10.1074/jbc.M114.560342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson P., Jeffery J., Al-Ejeh F., Schulz RB, Callen DF, Kumar R, KK Khanna. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J Biol Chem. 2014;289:18514–18525. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles—diagnostic and therapeutic implications. Int J Oncol. 2015;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;6956:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viticchiè G., Lena A.M., Latina A., Formosa A., Gregersen L.H., Lund A.H., Bernardini S., Mauriello A., Miano R., Melino G. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 15.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 16.Hidaka H., Seki N., Yoshino H., Yamasaki T., Yamada Y., Nohata N., Fuse M., Nakagawa M., Enokida H. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;1:44–57. doi: 10.18632/oncotarget.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahai S., Lee S.C., Lee D.Y., Yang J., Li M., Yang B.B. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;10 doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Croce C.M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nohata Nijiro, Hanazawa Toyoyuki, Enokida Hideki, Seki Naohiko. MicroRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang L., Deng Z., Shatseva T., Yang J., Peng C., Du W.W., Yee A.J., Ang L.C., Yang B.B. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 21.Zou C., Xu Q., Mao F., Li D., Bian C., Liu L.Z., Jiang Y., Chen X., Qi Y., Lai L. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–2145. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Q, Gumireddy K, Schrier M, Sage C le, Nagel R, R Nagel, Nair S, Egan DA, Li A, Argami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Khuu C, Utheim TP, Sehic A. The three paralogous microRNA clusters in development and disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica (Cairo) 2016 doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan W, Li Y, Lim SG, Tan TM. MiR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol. 2014;20:5962–5972. doi: 10.3748/wjg.v20.i20.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 27.Young JA, Sermwittayawong D, Kim HJ, Nandu S, An N, Erdjument-Bromage H, Tempst P, Coscoy L, Winoto A. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J Biol Chem. 2011;286:6521–6531. doi: 10.1074/jbc.M110.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Anderson M.R. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Cory Giles. Scratch-wound assay. Methods Mol Biol. 2013;769:25–30. doi: 10.1007/978-1-61779-207-6_2. [DOI] [PubMed] [Google Scholar]
- 31.Jan HJ, Lee CC, Shih YL, Hueng DY, Ma HI, Lai JH, Wei HW, Lee HM. Osteopontin regulates human glioma cell invasiveness and tumor growth in mice. Neuro Oncol. 2010;12:58–70. doi: 10.1093/neuonc/nop013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itahana K, Itahana Y, Dimri GP. Colorimetric detection of senescence-associated β galactosidase. Methods Mol Biol. 2013;965:143–156. doi: 10.1007/978-1-62703-239-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert C, Svejstrup JQ. RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2704s75. [Chapter 27: Unit 27.4] [DOI] [PubMed] [Google Scholar]
- 34.Shetty P, Bargale A, Patil BR, Mohan R, Dinesh US, Vishwanatha JK, Pramod BG, Vidya SP, Amsavardani TS. Cell surface interaction of annexin A2 and galectin-3 modulates epidermal growth factor receptor signaling in Her-2 negative breastcancer cells. Mol Cell Biochem. 2016;411:221–233. doi: 10.1007/s11010-015-2584-y. [DOI] [PubMed] [Google Scholar]
- 35.Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, Green MR. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell. 2009;36:219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT, Wang SP, Nesvizhskii AI, Chen YJ, Yang PC. GSK3β controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene. 2014;24:3172–3182. doi: 10.1038/onc.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aomatsu K, Arao T, Abe K, Kodama A, Sugioka K, Matsumoto K, Kudo K, Kimura H, Fujita Y, Nishio K. Slug is up regulated during wound healing and regulates cellular phenotypes in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2012;53:751–756. doi: 10.1167/iovs.11-8222. [DOI] [PubMed] [Google Scholar]
- 38.Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;3:715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Liu Z. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178–6190. doi: 10.18632/oncotarget.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang H, Wang F, Chu D, Zhang W, Liao Z, Fu Z, Yan X, Zhu H, Guo W, Chen X. miR-93 functions as an oncomiR for the downregulation of PDCD4 in gastric carcinoma. Sci Rep. 2016;6:23772. doi: 10.1038/srep23772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, Zhou L, Cao P, Gong H, Zhang Y. MicroRNA-93 regulates cyclin G2 expression and plays an oncogenic role in laryngeal squamous cell carcinoma. Int J Oncol. 2015;46:161–174. doi: 10.3892/ijo.2014.2704. [DOI] [PubMed] [Google Scholar]
- 42.Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E, Suraokar M, Wistuba I, Minna JD, Pertsemlidis A. MiR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;34:4307–4315. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;50:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, Park YY, Lee JS, Safe S. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2012;31:1034–1044. doi: 10.1038/onc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu MH, Han C, Srivastava AK, Cui T, Zou N, Gao ZQ, Wang QE. MiR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37:5645–5651. doi: 10.1007/s13277-015-4328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Lili, Wang Chanjuan, Lei Fangyong, Zhang Longjuan, Zhang Xin, Aibin Liu, Geyan Wu, Jinrong Zhu, Song Libing. MiR-93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget. 2015;6:8286–8299. doi: 10.18632/oncotarget.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbri E, Montagner G, Bianchi N, Finotti A, Borgatti M, Lampronti I, Cabrini G, Gambari R. MicroRNA miR-93-5p regulates expression of IL-8 and VEGF in neuroblastoma SK-N-AS cells. Oncol Rep. 2016;35:2866–2872. doi: 10.3892/or.2016.4676. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C, Quan L, Bai J, Xu N. MiR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol Carcinog. 2013;52:634–646. doi: 10.1002/mc.21899. [DOI] [PubMed] [Google Scholar]
- 49.Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu FQ, Wang MS. MiR-106a promotes growth and metastasis of non–small cell lung cancer by targeting PTEN. Int J Clin Exp Pathol. 2015;8:3827–3834. [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Gersbach E, Zhang X, Xu X, Dong R, Lee P, Liu J, Kong B, Shao C, Wei JJ. MiR-106a represses the Rb tumor suppressor p130 to regulate cellular proliferation and differentiation in high-grade serous ovarian carcinoma. Mol Cancer Res. 2013;11:1314–1325. doi: 10.1158/1541-7786.MCR-13-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu M, Zhang N, He S, Lui Y, Lu G, Zhao L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588:600–607. doi: 10.1016/j.febslet.2013.12.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Predicted miRNAs Targeting FBXO31
Supplementary Materials.