Abstract
Serial femtosecond crystallography (SFX) allows structures of proteins to be determined at room temperature with minimal radiation damage. A highly viscous matrix acts as a crystal carrier for serial sample loading at a low flow rate that enables the determination of the structure, while requiring consumption of less than 1 mg of the sample. However, a reliable and versatile carrier matrix for a wide variety of protein samples is still elusive. Here we introduce a hydroxyethyl cellulose-matrix carrier, to determine the structure of three proteins. The de novo structure determination of proteinase K from single-wavelength anomalous diffraction (SAD) by utilizing the anomalous signal of the praseodymium atom was demonstrated using 3,000 diffraction images.
Introduction
Serial femtosecond crystallography (SFX) using ultrashort pulses from X-ray free-electron lasers (XFELs) can overcome typical radiation damage to protein crystals via the “diffraction-before-destruction” approach1–7. This has been used to obtain crystal structures of interesting proteins at room temperature8–18. Liquid jet injection of small protein crystals with continuous flow at relatively high speed (~10 m sec−1) is frequently exploited for serial sample loading19, consuming 10~100 mg of the sample. To reduce sample consumption, micro-extrusion techniques of specimens using viscous media such as a lipidic cubic phase (LCP)20, grease21, Vaseline (petroleum jelly)22 and agarose23 have been developed. These media can maintain a stable stream at a lower flow rate of 0.02~0.5 μl min-1 allowing sample consumption of less than ~1 mg. More recently, synchrotron-based serial crystallography has also been developed22, 24, 25, demonstrating that the sample loading technique with a viscous media becomes even more important in serial crystallography. This method with viscous media is technically simple, but some media produce stronger X-ray scattering that increase background noise. For data collection from small crystals (~1 μm), at atomic resolution, and de novo phasing with weak anomalous signals, a crystal carrier with low background scattering is essential to improve the signal-to-noise ratio23. To reduce background scattering from the carrier media, we introduced a hyaluronic acid matrix in SFX26. At the SPring-8 Angstrom Compact Free Electron Laser (SACLA)27, we operate an injector system under a helium atmosphere at 1 atm during micro-extrusion of the matrices28. However, hyaluronic acid matrix is strongly adhesive, resulting in frequent clogging of the sample-vacuum nozzle which acts as a sample catcher22 in our injector system. In addition, the general adaptability of hydrogel matrices to de novo phasing with heavy atoms is still unclear.
Here we introduce hydroxyethyl cellulose (cellulose matrix) for serial sample loading. We demonstrate the cellulose matrix as a protein carrier for SFX using small and large sized crystals (1 × 1 × 1 to 20 × 20 × 30 μm). In addition, we demonstrate the successful de novo phasing in SFX by applying praseodymium (Pr)-SAD, single-isomorphous replacement (SIR) and SIR with anomalous scattering (SIRAS) phasing to determine the structure of proteinase K. Furthermore, to reduce background scattering, a novel grease matrix, Super Lube nuclear grade grease (nuclear grease), was introduced in this study.
Results and Discussion
Crystal structures for lysozyme and thaumatin
We performed SFX experiments using femtosecond X-ray pulses from SACLA. Using lysozyme (1 × 1 × 1 μm) and thaumatin (2 × 2 × 4 μm) crystals (Supplementary Fig. 1a,b) dispersed in a cellulose matrix, we were able to collect 100,000–150,000 images in approximately 60–80 min at a wavelength of 1.24 Å (Table 1). At a flow rate of 0.43 and 0.47 μl min−1, a total sample volume of about 30–40 μl was used with a crystal number density of 5.8 × 108 crystals ml−1 for lysozyme, and 4.3 × 108 crystals ml−1 for thaumatin. We indexed and integrated 30,000–40,000 images for both the lysozyme (space group P43212) and thaumatin (space group P41212) crystals. The lysozyme and thaumatin crystals yielded data sets at 1.8-Å and 1.55-Å resolution with a completeness of 100% and a CC1/2 of 0.992 and 0.988, respectively. We determined and refined the crystal structures of lysozyme [Protein Data Bank (PDB) ID: 5wr9] and thaumatin (PDB ID: 5wr8) at 1.8-Å and 1.55-Å resolution (Fig. 1a,b), respectively. For the larger lysozyme crystals of the size 20 × 20 × 30 μm, the X-ray wavelength was kept at 0.95 Å. The microcrystals were used to acquire data sets at 1.45-Å resolution with a completeness of 100% and a CC1/2 of 0.995 (PDB ID: 5wra, Table 1).
Table 1.
Crystallographic statistics. Values in parentheses are for the outermost shell.
| Protein | Lysozyme | Thaumatin | |||
|---|---|---|---|---|---|
| Carrier | 16% cellulose | 11% cellulose | Nuclear grease | 22% cellulose | |
| Crystal density (crystals/ml) | 5.8 × 108 | 1.7 × 107 | 2.4 × 108 | 4.3 × 108 | |
| Crystal size (μm) | 1 × 1 × 1 | 20 × 20 × 30 | 5 × 5 × 5 | 2 × 2 × 4 | |
| Nozzle size (μm) | 50 | 130 | 100 | 50 | |
| Flow rate (μl/min) | 0.43 | 0.75 | 0.42 | 0.47 | |
| Data collection | |||||
| wavelength (Å) | 1.24 | 0.95 | 1.77 | 1.24 | |
| Space group | P43212 | P43212 | P43212 | P41212 | |
| Unit-cell parameter | |||||
| a = b (Å) | 80.0 | 79.6 | 79.6 | 58.5 | |
| c (Å) | 38.4 | 38.3 | 38.2 | 151.6 | |
| Number of collected images | 149,938 | 107,856 | 105,769 | 101,383 | |
| Number of hits | 41,575 | 58,321 | 30,929 | 55,751 | |
| Number of indexed images | 29,593 | 40,787 | 19,271 | 43,350 | |
| Indexing rate from hits (%) | 71.2 | 69.9 | 62.3 | 77.8 | |
| Number of merged images | 29,593 | 40,787 | 19,271 | 43,350 | |
| Number of total reflections | 4,823,284 | 21,187,517 | 3,440,102 | 24,822,961 | |
| Number of unique reflections | 12,068 | 22,415 | 8,750 | 38,328 | |
| Resolution range (Å) | 30–1.8 (1.86–1.80) | 30–1.45 (1.50–1.45) | 30–2.0 (2.07–2.00) | 30–1.55 (1.60–1.55) | |
| Completeness (%) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | |
| Multiplicity | 399.7 (283.0) | 945.2 (677.3) | 393.2 (81.9) | 647.6 (668.5) | |
| R split (%)† | 7.1 (51.0) | 5.1 (50.4) | 8.0 (53.0) | 8.6 (33.4) | |
| CC1/2 | 0.992 (0.764) | 0.995 (0.735) | 0.988 (0.654) | 0.988 (0.760) | |
| <I/σ(I)> | 10.2 (2.2) | 13.4 (2.2) | 10.5 (2.0) | 7.7 (2.0) | |
| Total amounts of protein used (mg) | 0.7 | 0.9 | 0.5 | 0.5 | |
| Refinement | |||||
| R/R free (%) | 17.5/18.4 | 18.1/19.6 | 18.1/20.2 | 12.7/15.1 | |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 | 0.006 | |
| Bond angles (°) | 1.059 | 1.071 | 1.070 | 0.984 | |
| PDB code | 5wr9 | 5wra | 5wrb | 5wr8 | |
| Protein | proteinase K | ||||
| Carrier | 16% cellulose (Pr) | 16% cellulose (native) | |||
| Crystal density (crystals/ml) | 9.3 × 107 | 4.9 × 107 | |||
| Crystal size (μm) | 4 × 4 × 4–5 × 5 × 7 | 8 × 8 × 8–12 × 12 × 12 | |||
| Nozzle size (μm) | 50 | 110 | |||
| Flow rate (μl/min) | 0.47 | 0.38 | |||
| Data collection | |||||
| wavelength (Å) | 1.24 | 0.95 | |||
| Space group | P43212 | P43212 | |||
| Unit-cell parameter | |||||
| a = b (Å) | 68.6 | 68.3 | |||
| c (Å) | 108.8 | 108.4 | |||
| Number of collected images | 180,000 | 145,000 | |||
| Number of hits | 40,503 | 59,246 | |||
| Number of indexed images | 30,930 | 47,503 | |||
| Indexing rate from hits (%) | 76.4 | 80.1 | |||
| Number of merged images | 30,000 | 3,000 | 1,000 | 32,000 | 1,000 |
| Number of total reflections | 16,961,902 | 1,540,467 | 520,503 | 18,624,772 | 545,845 |
| Number of unique reflections | 42,391 | 42,386 | 42,060 | 42,385 | 42,273 |
| Resolution range (Å) | 32.7–1.50 (1.53–1.50) | 27.2–1.50 (1.53–1.50) | |||
| Completeness (%) | 100 (100) | 100 (99.9) | 99.2 (93.3) | 100 (100) | 99.7 (99.6) |
| Multiplicity | 400.1 (151.3) | 36.3 (13.4) | 12.4 (4.8) | 439.4 (312.0) | 12.9 (9.1) |
| R split (%)† | 7.8 (44.5) | 24.4 (99.7) | 43.1 (120.8) | 7.1 (40.9) | 41.4 (189.7) |
| CC1/2 | 0.990 (0.776) | 0.896 (0.389) | 0.713 (0.272) | 0.992 (0.810) | 0.761 (0.124) |
| <I/σ(I)> | 10.2 (2.3) | 3.7 (1.3) | 2.5 (1.5) | 10.9 (2.8) | 2.3 (0.9) |
| Total amounts of protein used (mg) | 0.9 | 0.09 | 0.03 | 0.4 | 0.01 |
| Refinement | |||||
| R/R free (%) | 17.6/19.3 | ||||
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.009 | ||||
| Bond angles (°) | 1.004 | ||||
| PDB code | 5wrc | ||||
† .
Figure 1.
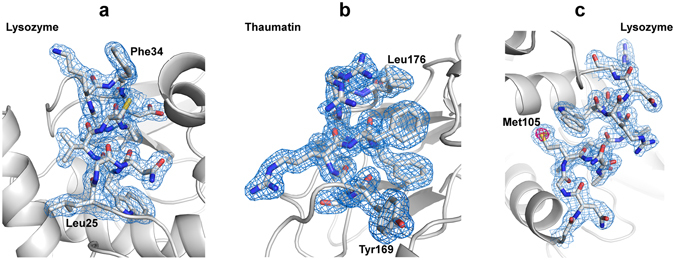
Electron density maps of lysozyme and thaumatin. Close-up views of (a) the lysozyme structure at 1.8-Å resolution and (b) the thaumatin structure at 1.55-Å resolution for the sample delivered in a cellulose matrix and (c) the lysozyme structure at 2.0-Å resolution for the sample delivered in a nuclear grease matrix with 2F o – F c electron density maps contoured at the 1.0 σ level (coloured blue). An anomalous difference Fourier map contoured at the 4.0 σ level (coloured magenta) shows the sulfur atom of Met105 in (c). These figures were drawn with PyMol (http://www.pymol.org).
In this study, 16% (w/v) and 22% (w/v) cellulose matrices were used for the small sized lysozyme (1 × 1 × 1 μm) and thaumatin (2 × 2 × 4 μm) crystals, respectively. The cellulose matrix with randomly oriented crystals was extruded through an injector nozzle with an inner diameter (i.d.) of 50 μm. On the other hand, for the larger lysozyme crystals (20 × 20 × 30 μm), an 11% (w/v) cellulose matrix was extruded through a 130-μm-i.d. nozzle. The cellulose matrix formed a stable flow for all protein samples (an example: Supplementary Fig. 2a). The matrix at low cellulose concentrations (less than ~5%) cannot be extruded from our injector system as a continuous sample column. On the other hand, a matrix at a cellulose concentration (~30%) becomes too hard for micro-extrusion. The cellulose concentration therefore was preferably ~10–20%. The sample preparation in our technique can be performed by simply mixing with matrix medium. Although the medium mixing technique using a syringe coupler may prevent crystal dehydration23, 29, the technique could cause mechanical damage to brittle crystals by physical contact between the crystals and the coupler interior surface, resulting in a deterioration of diffraction quality. In such cases, a simple, quick mixing with a spatula on a glass slide21 would be better to preserve the crystals. The cellulose matrix has lower background scattering (Supplementary Fig. 3a) compared to the conventional grease matrix, the synthetic grease Super Lube (Supplementary Fig. 3b) generated diffuse scatterings in the resolution range of 4–5 Å, and LCP14 (Fig. 2), while the cellulose matrix gives a slightly higher background scattering in the resolution range of ~3.5–2.5 Å. There were no significant differences between cellulose and hyaluronic acid matrices26, suggesting that polysaccharide hydrogels tend to have lower background scattering. However, the cellulose matrix is less adhesive than the hyaluronic acid matrix and prevents clogging of the sample-vacuum nozzle as a sample catcher22 (Supplementary Fig. 2) and adhesion of the matrix to the injector nozzle surface in our injector system. In addition, hyaluronic acid is more expensive compared to hydroxyethyl cellulose, up to ~1,000 times the price per gram. Hydrogels, LCP and Vaseline can be extruded as a continuous column with an approximately same diameter as a 50-μm-i.d. (or less) injector nozzle size. On the other hand, grease matrix tends to produce a column larger than the nozzle i.d. A sample column with a smaller diameter (~50 μm) contributes to the reduction of sample consumption and background scattering from the matrix26. A matrix with low background scattering is important to collect a high-resolution data set from ~1 μm (or less) crystals.
Figure 2.
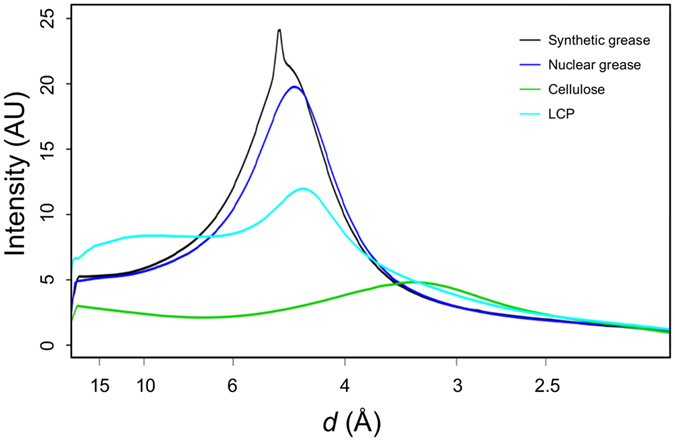
The average background scattering intensities of ~2,000 images from each matrix. Super Lube synthetic grease, Super Lube nuclear grease, 16% (w/v) hydroxyethyl cellulose solution and LCP are depicted by the black, blue, green and cyan lines, respectively.
De novo phasing
Crystallographic phasing for routine structure determination remains a challenge in SFX. In this study, using the cellulose matrix, we attempted the de novo phasing of proteinase K. We collected ~180,000 images from the microcrystals (size 4 × 4 × 4–5 × 5 × 7 μm) of Pr-derivatized proteinase K (Supplementary Fig. 1c) at a wavelength of 1.24 Å (Table 1). We successfully indexed and integrated approximately 31,000 images in space group P43212. The dataset extended to 1.5-Å resolution with a completeness of 100% and a CC1/2 of 0.990. The overall <I/σ(I)> of the merged observations was 10.2. Substructure determination and phasing were performed by SHELXD and SHELXE 30. We succeeded in locating two Pr ions in the asymmetric unit and could solve the substructure at 2.0-Å resolution, but not at 2.2-Å resolution. The two Pr-binding sites were identical to those of the calcium ions in the native structure (Fig. 3), indicating that the two calcium atoms were replaced by the Pr atoms31. The coordinates of the heavy atoms were employed for both the refinement and the phase calculation at 1.8-Å resolution in SHEXLE. A polyalanine model of proteinase K was automatically traced by SHELXE. Subsequently, 99% (277 of 279 residues) of the structure was automatically modelled with side chains by Buccaneer 32. Finally, we refined the structure at 1.5-Å resolution to an R/R free of 17.6/19.3% (PDB ID: 5wrc). The expected magnitude of the anomalous signal (<|ΔF ano|>/<|F|>) is ~4.8% at 10 keV based on the formula in Hendrickson & Teeter33 and Dauter et al.34.
Figure 3.
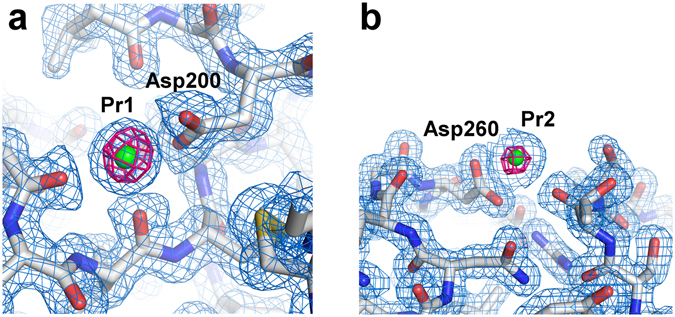
Electron density maps of proteinase K. (a,b) Close-up views of Pr ion binding sites with 2F o – F c electron density maps contoured at the 1.0 σ level (coloured blue). Bound Pr ions are depicted as a green sphere. The anomalous difference Fourier maps using 3,000 images (contoured at the 6.0σ level) are shown in magenta. These figures were drawn with PyMol (http://www.pymol.org).
We found that 3,000 indexed images were sufficient for SAD phasing of proteinase K crystals. In this phasing, we used the first 3,000 of 30,930 indexed images, without deliberate selection of the best images. SHELXD located only one Pr atom in the asymmetric unit, when 3,000 indexed images were used. A polyalanine model from SHELXE at 1.7-Å resolution was completed in Buccaneer. We obtained 99% of the complete model. The final anomalous difference Fourier maps using 3,000 images in Fig. 3 display significant anomalous peak heights (17.1 and 11.2σ, obtained from ANODE 35) of the two Pr atoms.
Next, we employed single-isomorphous replacement (SIR) and SIR with anomalous scattering (SIRAS) for phasing. We obtained a data set (32,000 indexed images) from native crystals of proteinase K at a wavelength 0.95 Å36, at a different beam time using different crystallization batches, at 1.5-Å resolution with a completeness of 100%, a CC1/2 of 0.992. Only 2,000 images in total (native/derivative: 1,000/1,000) were sufficient for SIR and SIRAS phasing of proteinase K, while SAD phasing required 3,000 images. The CC1/2 value of the 1,000-image derivative dataset was only 71.3% (27.2% for 1.53–1.50 Å), while that of the full dataset was 99.0% (77.6% for 1.53–1.50 Å) (Supplementary Fig. 4). As shown in Fig. 4, a combination of the native dataset with the derivative dataset boosted the peak heights in the anomalous difference map and allowed phasing from fewer images than using derivative images alone. This is in good agreement with the result from the previously reported I-SAD phasing of a membrane protein bacteriorhodopsin using an iododetergent37.
Figure 4.
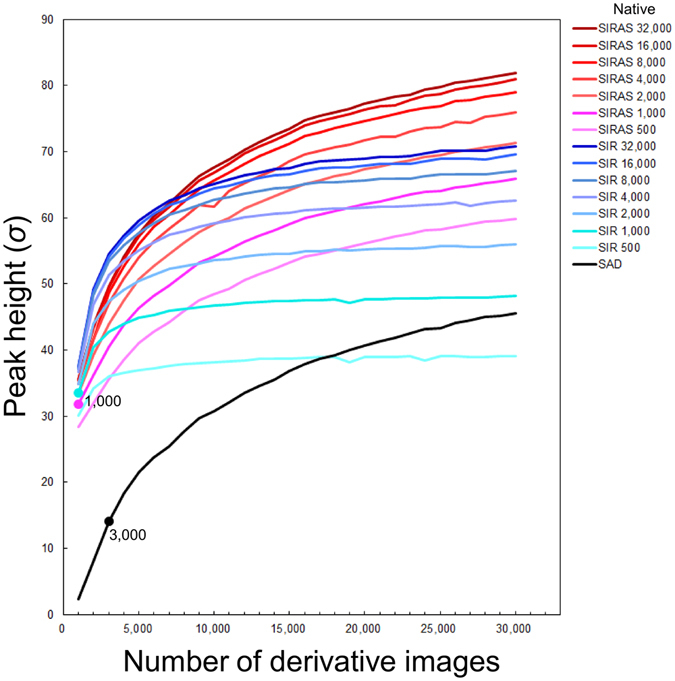
Improvement of anomalous difference peak heights with the number of derivative images. The plot of the sum of the anomalous peak heights from two Pr atoms. SAD, SIR and SIRAS phasing are shown in the black, blue and red lines, respectively. For SIR and SIRAS phasing, the number of native images were varied from 500 to 32,000. Filled circles indicate the minimum number of derivative images necessary for each phasing method and number of native images.
In SFX, de novo phasing for heavy atom-derivatized proteions has been demonstrated16, 37–42. In addition, native sulfur SAD phasing was also achieved40, 43, 44. These results indicate that de novo phasing is now routinely available for SFX. Our cellulose matrix with low background scattering noise is compatible with the accurate measurement of weak anomalous signals essential for de novo phasing from SFX data.
A novel grease matrix with low background scattering
To reduce background scattering from conventional grease matrix21, 26, we introduced a novel grease matrix, Super Lube nuclear grade approved grease (nuclear grease). For lysozyme crystals (5 × 5 × 5 μm), we were able to collect ~100,000 images in approximately 1 hour at a wavelength 1.77 Å (Table 1). We indexed and integrated ~19,000 images for the lysozyme crystals. The crystals yielded data sets at 2.0-Å resolution with a completeness of 100% and a CC1/2 of 0.988. We determined and refined the crystal structure of lysozyme (PDB ID: 5wrb) at 2.0-Å resolution.
The conventional grease matrices (mineral-oil based AZ grease and untreated Super Lube synthetic grease without grinding treatment) extruded through a 110-μm-i.d. nozzle tended to produce a larger-diameter grease column (approximately ~210 μm) about the size of the outer diameter (o.d.) of the nozzle21, 26. On the other hand, the nuclear grease matrix was extruded as a continuous column with a diameter of ~100 μm through a 100-μm-i.d. nozzle (Supplementary Fig. 2b). The Super Lube synthetic grease tended to give a stronger diffraction ring at ~4.8-Å resolution in about 30% of all diffraction images (Fig. 2 and Supplementary Fig. 3b)26. Weaker background scattering was noted when using nuclear grease compared with Super Lube synthetic grease (Fig. 2 and Supplementary Fig. 3c). In the lysozyme structure with the nuclear grease matrix, we observed a weak anomalous scattering signal from sulfur atoms (e.g. the sulfur atom of Met105, Fig. 1c). On the other hand, an anomalous signal from the sulfur atoms in the proteinase K structure from ~20,000 indexed images was not discernible when using the conventional Super Lube synthetic grease matrix26. Using a wide variety of proteins, the adaptability of grease matrix has been demonstrated in SFX15, 16, 18, 21, 26, 37, 39, 43, 45. These results suggest that grease has potential as a versatile matrix carrier, but some crystals are incompatible with the grease matrix. The cellulose and hyaluronic acid matrices provide alternatives for grease-sensitive protein crystals. Grease and hydrogel crystal carriers are thus complementary (Table 2).
Table 2.
Crystal carrier media for serial sample loading.
| Media | Advantages | Disadvantages | References |
|---|---|---|---|
| Oil | general versatility | higher background scattering | |
| Nuclear grade grease | lower background scattering among grease matrices | salt-like impurities in grease | this study |
| Synthetic grease | higher versatility | gives a stronger diffraction ring at ~4.8 Å | 26 |
| Mineral-oil based grease | higher versatility | a larger diameter sample column | 21 |
| Vaseline (petroleum jelly) | a smaller diameter sample column (~40 μm) | gives stronger diffraction rings at 4.2 and 3.8 Å | 22 |
| Hydrogel | lower background scattering | damage to crystals by osmotic shock | |
| Hydroxyethyl cellulose | simple preparation | adhesive | this study |
| Hyaluronic acid | simple preparation | strongly adhesive, expensive | 26 |
| Agarose | compatible with proteins | requires heat treatment at temperatures higher than 85 °C as a pre-preparation | 23 |
| Other | |||
| LCP (e.g., monoolein) | applicable to soluble and membrane proteins | higher background scattering, but lower than grease in the resolution range of 4–5 Å | 20, 22, 23 |
Using the cellulose matrix as a general protein carrier, we obtained the structures of soluble proteins beyond 1.8-Å resolution at room temperature. We have successfully applied Pr-SAD, SIR and SIRAS phasing to SFX, using 3,000 indexed images for SAD and 2,000 images for SIR and SIRAS, demonstrating that we can accurately measure anomalous signals. Matrix carriers with a stable sample flow and a small diameter sample column have various application in SFX experiments such as femtosecond to millisecond time-resolved studies of light-driven structural changes, and chemical dynamics using pump-probe techniques14, 18, 46–50.
Materials and Methods
Sample preparation
Using a 20 mg ml−1 lysozyme solution, the crystals with a size of 1 × 1 × 1 μm, 5 × 5 × 5 μm and 20 × 20 × 30 μm were prepared following previously reported protocols21, except for the incubation temperature during crystallization at 12, 17 and 26 °C for 10 min, respectively. Thaumatin I was purified from crude thaumatin powder as described previously51. Thaumatin crystallization was performed using the batch method. Microcrystals (2 × 2 × 4 μm) were obtained by mixing in an ice bath an equal volume of the 40 mg ml−1 protein solutions and the reservoir solution, which consisted of 20 mM N-(2-acetamido) iminodiacetic acid (ADA) and 2.0 M potassium sodium tartrate (pH 7.3). Proteinase K from Engyodontium album (No. P2308, Sigma) at a concentration of 40 mg ml−1 was crystalized by previously reported protocols26. For Pr-derivatized proteinase K, a 100 μl sample of the crystal solution was added to a 100 μl heavy-atom solution comprised of 50 mM PrCl3, 0.5 M NaNO3 and 50 mM MES–NaOH (pH 6.5). The solution was then incubated at 20 °C for 90 min. To determine a crystal number density of the crystal solution, we counted the number of crystals in the solution using a hemocytometer (OneCell, cat. no. OC-C-S02) under a Hirox digital microscope (Hirox, KH-8700). The crystal number density was adjust to an approximately 107–108 crystals ml−1.
In this study, we used hydroxyethyl cellulose (mw ~250,000, No. 09368, Sigma) as the crystal carrier matrix. Protein microcrystals were prepared according to the following procedures. For lysozyme and proteinase K crystals, after a 100-μl sample of storage solution was centrifuged at ~1,300–3,000 × g for 10 sec using a compact tabletop centrifuge, a 40-μl aliquot of supernatant solution was dispensed into 50 μl of 32% (w/v) hydroxyethyl cellulose aqueous solution for lysozyme (1 × 1 × 1 μm) and proteinase K, or 22% (w/v) hydroxyethyl cellulose aqueous solution for lysozyme (20 × 20 × 30 μm) on a glass slide and then mixed with a spatula for ~15 sec. After a 50-μl aliquot of the remaining supernatant solution was removed, a 10-μl aliquot of the crystal solution was dispensed into 90 μl of the hydroxyethyl cellulose solution and then mixed for ~15 sec. For thaumatin crystals, after a 100-μl sample of storage solution was centrifuged at ~1,300–3,000 × g for 10 sec using a compact tabletop centrifuge, a 90-μl aliquot of supernatant solution was removed. A 10-μl aliquot of the crystal solution was dispensed into 90 μl of 24% (w/v) hydroxyethyl cellulose aqueous solution on a glass slide and then mixed for ~15 sec. For the grease matrix, the lysozyme crystals (5 × 5 × 5 μm) were mixed with the Super Lube nuclear grade grease (No. 42150, Synco Chemical Co.) using the same procedure reported by Sugahara et al.21 The grease was filtered through 10 μm mesh (No. 06-04-0041-2314, CellTrics) before mixing with protein crystals to remove salt-like impurities in the grease. We performed this matrix preparation immediately before SFX experiments.
Data collection
We carried out the experiments using femtosecond X-ray pulses from SACLA27. The X-ray wavelength was 0.95, 1.24 or 1.77 Å (13, 10 or 7 keV) with a pulse energy of ~200 μJ. Each X-ray pulse delivers ~7 × 1010 photons within a 10-fs duration (FWHM) at a wavelength of 1.77 Å (7 keV) to the matrices. Data were collected using focused X-ray beams of 1.5 × 1.5 μm2 by Kirkpatrick-Baez mirrors52. The crystals in a cellulose or grease matrix were serially loaded using a high viscosity micro-extrusion injector system installed in a helium ambiance, diffraction chamber. The experiments were carried out using a Diverse Application Platform for Hard X-ray Diffraction in SACLA (DAPHNIS)28 at BL353. The microcrystals embedded in the matrix were kept at a temperature of approximately 20 °C in the micro-extrusion injector. The sample chamber was kept at a temperature of ~26 °C and a humidity greater than 50%. Diffraction images were collected using a custom-built 4M pixel detector with multi-port CCD sensors54. The matrix with randomly oriented crystals was extruded through injector nozzles with inner diameters (i.d.) of 50, 100, 110 or 130 μm (Table 1). Data collection was guided by realtime analysis by the SACLA data processing pipeline55.
Background intensity determination
The background intensities from Super Lube synthetic grease, Super Lube nuclear grease and hydroxyethyl cellulose through a 100-μm-i.d. nozzle at 1.77 Å and that from LCP14 through a 75-μm-i.d. nozzle at 1.61 Å were determined by a procedure similar to that used in Conrad et al.23 Details of the calculation have been described previously26. Diffraction images for LCP were retrieved from CXIDB56 (http://www.cxidb.org/) #53.
Structure determination
Diffraction images were filtered and converted by Cheetah 57 adapted55 for the SACLA data acquisition system58. Diffraction peak positions were determined using the built-in Zaefferer algorithm and passed on to DirAx 59 for indexing. No sigma cutoff or saturation cutoff were applied. Measured diffraction intensities were merged by process_hkl in the CrystFEL suite60 with scaling (–scale option). The structures of lysozyme and thaumatin were determined by difference Fourier synthesis using search models (PDB: 3WUL for lysozyme, and 3X3P for thaumatin). For Pr-derivatized proteinase K, substructure search, phasing and phase improvement were carried out using the SHELX C, D and E programs30. The autotraced model from SHELXE was fed into Buccaneer 32 from the CCP4 suite61. Manual model revision and structure refinement were performed using Coot 62 and PHENIX 63, respectively. Details of the data collection and refinement statistics are summarized in Table 1.
Electronic supplementary material
Acknowledgements
The XFEL experiments were carried out at the BL3 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal nos 2015A8026, 2015A8048, 2015B8029, 2015B8042, 2015B8046 and 2015B8047). This work was supported by the X-ray Free-Electron Laser Priority Strategy Program (MEXT), partly by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (KAKENHI No. 25650026), partly by the Research Acceleration Program of Japan Science and Technology Agency and partly by the Platform for Drug Discovery, Informatics, and Structural Life Science (MEXT). C.S. is supported by National Research Foundation of Korea (grants NRF-2015R1A5A1009962 and NRF-2016R1A2B3010980). The authors thank Mr. Kazunori Hata for expansion of the grease matrix method and the SACLA beamline staff for technical assistance. We are grateful for the computational support from the SACLA HPC system and the Mini-K super computer system.
Author Contributions
M. Sugahara conceived the research, M. Sugahara, T.M. K.N. and E.N. prepared the microcrystals, M. Sugahara, T.M. M. Suzuki, S. Inoue, C.S. R.T. T. Nakatsu, E.M. and F.Y. performed the data collection, M. Sugahara, T.M. and M. Suzuki performed data processing and solved the structure for lysozyme and thaumatin proteins, T. Nakane and O.N. performed data processing and solved the structure for proteinase K, developed the data processing pipeline and performed the background scattering data analysis, K.T. Y.J. T.K. T.H. and M.Y. developed the DAPHNIS and detectors, M. Sugahara, T. Nakane, T.M. M. Suzuki, S. Inoue, C.S. and K.N. wrote the manuscript with input from all of the coauthors and S. Iwata coordinated the project.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00761-0
Accession codes: The coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 5wr9, 5wra and 5wrb for lysozyme, 5wr8 for thaumatin and 5wrc for proteinase K. Diffraction images have been deposited to CXIDB under ID 45 (proteinase K, native), 48 (proteinase K, derivative), 49 (thaumatin) 47 (lysozyme, grease) and 50 (lysozyme, cellulose).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 3.Emma P, et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nat. Photonics. 2010;4:641–647. doi: 10.1038/nphoton.2010.176. [DOI] [Google Scholar]
- 4.Schlichting I, Miao J. Emerging opportunities in structural biology with X-ray free-electron lasers. J. Curr. Opin. Struct. Biol. 2012;22:613–626. doi: 10.1016/j.sbi.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barty A, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlichting I. Serial femtosecond crystallography: the first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuda Y, et al. Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography. J. Biochem. 2016;159:527–538. doi: 10.1093/jb/mvv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson LC, et al. Lipidic phase membrane protein serial femtosecond crystallography. Nat. Methods. 2012;9:263–265. doi: 10.1038/nmeth.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339:227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, et al. Serial femtosecond crystallography of G protein–coupled receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, et al. Architecture of the synaptotagmin–SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nango E, et al. A three dimensional movie of structural changes in bacteriorhodopsin. Science. 2016;354:1552–1557. doi: 10.1126/science.aah3497. [DOI] [PubMed] [Google Scholar]
- 15.Colletier JP, et al. Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein IrisFP. J. Phys. Chem. Lett. 2016;7:882–887. doi: 10.1021/acs.jpclett.5b02789. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda Y, et al. Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc. Natl. Acad. Sci. USA. 2016;113:2928–2933. doi: 10.1073/pnas.1517770113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stagno JR, et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017;541:242–246. doi: 10.1038/nature20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suga M, et al. Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL. Nature. 2017;543:131–135. doi: 10.1038/nature21400. [DOI] [PubMed] [Google Scholar]
- 19.DePonte DP, et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D Appl. Phys. 2008;41:195505. doi: 10.1088/0022-3727/41/19/195505. [DOI] [Google Scholar]
- 20.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods. 2015;12:61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 22.Botha S, et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta Cryst. 2015;D71:387–397. doi: 10.1107/S1399004714026327. [DOI] [PubMed] [Google Scholar]
- 23.Conrad CE, et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ. 2015;2:421–430. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stellato F, et al. Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ. 2014;1:204–212. doi: 10.1107/S2052252514010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogly P, et al. Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ. 2015;2:168–176. doi: 10.1107/S2052252514026487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugahara M, et al. Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Sci. Rep. 2016;6:24484. doi: 10.1038/srep24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa T, et al. A compact X-ray free-electron laser emitting in the sub-ångström region. Nat. Photonics. 2012;6:540–544. doi: 10.1038/nphoton.2012.141. [DOI] [Google Scholar]
- 28.Tono K, et al. Diverse application platform for hard X-ray diffraction in SACLA (DAPHNIS): application to serial protein crystallography using an X-ray free-electron laser. J. Synchrotron Rad. 2015;22:532–537. doi: 10.1107/S1600577515004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids. 1998;95:11–21. doi: 10.1016/S0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. 2010;D66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yazawa K, et al. Derivatization of Proteinase K with Heavy Atoms Enhances Its Thermal Stability. ACS Catalysis. 2016;6:3036–3046. doi: 10.1021/acscatal.6b00100. [DOI] [Google Scholar]
- 32.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. 2006;D62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Hendrickson WA, Teeter MM. Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulphur. Nature. 1981;290:107–113. doi: 10.1038/290107a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dauter Z, Dauter M, de La Fortelle E, Bricogne G, Sheldrick GM. Can anomalous signal of sulfur become a tool for solving protein crystal structures? J. Mol. Biol. 1999;289:83–92. doi: 10.1006/jmbi.1999.2743. [DOI] [PubMed] [Google Scholar]
- 35.Thorn A, Sheldrick GM. ANODE: anomalous and heavy-atom density calculation. J. Appl. Cryst. 2011;44:1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakane T, et al. Membrane protein structure determination by SAD, SIR or SIRAS phasing in serial femtosecond crystallography using a novel iododetergent. Proc. Natl. Acad. Sci. USA. 2016;113:13039–13044. doi: 10.1073/pnas.1602531113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barends TRM, et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505:244–247. doi: 10.1038/nature12773. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita K, et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Sci. Rep. 2015;5:14017. doi: 10.1038/srep14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nass K, et al. Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data. IUCrJ. 2016;3:180–191. doi: 10.1107/S2052252516002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colletier JP, et al. De novo phasing with X-ray laser reveals mosquito larvicide BinAB structure. Nature. 2016;539:43–47. doi: 10.1038/nature19825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter MS, et al. Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser. Nat. Commun. 2016;7:13388. doi: 10.1038/ncomms13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakane T, et al. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Cryst. 2015;D71:2519–2525. doi: 10.1107/S139900471501857X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batyuk A, et al. Native phasing of x-ray free-electron laser data for a G protein–coupled receptor. Sci. Adv. 2016;2:e1600292. doi: 10.1126/sciadv.1600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edlund P, et al. The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography. Sci. Rep. 2016;6:35279. doi: 10.1038/srep35279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nogly P, et al. Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography. Nat. Commun. 2016;7:12314. doi: 10.1038/ncomms12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barends TRM, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350:445–450. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- 47.Kern J, et al. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nat. Commun. 2014;5:4371. doi: 10.1038/ncomms5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuda T, Ohta K, Mikami B, Kitabatake N. High-resolution structure of the recombinant sweet-tasting protein thaumatin I. Acta Cryst. 2011;F67:652–658. doi: 10.1107/S174430911101373X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yumoto H, et al. Focusing of X-ray free-electron laser pulses with reflective optics. Nat. Photonics. 2013;7:43–47. doi: 10.1038/nphoton.2012.306. [DOI] [Google Scholar]
- 52.Tono K, et al. Beamline, experimental stations and photon beam diagnostics for the hard x-ray free electron laser of SACLA. New J. Phys. 2013;15:083035. doi: 10.1088/1367-2630/15/8/083035. [DOI] [Google Scholar]
- 53.Kameshima T, et al. Development of an X-ray pixel detector with multi-port charge-coupled device for X-ray free-electron laser experiments. Rev. Sci. Instrum. 2014;85:033110. doi: 10.1063/1.4867668. [DOI] [PubMed] [Google Scholar]
- 54.Nakane T, et al. Data processing pipeline for serial femtosecond crystallography at SACLA. J. Appl. Cryst. 2016;49:1035–1041. doi: 10.1107/S1600576716005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maia FRNC. The Coherent X-ray Imaging Data Bank. Nat. Methods. 2012;9:854–855. doi: 10.1038/nmeth.2110. [DOI] [PubMed] [Google Scholar]
- 56.Barty A, et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Cryst. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joti Y, Kameshima T, Yamaga M, Sugimoto T, Okada K. Data acquisition system for X-ray free-electron laser experiments at SACLA. J. Synchrotron Rad. 2015;22:571–576. doi: 10.1107/S1600577515004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duisenberg AJM. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Cryst. 1992;25:92–96. doi: 10.1107/S0021889891010634. [DOI] [Google Scholar]
- 59.White TA, et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Cryst. 2012;45:335–341. doi: 10.1107/S0021889812002312. [DOI] [Google Scholar]
- 60.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D66, 213–221 (2010). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


