Abstract
Genome-scale metabolic models (GSMMs) constitute a platform that combines genome sequences and detailed biochemical information to quantify microbial physiology at the system level. To improve the unity, integrity, correctness, and format of data in published GSMMs, a consensus IMGMD database was built in the LAMP (Linux + Apache + MySQL + PHP) system by integrating and standardizing 328 GSMMs constructed for 139 microorganisms. The IMGMD database can help microbial researchers download manually curated GSMMs, rapidly reconstruct standard GSMMs, design pathways, and identify metabolic targets for strategies on strain improvement. Moreover, the IMGMD database facilitates the integration of wet-lab and in silico data to gain an additional insight into microbial physiology. The IMGMD database is freely available, without any registration requirements, at http://imgmd.jiangnan.edu.cn/database.
Introduction
Genome-scale metabolic models (GSMMs) are a kind of a mathematical model that integrates multiple types of omics data, such as genomics, transcriptomics, proteomics, and metabolomics. GSMMs can clarify the relations among genes, proteins, and reactions. Models can be used to describe all biochemical reactions, metabolites, and genes involved in the metabolism of a specific organism. They have been used to decipher metabolic, regulatory, and signaling networks at the whole-organism level1–3. Since the first GSMM of Haemophilus influenzae Rd was constructed in 19994, more than 300 GSMMs for over 100 organisms have been built5, 6.
GSMMs have been developed for over 15 years, and four steps are involved in their construction: creation of a draft model, manual refinement, conversion to a mathematical format, and network evaluation6–9. Nonetheless, owing to differences in nomenclature10, integrity, and correctness11 as well as the format12 of published GSMMs, these models cannot be directly applied by other researchers. To reduce the manual labour needed for model construction and to increase the quality of GSMMs, some databases and tools, such as BIGG Models11 and MetaNetX13, have been developed. Nonetheless, they are limited by the quality and quantity of models. For example, BIGG Models includes only 80 high-quality GSMMs (as of 30 November 2016), which is far from the number of published models. In MetaNetX, only 24 (14.7%) models have been published, which were validated by experimental results (http://www.metanetx.org/cgi-bin/mnxweb/repository).
In this study, we built a database named In silico Microbial Genome-scale Metabolic Models (IMGMD) in the LAMP (Linux + Apache + MySQL + PHP) system. It provides a platform for integration and standardisation of all published microbial GSMMs. In IMGMD, users are not only able to browse and download standardised GSMMs but can also reconstruct GSMMs automatically. In addition to pathway mining and mutation library functions, users can access information that can guide pathway design and metabolic target identification.
Results and Discussion
Database content and web interface
IMGMD (http://imgmd.jiangnan.edu.cn/database/) has a user-friendly website for the following applications: (1) It can be used to download standardised GSMMs; this module integrates model-related information, such as gene–protein–reaction relations, genome information, and references (Fig. 1A). (2) It enables auto-reconstruction of GSMMs; this tool is based on homology alignments, and only sequences that meet a threshold are used for model construction. Additionally, transport proteins and sub-cellular location are identified for further model refinement (Fig. 1B). (3) It can be applied to explore potential pathways; using this function, users can explore the potential pathways from one metabolite to another in a certain GSMM (Fig. 1C). (4) It guides metabolic engineering; the mutation library includes in silico and in vivo metabolic engineering results, and accordingly, it provides guidance for target searches (Fig. 1D).
Figure 1.
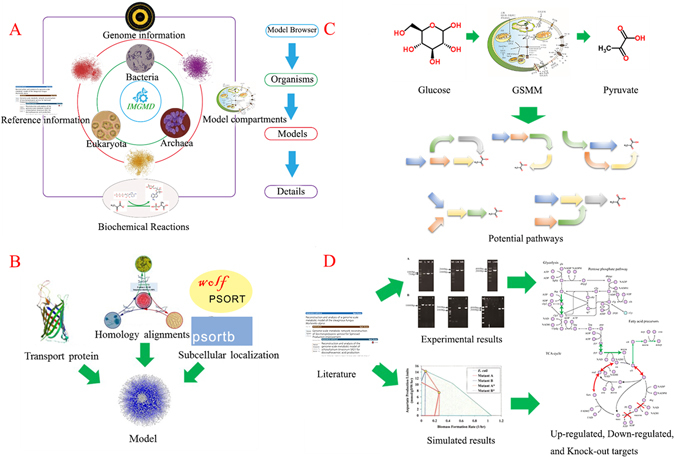
Summary of four functional modules in IMGMD. (A) The ‘model browser’ function; (B) the process of model auto-reconstruction; (C) pathway mining in a certain model organism; (D) gathering experimental and simulated results to identify metabolic engineering targets.
All web interfaces of the IMGMD database were tested in various browsers, such as Google Chrome, Mozilla Firefox, Internet Explorer, Opera, and Safari on Windows or Linux platforms. Despite minor differences in appearance, all tools functioned normally in all the tested browsers and on all platforms. Among the browsers tested, Google Chrome and Mozilla Firefox provided the best user experience. Hence, we recommend that users access the database using one of these two browsers.
The ‘model browse’ function in IMGMD
Using model browse, users can browse, search, and download almost all published microorganism models. From the main page of model browse, basic model information, such as the number of genes, reactions, and metabolites can be accessed. Using the search bar, models can be queried by organism name, model name, kingdom, or year of publication. We chose Saccharomyces cerevisiae as an example to demonstrate the use of ‘Search for organism’. All 8 S. cerevisiae models are returned. Then, by clicking on ‘Saccharomyces cerevisiae S288c’ for model iND750, a user can find detailed information about the organism (e.g., strain, genome information, and ORFs), model (e.g., model name, cell compartments, model download, and in silico media for simulation), and reference (e.g. reference name, journal name, and publication date; Fig. 2). The genome information is linked to the NCBI database14, which contains the genome assembly and annotation report for a microorganism. ORFs are linked to the protein sequence downloaded from the UniProt database15. The in silico media are linked to the MediaDB database16, a database of microbial growth conditions in defined media, which can be applied as the constraint condition for metabolic model growth. These standardised GSMMs in IMGMD can be further applied to many analyses using the COBRA Toolbox17–20. The ‘model browse’ module attempts to integrate scattered data on organisms, models, and literature, and promotes the establishment of GSMM standardisation.
Figure 2.
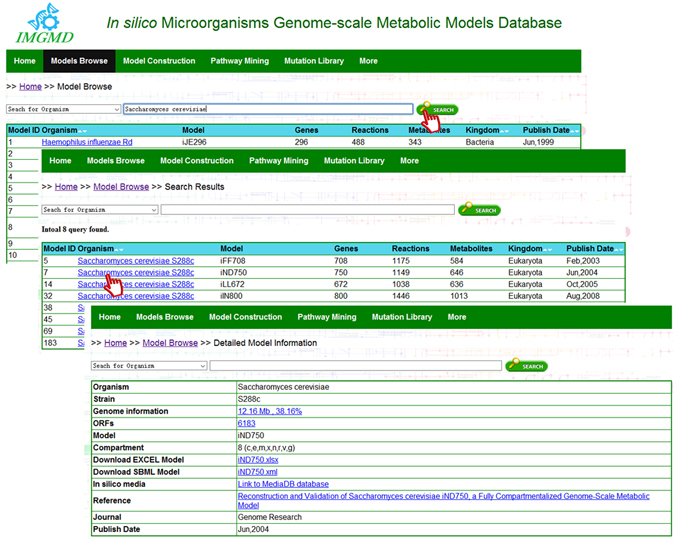
The ‘models browse’ module in IMGMD. Search results for Saccharomyces cerevisiae in model browse, and the detailed information on model iND750.
The ‘model auto-construction’ function in IMGMD
Five steps are needed to construct a model in IMGMD: (1) choosing three models for reference; (2) uploading the genome sequence; (3) choosing a threshold (eukaryotic: identity ≥40%, identity ≤10E-30; prokaryotic: identity ≥30%, identity ≤10E-6); (4) entering an e-mail address to receive results (optional); (5) submitting the job to the IMGMD database. Once the job is complete, the results contain three parts, including the model, transport proteins, and prediction of protein subcellular localisation (Fig. 3). Model construction is automatically implemented on the basis of the sequence alignment results. After protein sequences are submitted, the local BLASTP program will calculate the sequence similarity. Sequences that meet the established threshold are automatically screened using a Python script written in our lab. Based on the local Blast results, genes with high similarity are replaced in the reference models. Additionally, transport proteins are identified according to the alignment results, using the TCDB database21. For eukaryotic organisms, WoLF PSORT22 was chosen, whereas for prokaryotic organisms (gram-positive, gram-negative, or Archaea), PSORTb23 was employed to predict protein subcellular localisation.
Figure 3.
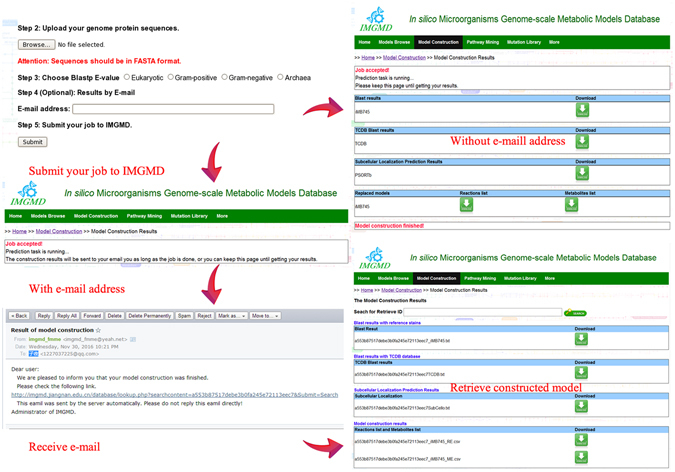
Flow chart for model construction in the IMGMD database.
Although some software or platforms for model auto-reconstruction have been developed, including ModelSEED24, RAVEN25, COBRA Toolbox26, SuBliMinal27, these tools have their advantages and disadvantages12. For instance, ModelSEED (http://modelseed.org/) is a Web service that includes the RAST genome annotation tool. Based on the annotation results, a model for a specific organism can be reconstructed automatically. Given that the RAST service (http://rast.nmpdr.org/rast.cgi) can annotate only prokaryotes, ModelSEED has limited applicability to eukaryotes. Besides, model construction by ModelSEED will take a long time, according to the job numbers. IMGMD is also a web platform that serves for model construction. It is based on the results of genome homologous alignment. Users can upload a target organism’s genome sequence and choose relevant parameters. After submission of the job to IMGMD, results will be returned within 1 day. Nonetheless, a model constructed by IMGMD is a draft model. It still needs to be further processed to obtain a GSMM. The COBRA Toolbox is based on the Matlab platform, which is commonly used for model construction. The COBRA Toolbox requires users to have basic Matlab knowledge and an advanced computer configuration for model analysis (Table 1).
Table 1.
Selected characteristics of software platforms for reconstruction and simulation of metabolic networks.
| Model SEED | RAVEN | COBRA Toolbox | SuBliMinal | IMGMD | |
|---|---|---|---|---|---|
| Input | Genome annotated in RAST | Annotated genome sequence | GSMM | Species name | Species genome sequence |
| Reference Database | SEED | KEGG | N/A | KEGG, MetaCyc | IMGMD |
| Interface | Web | Matlab | Matlab | Command Line | Web |
| License | Free | Free (requires a Matlab license) | Free (requires a Matlab license) | Free | Free |
| Output | SBML, Excel | SBML, Excel | SBML, Excel | SBML | Excel |
| Supports Simulations | Yes | Yes | Yes | No | No |
Pathway mining function in IMGMD
In this module, users can explore metabolic pathways at three levels. (1) According to the input metabolites as substrates and products, total pathways from a substrate to product in a GSMM can be output. For example, in the Mortierella alpina model iCY110628, 21 pathways exist from glucose to pyruvate, indicating that in addition to the basic glycolysis pathway in M. alpina (according to the KEGG pathway29), other pathways also could generate pyruvate. On the web page of pathway-mining results, information about the substrate and production can be linked to some metabolic databases, like KEGG29, ModelSEED24, ChEBI30, and PubChem31. Besides, on the page of detailed pathway information, reactions participating in a pathway are shown, including Reaction ID, Formula, Genes, Subsystem, and EC numbers (Fig. 4). (2) Comparisons between two or more GSMMs help to understand phenotypic characteristics based on metabolic pathway differences. When comparing the pathway differences between two Archaea, Methanococcus maripaludis (iMM518)32 and Methanosarcina barkeri (iMG746)33, there were 8 and 12 pathways from glucose to pyruvate, respectively. (3) Pathways that generate highly valuable products may exist in typical organisms. To mine these potential pathways, users can choose all collected models for the search, and then choose reactions in which species and corresponding genes can serve as references for a target strain to guide strain design. Considering these three levels, the function of pathway mining may be useful in synthetic biology and systems metabolic engineering.
Figure 4.
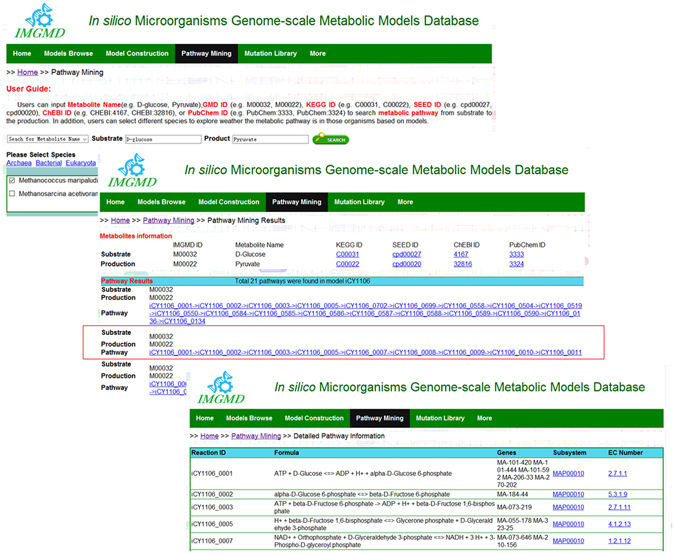
Pathway mining results for the iCY1106 model from glucose to pyruvate. A summary of all pathways found in the iCY1106 model from glucose to pyruvate, and detailed reaction information for a pathway in model iCY1106.
Mutation library function in IMGMD
The pathway prediction tool enables new pathway design for metabolic engineering; additionally, the mutation library function can be used for optimisation of the host strain. It can help to identify targets that couple cell growth with product formation, e.g., targets for gene upregulation, downregulation, and gene deletion34, 35. In IMGMD, a library that combines in vivo and in silico results to guide metabolic engineering was created.
Organisms, models, and genes can be used as keywords to search for mutation information. For example, in a search for mutation information with model iAF1260, 217 results can be found. The effect of a knockout of b4025, which encodes glucosephosphate isomerase (pgi, EC: 5.3.1.9) in E. coli, the growth rate and production rate can be viewed on another webpage (Fig. 5). According to the information on this new page, when galactose serves as a carbon source, the in silico growth decreases by 36.1%, while the in vivo growth rate increases by 12.0%36 (Table 2). Furthermore, the amino acid sequence and nucleic acid sequence of gene b4025 were also included (Fig. 5). The EC number of 5.3.1.9 is linked to BRENDA database for more detailed information.
Figure 5.
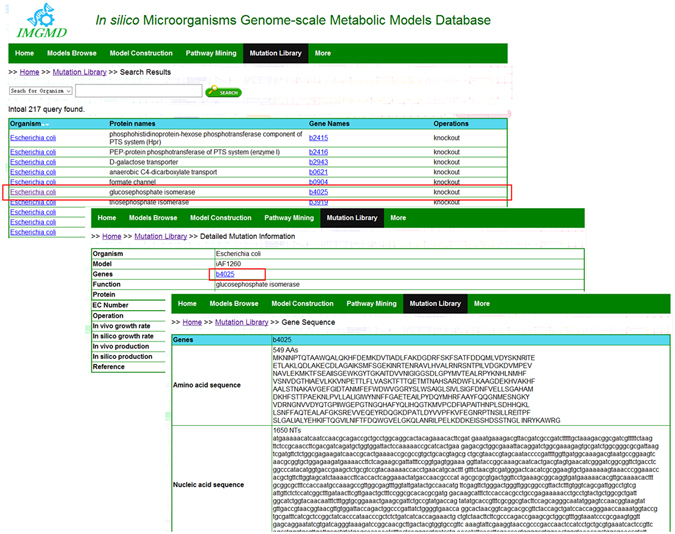
In silico and in vivo results on the b4025 deletion in the iAF1260 model, and detailed information on b4025 in E. coli.
Table 2.
Results of the b4025 deletion in E. coli collected by IMGMD36.
| strain | glucose as carbon source | galactose as carbon source | ||||||
|---|---|---|---|---|---|---|---|---|
| growth rate | H2 production (mol/mol) | growth rate | H2 production (mol/mol) | |||||
| Scaled | OD600 | Scaled | OD600 | |||||
| in silico | in vivo | in silico | in vivo | in silico | in vivo | in silico | in vivo | |
| wide type | 1.31 | 1.31 | 1.713 | 1.58 | 0.83 | 0.83 | 1.74 | 1.48 |
| Δ pgi | 1.19 | 0.02 | 1.740 | 0 | 0.53 | 0.93 | 1.835 | 1.16 |
pgi (b4025): encoding glucose-6-phosphate isomerase (EC: 5.3.1.9), which can catalyse d-glucose 6-phosphate into d-fructose 6-phosphate in glycolysis pathway.
In this module, 950 total mutation results were collected by literature mining. Additionally, 885 results (93.2%) were related to various knockout strategies, involving different algorithms, such as OptKnock19, GDLS37, ReacKnock38, DBFBA39, BAFBA40, and RobustKnock41. The remaining results are related to gene upregulation or downregulation42. Combined with the pathway mining and mutation library modules, IMGMD can be used to guide systems metabolic engineering, for both pathway screening and for target identification.
Methods
Data collection
GSMM data were collected from existing databases (http://systemsbiology.ucsd.edu/InSilicoOrganisms/OtherOrganisms, http://synbio.tju.edu.cn/GSMNDB/gsmndb.htm) and by scrutinizing the primary scientific literature (Web of Science, PubMed, and Google Scholar; Fig. 6). As of November 30th, 2016, 328 GSMMs covering 139 microorganisms have been collected. Of these models, bacteria, eukaryotes, and archaea account for 82.1%, 15.2%, and 2.7%, respectively. Additionally, based on literature mining results from 3,296 articles concerning GSMMs, a mutation library containing 950 mutations in 683 genes from 31 microorganisms was generated.
Figure 6.
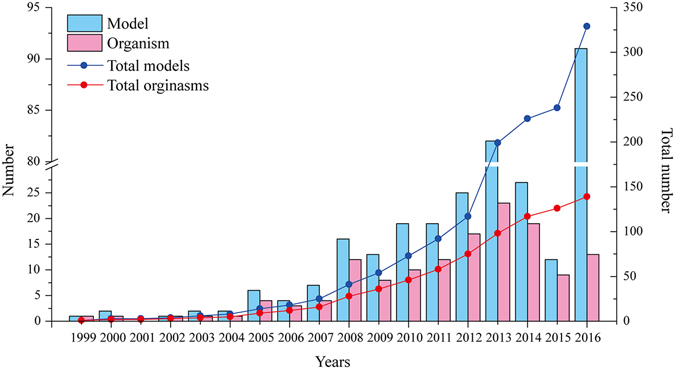
The development of GSMMs from 1999 to 2016.
Data processing
After collecting information on 328 models, 58 models could not be found, and 270 downloadable models were classified by format according to their written language, i.e., Systems Biology Markup Language (SBML), Microsoft Excel, Microsoft Word, or PDF. To read these models using the COBRA Toolbox, models in all formats were rewritten in the Excel format. Word and PDF files were manually transformed into Excel files. The SBML models were transformed into Excel, using the COBRA Toolbox on the Matlab platform. Nonetheless, during this process, some models written in SBML could not be read using the COBRA Toolbox. Eventually, 265 of total 328 (80.8%) published models were standardised in the Excel and SBML formats and can be downloaded from our database.
GSMMs consist of metabolite lists and reaction lists. Since the GSMMs were constructed by different researchers, metabolites can be represented in various forms. For example, in E. coli model iAF126043, pyruvate was represented as pyr. In Saccharomyces cerevisiae model Yeast 1.044, and Yarrowia lipolytica model iNL89545, it was indicated by PYR and s_1277, respectively. In the IMGMD database, according to their unique IDs in various biochemical databases (KEGG, SEED, ChEBI, and PubChem), the metabolites from different models were unified using IMGMD metabolite IDs. Then, 8367 metabolites from these different models were integrated. Additionally, 77.65% of metabolites can be linked to at least one of these databases (Table 3).
Table 3.
Distribution of metabolites from different metabolite databases.
| KEGG | SEED | ChEBI | PubChem | Others* | Numbers | Of total metabolites (%) |
|---|---|---|---|---|---|---|
| √ | √ | √ | √ | 4133 | 49.40 | |
| √ | √ | √ | 4136 | 49.43 | ||
| √ | √ | √ | 4205 | 50.26 | ||
| √ | √ | √ | 4296 | 51.34 | ||
| √ | √ | √ | 4133 | 49.40 | ||
| √ | √ | 4373 | 52.26 | |||
| √ | √ | 4212 | 50.34 | |||
| √ | √ | 4447 | 53.15 | |||
| √ | √ | 4163 | 49.75 | |||
| √ | √ | 4306 | 51.46 | |||
| √ | √ | 4225 | 50.50 | |||
| √ | 4620 | 55.22 | ||||
| √ | 5649 | 67.52 | ||||
| √ | 4608 | 55.07 | ||||
| √ | 4708 | 56.27 | ||||
| √ | 1871 | 22.36 |
*Others indicates that metabolites from 235 models could not be found in any of the four databases.
A list of reactions, including the Gene–Protein–Reaction relations for the models, should contain 15 columns of information, e.g., a reaction description, formula, and genes26. For the formula column, metabolites are first replaced and rearranged according to their unified IMGMD database metabolite IDs. Additionally, because some information was lacking, data (e.g., gene data) were collected by referring to information such as EC numbers, reaction descriptions, and formulas, in the other columns for the models. Finally, 17 of 19 (89.5%) GSMMs were filled out, except for models of Streptomyces lividans (GMD-TK24) and Pseudomonas putida (PpuMBEL1071). For the Formula column, metabolites were first replaced with the unified IMGMD metabolite IDs. For example, the reaction catalysed by alcohol dehydrogenase (EC: 1.1.1.1) was unified as ‘M00442[c] + M00003[c] 〈=〉 M00083[c] + M00079[c] + M00004[c]’. In all models, to arrange metabolites on the left and right of ‘〈=〉 ’ in order, a MATLAB script developed in our lab was used. Lastly, the reaction was unified as ‘M00003[c] + M00442[c] 〈=〉 M00004[c] + M00079[c] + M00083[c]’. If we ignore the cell compartments and do not count transport and exchange reactions, the database contains 21436 reactions.
During the process of literature mining, mutation information is stored in an Excel file. Information such as organisms, models, genes, operations, in vivo or in silico production, and in vivo or in silico growth rate is collected. Additionally, amino acid and nucleic acid sequences of related genes collected from the KEGG database are also stored in this Excel file.
Database design and implementation
All processed data are stored in a MySQL database and are available through a Web server built in the standard LAMP (Linux + Apache + MySQL + PHP) system to provide fast and secure data access. XAMPP for Linux 5.6.15 (https://www.apachefriends.org/index.html) was installed on CentOS Linux 5.8 (https://www.centos.org/download/). BLAST 2.2.28 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release) and Python 2.7.11 (https://www.python.org/) were used for model auto-reconstruction, and a C++ script based on a depth optimisation algorithm was written to explore the metabolic pathways in a particular GSMM.
Conclusion
The IMGMD database (http://imgmd.jiangnan.edu.cn/database) provides a platform that integrates the names of metabolites and metabolic reactions from common biochemical databases and existing model repositories. This database includes 328 models for 139 microorganisms and provides 265 standardised models for downloading. Based on a homologous sequence alignment method, models can be reconstructed automatically in the IMGMD database, which can accelerate the process of model construction. Furthermore, IMGMD provides a pathway mining tool for pathway design and a mutation library for strain optimisation.
Compared with other GSMM databases, the IMGMD database is specific for microorganisms. It is user-friendly and feature-rich; accordingly, the scientific community can easily use and extend the knowledge base. Thus, IMGMD will be a useful database for the design phase of systems metabolic engineering. Future developments include integration of the COBRA Toolbox, which will allow users to directly simulate gene deletion or over-expression, on the IMGMD platform. Besides, the IMGMD database is maintained by our lab and will be updated annually, to keep pace with the advances of GSMMs.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (No. 2014AA021501, 2014AA021701), the National Natural Science Foundation of China (No. 21422602, 31660320), the Research Program for State Key Laboratory of Food Science and Technology of Jiangnan University (SKLF-ZZA-201602).
Author Contributions
C.Y., N.X., C.D. and Y.Y. designed and developed database, X.Z. and X.C. performed data collecting and analysis. C.Y., F.G. and L.L. wrote the manuscript with contributions of all authors. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu LM, Agren R, Bordel S, Nielsen J. Use of genome-scale metabolic models for understanding microbial physiology. FEBS Lett. 2010;584:2556–2564. doi: 10.1016/j.febslet.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 2.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien EJ, Monk JM, Palsson BO. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards IJ. Systems Properties of the Haemophilus influenzae Rd Metabolic Genotype. J. Biol. Chem. 1999;274:17410–17416. doi: 10.1074/jbc.274.25.17410. [DOI] [PubMed] [Google Scholar]
- 5.Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat. Biotechnol. 2014;32:447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- 6.Kim TY, Sohn SB, Bin Kim Y, Kim WJ, Lee SY. Recent advances in reconstruction and applications of genome-scale metabolic models. Curr. Opin. Biotechnol. 2012;23:617–623. doi: 10.1016/j.copbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fondi, M. & Liò, P. In Bacterial Pangenomics: Methods and Protocols (eds Alessio Mengoni, Marco Galardini & Marco Fondi) 233–256 (Springer New York, 2015). [DOI] [PubMed]
- 9.Nogales, J. In A Practical Protocol for Genome-Scale Metabolic Reconstructions 1–25 (Humana Press, 2014).
- 10.Ganter M, Bernard T, Moretti S, Stelling J, Pagni M. MetaNetX.org: a website and repository for accessing, analysing and manipulating metabolic networks. Bioinformatics. 2013;29:815–816. doi: 10.1093/bioinformatics/btt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King ZA, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–D522. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikrishnan A, Raman K. Critical assessment of genome-scale metabolic networks: the need for a unified standard. Brief. Bioinform. 2015;16:1057–1068. doi: 10.1093/bib/bbv003. [DOI] [PubMed] [Google Scholar]
- 13.Moretti S, et al. MetaNetX/MNXref - reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016;44:D523–D526. doi: 10.1093/nar/gkv1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwala R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apweiler R, et al. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards, M. A. et al. MediaDB: A Database of Microbial Growth Conditions in Defined Media. Plos One9 (2014). [DOI] [PMC free article] [PubMed]
- 17.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 2007;282:28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 19.Burgard AP, Pharkya P, Maranas CD. OptKnock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 20.Ranganathan, S., Suthers, P. F. & Maranas, C. D. OptForce: An Optimization Procedure for Identifying All Genetic Manipulations Leading to Targeted Overproductions. PLoS Comput. Biol. 6 (2010). [DOI] [PMC free article] [PubMed]
- 21.Saier MH, et al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2015;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nancy YY, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agren R, et al. The RAVEN Toolbox and Its Use for Generating a Genome-scale Metabolic Model for Penicillium chrysogenum. PLoS Comput. Biol. 2013;9:e1002980. doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swainston N, Smallbone K, Mendes P, Kell D, Paton N. The SuBliMinaL Toolbox: automating steps in the reconstruction of metabolic networks. J. Integr. Bioinform. 2011;8:186–202. doi: 10.2390/biecoll-jib-2011-186. [DOI] [PubMed] [Google Scholar]
- 28.Ye C, et al. Reconstruction and analysis of a genome-scale metabolic model of the oleaginous fungus Mortierella alpina. BMC Syst. Biol. 2015;9:1–11. doi: 10.1186/s12918-014-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings J, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal N, Widiastuti H, Karimi IA, Zhou Z. A genome-scale metabolic model of Methanococcus maripaludis S2 for CO2 capture and conversion to methane. Mol. Biosyst. 2014;10:1043–1054. doi: 10.1039/C3MB70421A. [DOI] [PubMed] [Google Scholar]
- 33.Gonnerman MC, Benedict MN, Feist AM, Metcalf WW, Price ND. Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotech. J. 2013;8:1070–1079. doi: 10.1002/biot.201200266. [DOI] [PubMed] [Google Scholar]
- 34.Dai Z, Nielsen J. Advancing metabolic engineering through systems biology of industrial microorganisms. Curr. Opin. Biotechnol. 2015;36:8–15. doi: 10.1016/j.copbio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Kim B, Kim WJ, Kim DI, Lee SY. Applications of genome-scale metabolic network model in metabolic engineering. J. Ind. Microbiol. Biotechnol. 2015;42:339–348. doi: 10.1007/s10295-014-1554-9. [DOI] [PubMed] [Google Scholar]
- 36.Seppala JJ, et al. Prospecting hydrogen production of Escherichia coli by metabolic network modeling. Int. J. Hydrogen. Energ. 2013;38:11780–11789. doi: 10.1016/j.ijhydene.2013.07.002. [DOI] [Google Scholar]
- 37.Kim J, Reed JL. Refining metabolic models and accounting for regulatory effects. Curr. Opin. Biotechnol. 2014;29:34–38. doi: 10.1016/j.copbio.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Xu, Z., Zheng, P., Sun, J. & Ma, Y. ReacKnock: Identifying Reaction Deletion Strategies for Microbial Strain Optimization Based on Genome-Scale Metabolic Network. Plos One8 (2013). [DOI] [PMC free article] [PubMed]
- 39.Choon, Y. W. et al. Differential Bees Flux Balance Analysis with OptKnock for In Silico Microbial Strains Optimization. Plos One9 (2014). [DOI] [PMC free article] [PubMed]
- 40.Choon YW, Mohamad MS, Deris S, Illias RM. A hybrid of bees algorithm and flux balance analysis (BAFBA) for the optimisation of microbial strains. Int. J. Data Min. Bioin. 2014;10:225–238. doi: 10.1504/IJDMB.2014.064016. [DOI] [PubMed] [Google Scholar]
- 41.Tepper N, Shlomi T. Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics. 2010;26:536–543. doi: 10.1093/bioinformatics/btp704. [DOI] [PubMed] [Google Scholar]
- 42.Bhan N, Xu P, Khalidi O, Koffas MA. Redirecting carbon flux into malonyl-CoA to improve resveratrol titers: proof of concept for genetic interventions predicted by OptForce computational framework. Chem. Eng. Sci. 2013;103:109–114. doi: 10.1016/j.ces.2012.10.009. [DOI] [Google Scholar]
- 43.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007;3:121–128. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobson, P. D. et al. Further developments towards a genome-scale metabolic model of yeast. BMC Syst. Biol. 4 (2010). [DOI] [PMC free article] [PubMed]
- 45.Loira N, Dulermo T, Nicaud JM, Sherman DJ. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst. Biol. 2012;6:35. doi: 10.1186/1752-0509-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]


