Abstract
Agricultural soil is often subjected to waterlogging after heavy rainfalls, resulting in sharp and explosive increases in the emission of nitrous oxide (N2O), an important greenhouse gas primarily released from agricultural soil ecosystems. Previous studies on waterlogged soil examined the abundance of denitrifiers but not the composition of denitrifier communities in soil. Also, the PCR primers used in those studies could only detect partial groups of denitrifiers. Here, we performed pyrosequencing analyses with the aid of recently developed PCR primers exhibiting high coverage for three denitrification genes, nirK, nirS, and nosZ to examine the effect of short-term waterlogging on denitrifier communities in soil. We found that microbial communities harboring denitrification genes in the top 5 cm of soil distributed according to soil depth, water-soluble carbon, and nitrate nitrogen. Short-term waterlogging scarcely affected abundance, richness, or the alpha-diversities of microbial communities harboring nirK, nirS, and nosZ genes, but significantly affected their composition, particularly in microbial communities at soil depths of 0 to 1 cm. Our results indicated that the composition of denitrifying microbial communities but not the abundance of denitrifiers in soil was responsive to short-term waterlogging of an agricultural soil ecosystem.
Introduction
Nitrous oxide (N2O) is a greenhouse gas that is mainly released from soil ecosystems1. Thus it has received considerable attention from soil scientists in recent decades2–4. Agricultural soil produces N2O during nitrification and denitrification processes5, 6. During denitrification, N2O emission from soil is affected by oxygen levels, soil moisture, and substrate availability7. The oxygen level in soil is often the most critical factor, because true denitrification occurs under anaerobic conditions7. Moisture affects denitrification by altering oxygen supply to the soil8. For example, large N2O emissions are observed immediately following irrigation or rainfall9–16. Specifically, Akiyama et al. observed large N2O emissions (up to 1.59 kg N ha−1 day−1) following heavy rainfalls in an upland converted paddy field, accounting for 55% to 80% of the annual N2O emission from that field15. Additionally, we observed large N2O emissions using temporarily (24 h) waterlogged intact soil cores sampled from the same field17. Knowledge of the composition of corresponding microbial communities is required to link rapid increases in N2O emissions with environmental changes18. Therefore, to understand the mechanisms underlying N2O emission following short-term waterlogging by rainfall, it is necessary to investigate the community composition of denitrifying microbes in soils and their responses to short-term waterlogging.
Molecular tools can be used to elucidate relationships between soil microbes and N2O emissions19. The information obtained from soil microbial DNA reveals the community composition and abundance of denitrifiers contributing to N2O emission in soil18, 20. To understand the composition of and variations in denitrifier communities in soil, pyrosequencing of PCR amplicons from denitrification genes generated from soil nucleic acids can be used. Similar to other PCR-based detection techniques, PCR primers are key factors that determine the reliability of pyrosequencing results. Recently, substantial efforts have been devoted to developing primers for denitrification genes in order to detect more microbial taxa21–30. Among these primers, those recently developed for nirK (encoding a copper-containing nitrite reductase)28, nirS (encoding a cytochrome cd1-containing nitrite reductase)28, and nosZ (encoding a nitrous oxide reductase)29, 31 exhibit significantly higher coverage of microbial taxa compared with others. Therefore, in this study, we used conventional PCR, quantitative PCR (qPCR), and pyrosequencing techniques, as well as recently developed PCR primers, to investigate communities of microbes harboring nirK, nirS, and nosZ genes in soil from an upland converted paddy field that exhibited active N2O emission immediately following heavy rainfalls.
Results
Detection of denitrification genes in soil cores
Existence of denitrification genes in raw soil samples was examined using conventional PCR and primers targeting the nirK gene in clusters I, II, III, and IV, the nirS gene in clusters I, II, and III, and the nosZ gene in clades I and II. Our results indicated detection of nirK in cluster II, nirS in cluster I, and nosZ in clades I and II (see Supplementary Fig. S1), suggesting that these sequences were dominant in these soil samples. Therefore, in this study, we used these four sets of PCR primers to analyze microbial communities harboring these denitrification genes in soil samples.
qPCR was performed to examine the abundance of denitrification genes in the soil samples (Fig. 1). As shown in Fig. 2, no gene showed significant alterations in copy number, suggesting no significant effect of waterlogging on the abundance of microbes harboring these genes. The copy number of nirK in cluster II was ~10- to ~70-fold higher than that of nirS in cluster I (Fig. 2a and b), and because the enzymes encoded by the nirK and nirS genes catalyze the same chemical reaction (reduction of nitrite to nitric oxide) during the denitrification process, these data suggested that nirK, but not nirS, played a key role in denitrification in these soil samples. The copy number of nosZ in clade II, which is a newly described cluster of nosZ sequences, was ~1- to ~7-fold higher than that of nosZ in clade I (Fig. 2c and d), suggesting that the microbes harboring nosZ in clade II may contribute to denitrification to a greater degree than those harboring nosZ in clade I. These results were consistent with those of a recent metagenomic analysis of agricultural soils32.
Figure 1.
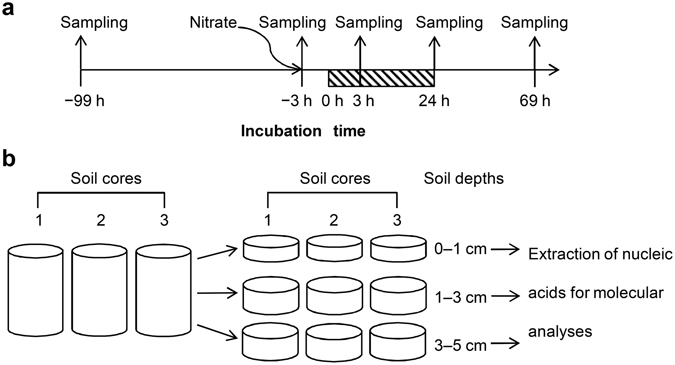
Sampling time (a) and dissection (b) of soil cores. The shadowed region indicates the waterlogging period. Arrows indicate the time of soil sampling and addition of nitrate.
Figure 2.
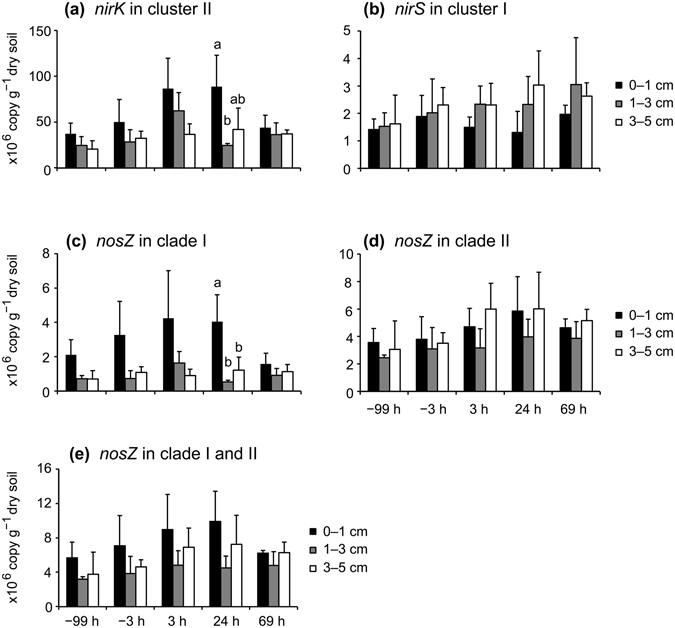
Quantification of the abundance of nirK in cluster II (a), nirS in cluster I (b), nosZ in clade I (c), nosZ in clade II (d), and the total quantity of nosZ (nosZ in clades I and II) (e) in soil by qPCR. Error bars represent standard deviations (n = 3). Different lowercase letters above columns represent significant differences among different soil depths at each sampling time.
The alpha-diversities and community compositions of denitrifiers in soil
Across all soil samples, 42,223 (nirK in cluster II), 117,270 (nirS in cluster I), 146,651 (nosZ in clade I), and 44,173 (nosZ in clade II) sequences with average sequence lengths of 412 ± 4 base pairs (nirK in cluster II), 367 ± 3 base pairs (nirS in cluster I), 413 ± 9 base pairs (nosZ in clade I), and 287 ± 15 base pairs (nosZ in clade II) were obtained. The Good’s library coverages were >96% (nirK in cluster II), >99% (nirS in cluster I and nosZ in clade I), and >94% (nosZ in clade II) (see Supplementary Table S6), suggesting that the numbers of obtained sequences were sufficient for diversity analysis. Rarefaction curves are shown in Supplementary Fig. S2.
The operational taxonomic unit (OTU) richness of all samples did not differ significantly according to one-way analysis of variance (ANOVA). Similar results were observed for the alpha-diversity indices, except for those of nirK in cluster II (see Supplementary Tables S6 and S7). Nearly 96% of nirK sequences in cluster II were affiliated with bacterial taxa, whereas >4% were affiliated with eukaryotes. The dominant taxa (the read number in a total of 18 soil samples was >1%) included alpha-, beta-, and gamma-proteobacteria, Chloroflexi, Verrucomicrobia, and Amoebozoa (Fig. 3a). With the exception of 32 sequences from fungi (Fusarium), all of the eukaryotic sequences were affiliated with genus Hartmannella of Amoebozoa (Fig. 3a). All of the dominant taxa for nirS in cluster I and nosZ in clade I were classified as alpha-, beta-, and gamma-proteobacteria (Figs 3b and 4a), suggesting low diversity at the phylum level. The dominant taxa for nosZ in clade II were classified into diverse phyla (Fig. 4b) belonging to two different kingdoms, Archaea and Bacteria, despite the archaeal sequences existing in <2% of 18 soil samples.
Figure 3.
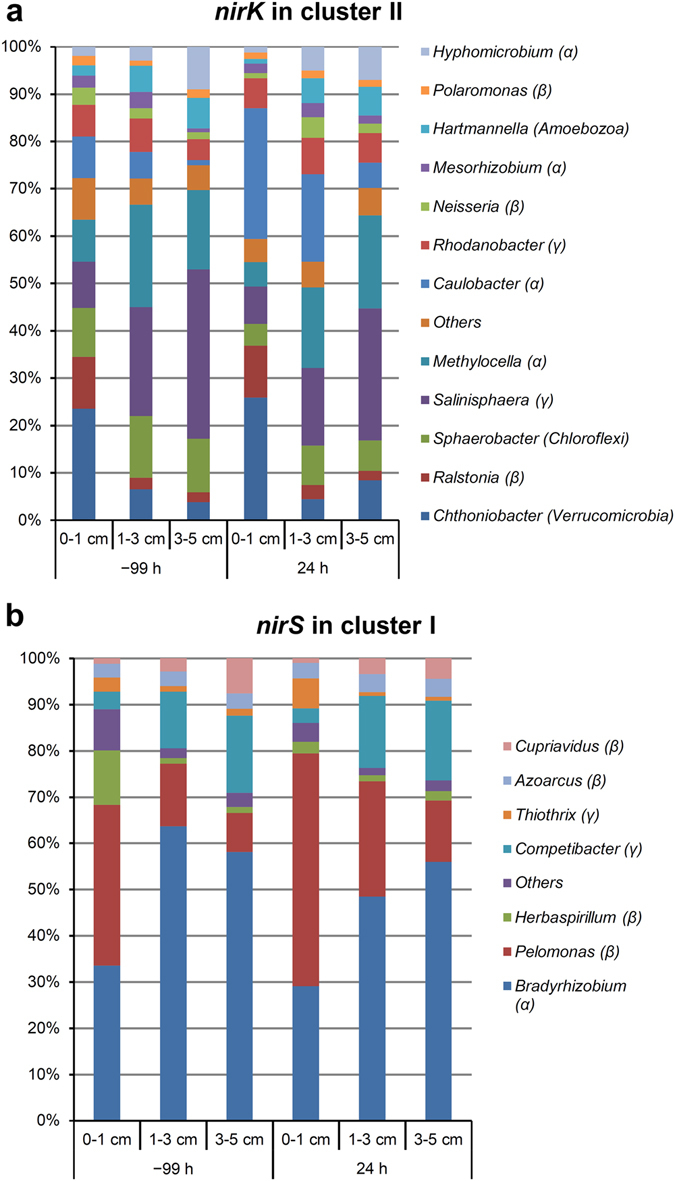
Average relative abundances of nirK in cluster II (a) and nirS in cluster I (b) sequences affiliated with taxonomic groups at the genus level. Each group is labeled with its genus name, with its phylum or subphylum name in parenthesis. Groups with <1% sequence number in the 18 soil samples were merged into the “Others” group. α, β, and γ represent alpha-, beta-, and gamma-proteobacteria, respectively.
Figure 4.
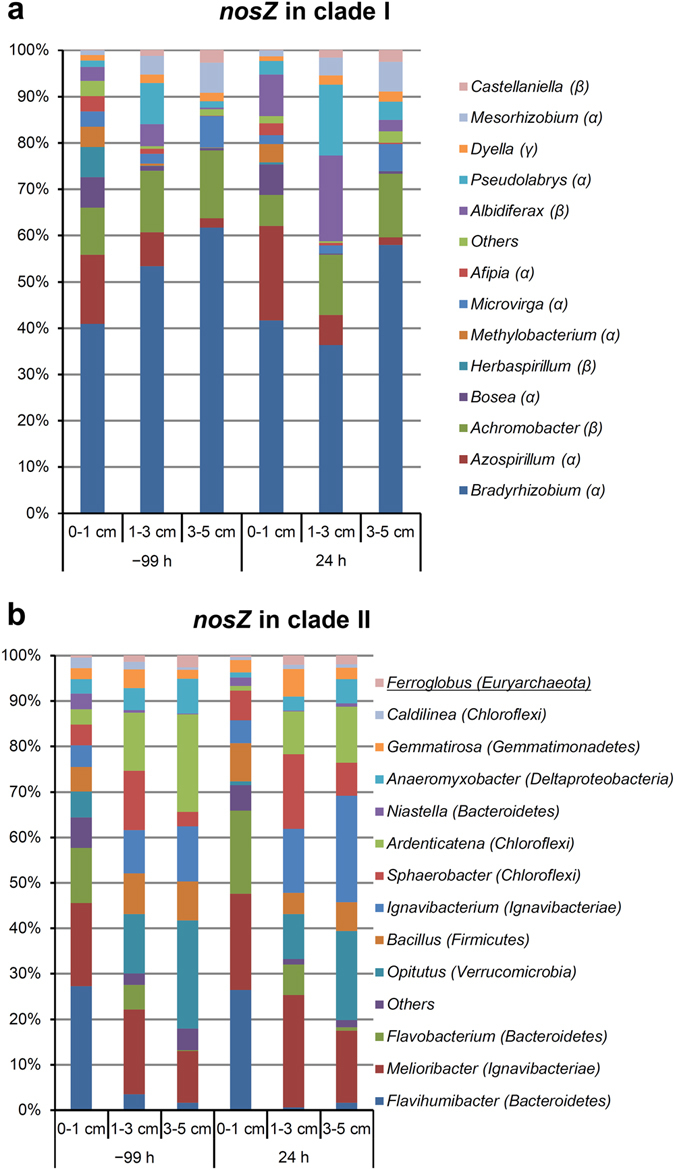
Average relative abundances of nosZ in clade I (a) and nosZ in clade II (b) sequences affiliated with taxonomic groups at the genus level. Each group is labeled with its genus name, with its phylum or subphylum name in parenthesis. Groups with <1% sequence number in the 18 soil samples were merged into the “Others” group. α, β, and γ represent alpha-, beta-, and gamma-proteobacteria, respectively. The underlined taxonomic name in (b) indicates an Archaea group.
Environmental factors shaping the alpha- and beta-diversities of microbes in soil
To characterize microbial communities in the soil prior to waterlogging (−99 h; Fig. 1), we examined correlations between the alpha-diversities of microbes in the −99-h soil cores and five environmental factors, including vertical distance (soil depth), horizontal distance between samples (soil core), water-soluble carbon, soil nitrogen in the form of nitrate (nitrate-N), and soil pH (see Supplementary Table S8). Spearman’s rank correlation test indicated that alpha-diversity indices of microbes harboring nirK in cluster II, but not other genes, positively correlated with water-soluble carbon, but negatively correlated with soil depth (see Supplementary Table S9), suggesting a decreased alpha-diversity of microbes harboring nirK in cluster II, with the increase in soil depth determined by water-soluble carbon.
We then performed constrained (or canonical) correspondence analysis (CCA) to determine the effect of environmental factors on the beta-diversities of denitrifiers harboring nirK in cluster II, nirS in cluster I, and nosZ in clades I and II in the soil samples (Fig. 5). The first two axes of the CCA explained 59% to 88% of the variations in these denitrification genes. Permutation tests on CCA results indicated that the effect of environmental factors on the beta-diversities of the denitrifiers was significant (number of permutations = 1000 in all cases; nirK in cluster II, F = 3.03, p = 0.001; nirS in cluster I, F = 6.82, p = 0.001; nosZ in clade I, F = 3.43, p = 0.001; nosZ in clade II, F = 2.62, p = 0.001). Consistent with the CCA data, permutational multivariate ANOVA (PERMANOVA; two-way ANOVA) results suggested that soil depth, water-soluble carbon, and nitrate-N significantly affected the beta-diversities of microbes harboring these genes (see Supplementary Table S10). According to partitioning analysis, the combined effect of soil depth, water-soluble carbon, and nitrate-N explained 76% (nirK in cluster II), 65% (nirS in cluster I), 43% (nosZ in clade I), and 51% (nosZ in clade II) of the observed variations (see Supplementary Fig. S3), suggesting that these environmental factors were important for shaping the beta-diversities of microbes harboring these genes in soil. Soil depth was particularly important, because its effect explained 63% (nirK in cluster II), 44% (nirS in cluster I), 34% (nosZ in clade I), and 35% (nosZ in clade II) of the observed variations (see Supplementary Fig. S3).
Figure 5.
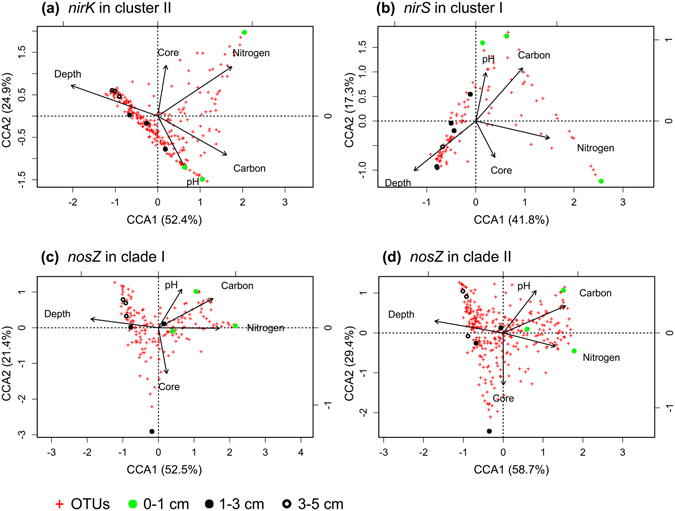
Evaluation of the effect of environmental factors on microbial communities harboring nirK in cluster II (a), nirS in cluster I (b), nosZ in clade I (c), and nosZ in clade II (d) in −99-h soil cores by constrained correspondence analysis.
To determine the importance of soil depth directly, we presented the beta-diversities as nonmetric multidimensional scaling (nMDS) plots (Fig. 6). For each gene, all soil samples shared >20% similarity (Fig. 6). When grouping samples at 40% similarity (Fig. 6), soil samples could be grouped into two clusters: a cluster dominated by samples originating from surface soil (0–1 cm) and a cluster composed of samples from deeper soil (1–3 cm and 3–5 cm) (Fig. 6 and Supplementary Fig. S4), suggesting differences between surface and deeper soil in microbial communities. The microbial communities at soil depths of 3 to 5 cm showed high degrees of similarity and could be grouped together, even with similarity as high as 60% (Figs 6 and S4). Our results indicated that community composition of denitrifiers varied according to soil depth.
Figure 6.
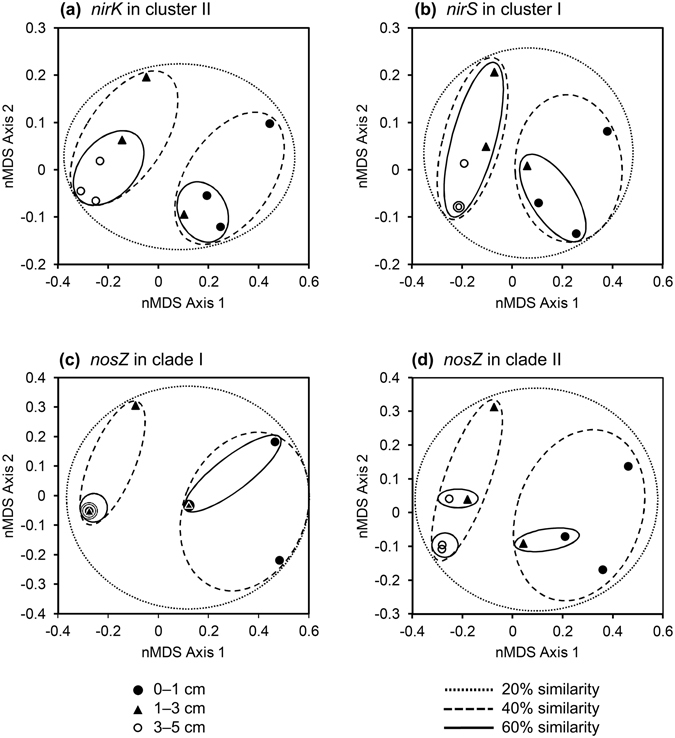
Nonmetric multidimensional scaling plot of Hellinger-transformed Bray-Curtis dissimilarity matrices describing microbial communities harboring nirK in cluster II (a), nirS in cluster I (b), nosZ in clade I (c), and nosZ in clade II (d) in −99-h soil cores. Overlapped symbols indicate equivalent values of data.
We then evaluated the identity of denitrifiers showing significant variations according to soil depth. For each gene, analysis using LEfSe algorithm33 identified OTUs enriched at each soil depth. Among these enriched OTUs, the dominant OTUs (OTU1–OTU20 for each gene) showing abundance >1% are summarized in Supplementary Table S11. Because these enriched OTUs were also dominant OTUs, the results presented in Supplementary Table S11 were almost in accord with data presented in Figs 3 and 4, except for OTUs affiliated with the genera Bradyrhizobium and Melioribacter, which covered multiple dominant OTUs.
Effect of waterlogging on denitrifier communities
Because short-term waterlogging did not increase the copy numbers of denitrification genes at all examined soil depths (Fig. 2), despite the large episodic N2O peaks observed17, we determined whether the richness, biodiversity, and community composition of microbes harboring denitrification genes in soil changed in response to waterlogging. The richness and alpha-diversities of microbial communities between −99-h and 24-h soil cores did not exhibit significant differences (Supplementary Table S6). This result suggested that waterlogging had no effect on the biodiversity of the microbes harboring nirK in cluster II. To compare the microbial-community compositions between −99-h and 24-h soil cores, we performed multiple analyses, including analysis of molecular variance (AMOVA), homogeneity of molecular variance (HOMOVA), and weighted and unweighted UniFrac analyses as previously described34. Among these analyses, only weighted UniFrac showed significant differences in most comparisons (see Supplementary Table S12). According to a previous report34, these data indicated the presence of a pivot between the communities, i.e., the two communities shared a large portion of microbial species, although there were a portion of unique species in each community. To verify this hypothesis, we generated Venn diagrams (see Supplementary Fig. S5). Although some OTUs were not shared between −99-h and 24-h samples, for each gene, the shared OTUs exhibited relative abundance ranging from 94.3% to 99.7% at all soil depths, thus forming pivots between communities in −99-h and 24-h soil cores (see Supplementary Fig. S5). Either nirS in cluster I or nosZ in clade I exhibited higher relative abundance (≥99.0%) of shared OTUs in the −99-h and 24-h samples as compared with that observed for nirK in cluster II (94.3–98.3%) and nosZ in clade II (94.5–97.4%), suggesting that nirS in cluster I and nosZ in clade I exhibited weaker responses to waterlogging relative to those observed from nirK in cluster II and nosZ in clade II.
Despite differences in OTU identities, we then determined which OTU showed significant changes in abundance after waterlogging. Analysis using the LEfSe algorithm identified these OTUs at each soil depth. Among these OTUs, dominant OTUs (OTU1–OTU20 for each gene) showing significant changes in their abundance are summarized in Supplementary Table S13. Either nirS in cluster I or nosZ in clade I contained fewer significantly changed OTUs (five OTUs for nirS in cluster I and six OTUs for nosZ in clade I) as compared with nirK in cluster II (12 OTUs) and nosZ in clade II (10 OTUs), and the abundance of the most dominant OTUs (OTU1–OTU3) of nirS in cluster I and nosZ in clade I did not change significantly. These findings were consistent with those from the Venn diagrams, suggesting that the microbial communities harboring nirS in cluster I and nosZ in clade I showed weaker responses to waterlogging as compared with responses from communities harboring nirK in cluster II and nosZ in clade II, which are major players in the denitrification process in this soil.
Discussion
Similar to other PCR-based detection methods, the reliability of pyrosequencing denitrification genes depends greatly upon the reliability of PCR primers. To collect intact and reliable information for denitrification genes from soil, the coverage for microbial-taxa sequences by PCR primers should be as high as possible. For this purpose, we used PCR primers targeting four clusters of nirK, three clusters of nirS, and two clades of nosZ to detect denitrifiers in soil. In our soil samples, both clades of nosZ were detected, whereas only one cluster of sequences was clearly detected for nirK and nirS (see Supplementary Fig. S1). Therefore, we used four sets of primers targeting these sequences to study the biodiversity of microbes harboring denitrification genes in soil.
Previous reports indicated positive correlations between nirK abundance and N2O emissions20 and negative correlations between nirS copy number and N2O/(N2O + N2) ratio35. Because the nirK copy number was higher than that of nirS in the soil used in this study (Fig. 2), we hypothesized that this soil exhibited high potential for releasing N2O during microbial denitrification. This hypothesis was supported by our previous observations in the field15 and soil-incubation experiments17, which showed sharp and explosive emissions of N2O from soil immediately after waterlogging.
According to a recent survey of microbial genomes harboring denitrification genes, the nosZ gene exhibits a higher frequency of co-occurrence with nirS as compared with nirK 36. Therefore, a higher ratio of nirK/nirS might suggest a lower abundance of the nosZ gene and a higher level of N2O emission. However, the nosZ gene in clade II does not co-occur with either nirK or nirS 37; therefore, the ratio of nirK/nirS may not always accurately predict the potential of soil to release N2O, particularly in clade II nosZ-dominated soils. Calculation of this ratio (nosZ in clades I and II)/(nirK + nirS) could account for nosZ in clade II and should constitute a better predictive approach. In this study, we observed no significant difference in copy numbers of denitrification genes after waterlogging. Therefore, the large N2O emissions observed in the upland converted paddy field after waterlogging15 may have been augmented by the low ratio of nosZ in clades I and II to nirK + nirS (~1:7 in the raw soil) in the soil rather than by the increased abundance of denitrifiers.
Using the newly developed PCR primers, we detected a wide range of microbial taxa in the soils. For the nirK gene, we detected sequences affiliated with 11 bacterial phyla/subphyla, as well as phyla of Fungi (Ascomycetes, grouped into the “Others” group) and Protists (Amoebozoa), with abundances of 0.1% and 4.4% in a total of 18 samples, respectively (Fig. 3a). This suggested that in this soil, aside from bacterial denitrifiers, Fungi and Protists may also contribute to denitrification, although this contribution might be minor. In contrast to the highly diverse sequences of the nirK gene, all of the dominant taxa associated with the denitrifiers harboring nirS in cluster I sequences belonged to the Proteobacteria phylum (Fig. 3b). Similar to the nirS gene, >99% of clade I sequences for the nosZ gene were also affiliated with the Proteobacteria phylum (Fig. 4a). The clade II sequences of the nosZ gene were affiliated with a wide range of bacterial and archaeal phyla (Fig. 4b), including 12 phyla/subphyla of bacteria and two phyla of archaea (Crenarchaeota and Euryarchaeota). These data were consistent with the primer design, because the primers targeting nirS in cluster I and nosZ in clade I mainly covered alpha-, beta-, and gamma-proteobacteria 28, 29, 31, whereas the primers targeting nirK in cluster II and nosZ in clade II covered a wide range of microbial phyla28, 29.
Distribution of microbial taxa according to soil depth was inconsistent. For microbes harboring nirK in cluster II, Chthoniobacter and Ralstonia were enriched in 0 to 1 cm of soil, whereas Salinisphaera and Hyphomicrobium were enriched in 3 to 5 cm of soil (Fig. 3a). Additionally, the copiotrophic taxon Bacteroidetes (Flavihumibacter, Flavobacterium, and Niastella) harboring nosZ in clade II was enriched in 0 to 1 cm of soil (Fig. 4b), which contained higher levels of nitrate-N as compared with deeper soil levels. This finding was consistent with that of a previous report indicating that soil with high levels of nitrogenous fertilizer favors a copiotrophic microbial community38.
The biogeographical distribution of soil microbial communities at the macroscale, including the continental scale39–42 and the field scale43–50, is well documented. Among the environmental factors investigated, soil pH is the most important factor in determining the biogeographical distribution of soil microbial communities39–46, 51. However, it is unclear whether soil pH is also the main factor at the microscale level (e.g., at the centimeter level). Here, we examined the effects of five environmental factors on the distribution of microbes harboring denitrification genes in soil at the microscale level, finding that soil depth, water-soluble carbon, and nitrate-N, but not soil pH, had significant effects on this distribution (Fig. 5 and Supplementary Table S10). Compared to the wide pH ranges spanning several pH units investigated at the continental and field scales, the pH range of our soil samples was narrow, spanning <0.5 pH units (see Supplementary Table S8). Therefore, it is possible that the effect of pH was too weak to be detected. Similar to our result, Yuan et al. recently reported that total nitrogen may be a primary factor that directly determines depth-related changes (0–20 cm of soil with pH ranges spanning 0.1–0.6 pH units) in soil bacterial-community composition47. Although further investigation using various types of soil is required, based on the available information, we believe that soil pH may not be the main factor at the microscale level for determining biogeographical distribution of soil microbial communities, given that pH ranges at this level may span <1 pH unit.
Microbial-community composition is often sensitive to the addition of nitrogen52, 53. Fierer et al. reported that at the field scale, long-term nitrogen fertilization had no significant effect on bacterial alpha-diversity, but a significant effect on bacterial-community composition38. However, the effect of soil nitrogen on microbes harboring denitrification genes at the microscale level was still unclear. In this study, we found that the alpha-diversity of microbes harboring the nirK gene in cluster II decreased along with the increases in soil depth to within 5 cm (see Supplementary Tables S6 and S7), and that this variation was positively correlated with water-soluble carbon, but was not correlated with nitrate-N (see Supplementary Table S9) levels, despite nitrate-N levels exhibiting a clear gradient based on soil depth in the analyzed samples. Additionally, by examining the effect of environmental factors on the beta-diversities of microbes harboring denitrification genes, we found that the beta-diversities of microbes harboring nirK in cluster II, nirS in cluster I, and nosZ in clades I and II were shaped by soil depth and water-soluble carbon, as well as nitrate-N (Fig. 5 and Supplementary Fig. S3). Moreover, we determined that the abundance of some OTUs, particularly dominant OTUs (see Supplementary Table S11), exhibited significant changes according to soil depth, suggesting variations in community composition based on soil depth. These findings indicated that soil nitrogen (nitrate-N) levels had no significant effect on the alpha-diversity of microbes harboring denitrification genes, but did exert a significant effect on beta-diversity and community composition, which were similar to findings associated with total bacterial composition in soil38.
According to a previous review discussing the creation and maintenance of microbial biogeographic patterns54, we hypothesized that selection and dispersal might be the two major processes shaping the beta-diversities of microbes harboring denitrification genes in soil. Because the water content of the field soil used in this study was low prior to waterlogging [water-filled pore space (WFPS) = 27 ± 7%], dispersal could be limited, which should have restricted microbial distribution. Selection eliminates microbes unable to adapt to local environments and should be reflected by differences in community compositions according to the gradients of nutrients, such as carbon and nitrogen. Therefore, a combination of nutrient selection and poor dispersal may have resulted in differences in microbial-community compositions. Because the level of both water-soluble carbon and nitrate-N showed clear gradients based on soil depth (see Supplementary Table S8), soil depth might be a proxy for various nutrients in soil as suggested in coastal ecosystems55. For each gene, our identification of different enriched OTUs at each soil depth (see Supplementary Table S11) might be explained as the result of selection and limited dispersal.
The field from which the soil samples used in this study originated is located in the East Asian monsoon region of Japan. In Japan, there are two rainy seasons from mid-May to late July and from mid-September to late October, with a dry period from late July to mid-September56. Even during the rainy season, rain and dry weather alternate frequently15. Therefore, from the beginning of the first rainy season to the end of the second rainy season, the field soil repeatedly experienced wet/dry cycles. Under such a climate, Akiyama et al. observed large N2O emissions following heavy rainfall from an upland converted paddy field15. Nitrous oxide emissions following heavy rainfall are common, and account for 55% to 80% of the annual N2O emission from this field15. Therefore, it was worth exploring the mechanisms underlying this phenomenon. As a result, we focused on the response of microbial communities, harboring denitrification genes, to short-term waterlogging.
The rewetting of dried soil upregulates expression of the rpoB gene, which encodes the β subunit of bacterial RNA polymerase, suggesting an active bacterial response to soil rewetting57. However, the active response of microbes in short-term waterlogged soil was unrelated to increased microbial populations as revealed by qPCR (Fig. 2), although the size of denitrifier community increased after a long-term soil wetting (one week or longer)19. Instead, we detected significant changes in microbial-community composition (see Supplementary Fig. S5 and Supplementary Tables S12 and S13). These data agreed with those from a previous report showing that simulated rainfall did not alter levels of microbial biomass carbon and total phospholipid fatty acid (PLFA), but did significantly alter microbial-community composition in two out of four soils according to PFLA profiles58.
In the soil samples examined in this study, water-soluble carbon was enriched at soil depths of 0 to 1 cm. According to a previous study59 and a review on irrigation60, this could be caused by repeated wet-dry cycles, which increase levels of dissolved organic carbon more quickly in surface soil than in deeper soil. The levels of nitrate-N were also higher at depths of 0 to 1 cm when compared with levels observed in deeper regions of these soil samples, which is a common phenomenon in soil subjected to a drying process61, 62. Because water-soluble carbon and nitrate-N were enriched at depths of 0 to 1 cm, N2O emissions from this surface soil should be greater than those from deeper soil as reported in previous studies60, 63. According to these results, we expected that the response to short-term waterlogging by rainfall would involve increased changes in the community composition of microbes harboring denitrification genes at soil depths of 0 to 1 cm relative to those observed in deeper soil. As shown in Supplementary Table S12, the weighted UniFrac distances for the microbial communities between the −99-h and 24-h samples were highest at soil depths of 0 to 1 cm, suggesting that the largest changes in microbial composition occurred in this region of the soil.
Conclusion
Our study had some limitations. Although pyrosequencing of PCR amplicons is a powerful technique used to investigate microbial communities in soil, some pitfalls limiting its power could be introduced during the pre-requisite PCR process, which involve bias during amplification64. Furthermore, the primers previously used for PCR amplification of denitrification genes did not cover all possible sequences available in public databases28, 29, 32, 37. In order to detect as many of our target sequences as possible, we performed conventional PCR, qPCR, and pyrosequencing using recently developed PCR primers28, 29, 31. However, because the development of these new primers was based on collected sequence information from current databases, continuous updates to this sequence information will require frequent generation of new primers. From this viewpoint, we note that the detected sequence information in this study may not cover 100% of the sequences in nature. Additionally, we only examined one type of soil (Fluvisol) lacking aboveground plants. Because microbial-community responses to soil rewetting are modified in the presence of plants56, 62, it is necessary to examine other types of soil in the presence and absence of different aboveground plants to form a general consensus regarding their roles in this process. In this study, the denitrifier communities we detected represent the total community in the soil. To acquire the information of the active denitrifier communities responsive to waterlogging, sequencing of mRNA extracted from soil should be conducted.
In summary, our results indicated that the copy numbers of all denitrification genes examined did not change significantly according to soil depth and were unaffected by short-term waterlogging. We also found that the alpha-diversity of microbes harboring nirK in cluster II varied according to soil depth in soil cores prior to waterlogging (−99 h) and was positively correlated with levels of water-soluble carbon in the soil, whereas the beta-diversities of microbes harboring the nirK, nirS, and nosZ genes were affected by soil depth and water-soluble carbon, as well as nitrate-N levels. Furthermore, short-term waterlogging scarcely affected the abundance, richness, or alpha-diversities of microbes harboring denitrification genes, but had significant effects on their community compositions. Our results provided insight into the communities of microbes harboring nirK, nirS, and nosZ genes in soil ecosystems. Because short-term soil waterlogging causes large emissions of the greenhouse gas N2O and occurs frequently worldwide, these data could be helpful for investigating the mechanisms underlying N2O emissions caused by soil waterlogging and understanding the contribution of the denitrification process to the global nitrogen cycle.
Methods
Preparation of soil samples
Because our previous study showed that nitrate was dominantly distributed in the top soil and N2O was also mainly generated from the top soil17, we sampled soil cores to a depth of 5 cm beneath the ground’s surface in this study. Intact soil cores (100 cm3; 5-cm internal diameter, 5-cm length) were extracted from an experimental field at the Institute for Agro-Environmental Sciences, NARO, Japan (36.025° N, 140.107° E), on September 2012. The field was a former rice paddy that was tile-drained and had been used to grow soybeans for 6 years, followed by 1 year of being fallow. The soil in this field was a gray lowland soil (Fluvisol) and exhibited a water content of 27 ± 7% WFPS, a bulk density of 1.17 ± 0.10 Mg m−3, and was determined to be sandy clay loam soil according to the United States Department of Agriculture classification system15. During the experiments, all soil cores were incubated at 25 °C, at which the largest peak of N2O emission was observed in the field15, and short-term waterlogging was performed for 24 h, a period typical of a heavy rainfall event that occurs in the field. Addition of nitrate represented simulation of fertilizer management in the field and occurred at −3 h. One milliliter of KNO3 (9 g N L−1) was applied to the soil-core surface to achieve 9 mg N core−1, which corresponded to a rate of 46 kg N ha−1. During waterlogging, from 0 h to 24 h (Fig. 1a), the soil cores were water saturated by keeping the water table level no higher than the soil surface. Under this condition, an anaerobic condition was successfully established as reported previously17. Triplicate soil cores taken at −99 h (the raw soil), −3 h (prior to waterlogging), 3 h (shortly after commencement of waterlogging), 24 h (after completion of waterlogging), and 69 h (recovery from waterlogging) were used for DNA extraction (Fig. 1a). These soil cores were separated into three layers: 0 to 1 cm, 1 to 3 cm, and 3 to 5 cm (Fig. 1b). The cores were then snap-frozen in liquid nitrogen and stored at −80 °C until use.
DNA extraction from soil
DNA was extracted from 0.4 g soil taken from each layer of the soil cores obtained at −99 h, −3 h, 3 h, 24 h, and 69 h of incubation (n = 3) using a FastDNA spin kit for soil (MP Biomedical, Santa Ana, CA, USA). To remove humic substances capable of potentially inhibiting enzymatic reactions in downstream analyses65, additional purification was performed using a MicroSpin S-400 HR column (GE Healthcare, Little Chalfont, UK) and a DNA clean & concentrator-5 kit (Zymo Research, Irvine, CA, USA). The concentrations of extracted DNA were measured using a NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Detection of denitrification genes using conventional and qPCR
Conventional PCR was performed to determine the existence of target sequences in DNA extracted from −99-h soil cores. A 25-μL reaction mixture contained 15 ng of soil DNA, primers (see Supplementary Table S1) at a final concentration of 0.2 μM (nirS and nirK) or 0.8 μM (nosZ), bovine serum albumin at a final concentrations of 0.48 mg mL−1 (nirS and nirK) or 1 mg mL−1 (nosZ), nuclease-free water, and the Premix Ex Taq polymerase (hot-start version; Takara Bio, Kusatsu, Shiga, Japan). Dimethyl sulfoxide was added to a final concentration of 5% to amplify cluster III sequences of nirK. A Veriti thermal cycler (Thermo Fisher Scientific) was used to perform conventional PCR. Cycling parameters for conventional PCR are listed in Supplementary Table S2.
The genes detected by conventional PCR in DNA extracted from −99-h soil cores were subjected to qPCR analysis using a QuantiTect SYBR Green PCR kit (Qiagen, Hilden, Germany) on a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). A 1-μL sample of 2-fold diluted soil DNA was used as a template in a 20-μL reaction mixture. The final concentration of each primer in the PCR mixture was 1 μM. Cycling parameters are listed in Supplementary Table S3.
Pyrosequencing of PCR amplicons of nirS and nosZ genes
To investigate denitrifier communities in the soil before and after waterlogging, DNA extracted from the −99-h (raw soil, prior to waterlogging) and 24-h (immediately after waterlogging) soil cores was used to generate DNA fragments for pyrosequencing. Gene-specific primers are listed in Supplementary Table S4. A 26-nucleotide adapter, a 4-nucleotide key (TCAG), and a 10-nucleotide multiplex identifier (MID) tag (also known as a barcode, which is absent in the reverse primers) specific to each sample were successively added to the 5′-end of the pyrosequencing primer sequences. Pyrosequencing primers with different MID tags were screened in silico using OligoAnalyzer (http://sg.idtdna.com/calc/analyzer), followed by experimental verification. To reduce bias generated during PCR amplification, two-step PCR was performed according to a previous study66. One microliter of the first-step PCR-reaction result (20 cycles with the primers used in the conventional PCR) was used as the template for the second-step PCR (15 cycles with pyrosequencing primers). The PCR-cycling parameters were the same as those used for conventional PCR, except for the number of cycles. Each of the PCR products was purified using a PureLink PCR purification kit (Thermo Fisher Scientific), followed by further purification with agarose gels using a QIAquick gel-extraction kit (Qiagen), and verified as single sharp peaks according to high-sensitivity DNA assays using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). After quantification using a TBS-380 mini-fluorimeter (Turner BioSystems, Sunnyvale, CA, USA) and a Quanti-iT PicoGreen dsDNA assay kit (Thermo Fisher Scientific), the purified PCR products for the same gene were diluted and mixed to equimolar ratios. Pyrosequencing of these purified PCR products on a GS Junior system (Roche, Basel, Switzerland) was conducted according to manufacturer protocol. A total of 72 DNA samples (three cores × three layers × two treatments × four genes) were sequenced using four Picotiter plates (one plate per run of the sequencer; Roche).
Processing and analyzing pyrosequencing data
Pyrosequencing data were processed and analyzed using Mothur version 1.36.167. The sequence reads were assigned to each sample according to sample-specific barcodes, with PCR primer and barcode sequences located at ends of the sequence reads subsequently trimmed. The criteria for retaining high-quality sequences were as follows: no base ambiguity, average quality score >25, homopolymer <8, one mismatch in the MID sequence, two mismatches in the primer sequence, minimum sequence lengths of 340 bp (nirK in cluster II), 350 bp (nirS in cluster I and nosZ in Clade I), and 420 bp (nosZ in clade II), and maximum sequence lengths of 460 bp (nirK in cluster II), 400 bp (nirS in cluster I), 450 bp (nosZ in clade I), and 540 bp (nosZ in clade II) (including the sequences of PCR primers and MIDs). The trimmed sequences were aligned against sequences downloaded from the FunGene database68. To reduce errors that may have occurred during pyrosequencing, 2% pre-clustering was performed as suggested69, and chimeric sequences identified by the UCHIME algorithm70 were removed. The remaining sequences were realigned using MAFFT71 and then clustered using the Furthest Neighbor algorithm, with cutoff values of 0.26 (nirK in cluster II), 0.17 (nirS in cluster I), and 0.18 (nosZ in clades I and II) corresponding to 74%, 83%, and 82% of the median similarity values of the denitrification gene sequences at the species level (see Supplementary Table S5), respectively, which were generated from ClustalW72 alignment of the sequences originating from the same genus. To acquire taxonomic identities of the OTUs, local BlastX73 searches against nirK, nirS, or nosZ sequences downloaded from the FunGene database were performed using the representative sequences of each OTU as queries.
To correct differences in sequencing depth among soil samples74, alpha- and beta-diversity analyses of each gene were performed using a randomly selected subset of 1841 (nirK cluster II), 5855 (nirS cluster I), 6281 (nosZ clade I), and 1826 (nosZ clade II) sequences per soil sample (the minimum number of sequences recovered among 18 samples per gene). Beta-diversities were presented as nMDS plots, which were generated from Hellinger-transformed Bray-Curtis matrices using the Vegan package in R (v3.2.5; http://www.R-project.org/), and Venn diagrams, which were generated from the VennDiagram package (v1.6.9)75 in R. CCA and variation-partition analyses were performed using the Vegan package in R to identify important environmental factors and their contributions to variations in microbial communities. LEfSe33, which was implemented in Mothur, was used to examine which OTU showed significantly varied abundance among samples.
Statistical analyses
One-way ANOVA and Tukey’s post-test were performed using R to examine variations in microbial richness and alpha-diversity indices. Spearman’s rank correlation test was performed using R to explore correlations between the alpha-diversity indices of the microbes harboring nirK in cluster II and environmental factors. An ANOVA-like permutation test and PERMANOVA were performed using the Vegan package in R to examine the significance of CCA results. Mothur was also used to test the global and pairwise differences among sample groups according to AMOVA76 and HOMOVA77, as well as weighted- and unweighted UniFrac analyses78.
Deposition of DNA sequences
All sequences acquired from pyrosequencing in this study were deposited to the DDBJ Sequence Read Archive under accession number DRA005237.
Electronic supplementary material
Acknowledgements
This work was financially supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry (28004 A), the Bio-oriented Technology Research Advancement Institution, Japan, the Funding Program for Next Generation World-Leading Researchers, Ministry of Education Culture, Sports, Science, and Technology ‘Program to Disseminate Tenure Tracking System’, and KAKENHI (B) (grant numbers 26310315 and 26292184). We appreciate the help of Dr. Takashi Okubo for his constructive opinion on data analysis.
Author Contributions
Y.W., Y.U., H.A., and M.H. designed the research. Y.W. performed DNA-extraction, PCR, and pyrosequencing experiments. Y.W. and Y.S. analyzed sequence data. Y.U. and H.A. conducted soil sampling, soil-core incubation, and measurement of soil physicochemical parameters. Y.W. and M.H. wrote the paper. All authors discussed the results and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yong Wang and Yoshitaka Uchida contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00953-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yong Wang, Email: ywangbioinfo@gmail.com.
Masahito Hayatsu, Email: hayatsu@affrc.go.jp.
References
- 1.Davidson EA. The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nat. Geosci. 2009;2:659–662. doi: 10.1038/ngeo608. [DOI] [Google Scholar]
- 2.Pathak H. Emissions of nitrous oxide from soil. Curr. Sci. India. 1999;77:359–369. [Google Scholar]
- 3.de Klein CAM, Sherlock RR, Cameron KC, van der Weerden TJ. Nitrous oxide emissions from agricultural soils in New Zealand - a review of current knowledge and directions for future research. J. Roy. Soc. New Zeal. 2001;31:543–574. doi: 10.1080/03014223.2001.9517667. [DOI] [Google Scholar]
- 4.Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S. Nitrous oxide emissions from soils: How well do we understand the processes and their controls? Phil. Trans. R. Soc. B. 2013;368:20130122. doi: 10.1098/rstb.2013.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrage N, Velthof GL, van Beusichem ML, Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 2001;33:1723–1732. doi: 10.1016/S0038-0717(01)00096-7. [DOI] [Google Scholar]
- 6.Hayatsu M, Tago K, Saito M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 2008;54:33–45. doi: 10.1111/j.1747-0765.2007.00195.x. [DOI] [Google Scholar]
- 7.Zhu X, Burger M, Doane TA, Horwath WR. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc Natl Acad Sci USA. 2013;110:6328–6333. doi: 10.1073/pnas.1219993110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul, E. A. & Clark, F. E. Soil Microbiology and Biochemistry (Academic Press, 1996).
- 9.Knowles R. Denitrification. Microbiol. Rev. 1982;46:43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flessa H, Dörsch P, Beese F. Seasonal variation of N2O and CH4 fluxes in differently managed arable soils in southern Germany. J. Geophys. Res. Atmos. 1995;100:23115–23124. doi: 10.1029/95JD02270. [DOI] [Google Scholar]
- 11.Bronson KF, Singh U, Neue HU, Abao EB. Automated chamber measurements of methane and nitrous oxide flux in a flooded rice soil: II. Fallow period emissions. Soil Sci. Soc. Am. J. 1997;61:988–993. doi: 10.2136/sssaj1997.03615995006100030039x. [DOI] [Google Scholar]
- 12.Jørgensen RN, Jørgensen BJ, Nielsen NE. N2O emission immediately after rainfall in a dry stubble field. Soil Biol. Biochem. 1998;30:545–546. doi: 10.1016/S0038-0717(97)00144-2. [DOI] [Google Scholar]
- 13.Pu GX, Saffigna PG, Strong WM. Potential for denitrification in cereal soils of northern Australia after legume or grass-legume pastures. Soil Biol. Biochem. 1999;31:667–675. doi: 10.1016/S0038-0717(98)00154-0. [DOI] [Google Scholar]
- 14.Allen DE, Kingston G, Rennenberg H, Dalal RC, Schmidt S. Effect of nitrogen fertilizer management and waterlogging on nitrous oxide emission from subtropical sugarcane soils. Agr. Ecosyst. Environ. 2010;136:209–217. doi: 10.1016/j.agee.2009.11.002. [DOI] [Google Scholar]
- 15.Akiyama H, et al. Nitrification, ammonia-oxidizing communities, and N2O and CH4 fluxes in an imperfectly drained agricultural field fertilized with coated urea with and without dicyandiamide. Biol. Fert. Soils. 2013;49:213–223. doi: 10.1007/s00374-012-0713-2. [DOI] [Google Scholar]
- 16.Hansen M, Clough TJ, Elberling B. Flooding-induced N2O emission bursts controlled by pH and nitrate in agricultural soils. Soil Biol. Biochem. 2014;69:17–24. doi: 10.1016/j.soilbio.2013.10.031. [DOI] [Google Scholar]
- 17.Uchida Y, Wang Y, Akiyama H, Nakajima Y, Hayatsu M. Expression of denitrification genes in response to a waterlogging event in a Fluvisol and its relationship with large nitrous oxide pulses. FEMS Microbiol. Ecol. 2014;88:407–423. doi: 10.1111/1574-6941.12309. [DOI] [PubMed] [Google Scholar]
- 18.Philippot L, Andert J, Jones CM, Bru D, Hallin S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Global Change Biol. 2011;17:1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x. [DOI] [Google Scholar]
- 19.Hamonts K, et al. Influence of soil bulk density and matric potential on microbial dynamics, inorganic N transformations, N2O and N2 fluxes following urea deposition. Soil Biol. Biochem. 2013;65:1–11. doi: 10.1016/j.soilbio.2013.05.006. [DOI] [Google Scholar]
- 20.Clark IM, Buchkina N, Jhurreea D, Goulding KWT, Hirsch PR. Impacts of nitrogen application rates on the activity and diversity of denitrifying bacteria in the Broadbalk Wheat Experiment. Phil. Trans. R. Soc. B. 2012;367:1235–1244. doi: 10.1098/rstb.2011.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry S, et al. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Meth. 2004;59:327–335. doi: 10.1016/j.mimet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Henry S, Bru D, Stres B, Hallet S, Philippot L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Gutiérrez JC, et al. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Meth. 2004;57:399–407. doi: 10.1016/j.mimet.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Throbäck IN, Enwall K, Jarvis Å, Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Qiu XY, et al. Detection and quantification of copper-denitrifying bacteria by quantitative competitive PCR. J. Microbiol. Meth. 2004;59:199–210. doi: 10.1016/j.mimet.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Braker G, Tiedje JM. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 2003;69:3476–3483. doi: 10.1128/AEM.69.6.3476-3483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bru D, Sarr A, Philippot L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007;73:5971–5974. doi: 10.1128/AEM.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei W, et al. Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 2015;9:1954–1965. doi: 10.1038/ismej.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones CM, Graf DRH, Bru D, Philippot L, Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013;7:417–426. doi: 10.1038/ismej.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallin S, Lindgren PE. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones CM, et al. Recently identified microbial guild mediates soil N2O sink capacity. Nat. Clim. Change. 2014;4:801–805. doi: 10.1038/nclimate2301. [DOI] [Google Scholar]
- 32.Orellana LH, et al. Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: Method development and implications for the nitrogen cycle. mBio. 2014;5:e01193–01114. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloss PD. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2008;2:265–275. doi: 10.1038/ismej.2008.5. [DOI] [PubMed] [Google Scholar]
- 35.Čuhel J, et al. Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 2010;76:1870–1878. doi: 10.1128/AEM.02484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graf DRH, Jones CM, Hallin S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One. 2014;9:e114118. doi: 10.1371/journal.pone.0114118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanford RA, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. USA. 2012;109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fierer N, et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RT, et al. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu HY, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 43.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 44.Baker KL, et al. Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol. Biochem. 2009;41:2292–2298. doi: 10.1016/j.soilbio.2009.08.010. [DOI] [Google Scholar]
- 45.Jenkins SN, et al. Actinobacterial community dynamics in long term managed grasslands. Anton. Leeuw. Int. J. G. 2009;95:319–334. doi: 10.1007/s10482-009-9317-8. [DOI] [PubMed] [Google Scholar]
- 46.Shen CC, et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013;57:204–211. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 47.Yuan YL, Si GC, Wang J, Luo TX, Zhang GX. Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol. Ecol. 2014;87:121–132. doi: 10.1111/1574-6941.12197. [DOI] [PubMed] [Google Scholar]
- 48.Sul WJ, et al. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol. Biochem. 2013;65:33–38. doi: 10.1016/j.soilbio.2013.05.007. [DOI] [Google Scholar]
- 49.Embarcadero-Jiménez S, Rivera-Orduña FN, Wang ET. Bacterial communities estimated by pyrosequencing in the soils of chinampa, a traditional sustainable agro-ecosystem in Mexico. J. Soil Sediments. 2016;16:1001–1011. doi: 10.1007/s11368-015-1277-1. [DOI] [Google Scholar]
- 50.Yang YF, et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014;8:430–440. doi: 10.1038/ismej.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 52.Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turlapati SA, et al. Chronic N-amended soils exhibit an altered bacterial community structure in Harvard Forest, MA, USA. FEMS Microbiol. Ecol. 2013;83:478–493. doi: 10.1111/1574-6941.12009. [DOI] [PubMed] [Google Scholar]
- 54.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 55.Gong J, et al. Depth shapes alpha- and beta-diversities of microbial eukaryotes in surficial sediments of coastal ecosystems. Environ. Microbiol. 2015;17:3722–3737. doi: 10.1111/1462-2920.12763. [DOI] [PubMed] [Google Scholar]
- 56.Qian W, Kang HS, Lee DK. Distribution of seasonal rainfall in the East Asian monsoon region. Theor. Appl. Climatol. 2002;73:151–168. doi: 10.1007/s00704-002-0679-3. [DOI] [Google Scholar]
- 57.Barnard RL, Osborne CA, Firestone MK. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013;7:2229–2241. doi: 10.1038/ismej.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steenwerth KL, Jackson LE, Calderón FJ, Scow KM, Rolston DE. Response of microbial community composition and activity in agricultural and grassland soils after a simulated rainfall. Soil Biol. Biochem. 2005;37:2249–2262. doi: 10.1016/j.soilbio.2005.02.038. [DOI] [Google Scholar]
- 59.Lundquist EJ, Jackson LE, Scow KM. Wet-dry cycles affect dissolved organic carbon in two California agricultural soils. Soil Biol. Biochem. 1999;31:1031–1038. doi: 10.1016/S0038-0717(99)00017-6. [DOI] [Google Scholar]
- 60.Trost B, et al. Irrigation, soil organic carbon and N2O emissions. A review. Agron. Sustain. Dev. 2013;33:733–749. doi: 10.1007/s13593-013-0134-0. [DOI] [Google Scholar]
- 61.Simpson JR. The mechanism of surface nitrate accumulation on a bare fallow soil in Uganda. J. Soil Sci. 1960;11:45–60. doi: 10.1111/j.1365-2389.1960.tb02200.x. [DOI] [Google Scholar]
- 62.Robinson JBD, Gacoka P. Evidence of the upward movement of nitrate during the dry season in the Kikuyu red loam coffee soil. J. Soil Sci. 1962;13:133–139. doi: 10.1111/j.1365-2389.1962.tb00690.x. [DOI] [Google Scholar]
- 63.Harrison-Kirk T, Beare MH, Meenken ED, Condron LM. Soil organic matter and texture affect responses to dry/wet cycles: Effects on carbon dioxide and nitrous oxide emissions. Soil Biol. Biochem. 2013;57:43–55. doi: 10.1016/j.soilbio.2012.10.008. [DOI] [Google Scholar]
- 64.Wu JY, et al. Effects of polymerase, template dilution and cycle number on PCR based 16S rRNA diversity analysis using the deep sequencing method. BMC Microbiol. 2010;10:255. doi: 10.1186/1471-2180-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Fujii T. Evaluation of methods of determining humic acids in nucleic acid samples for molecular biological analysis. Biosci Biotechnol Biochem. 2011;75:355–357. doi: 10.1271/bbb.100597. [DOI] [PubMed] [Google Scholar]
- 66.Berry D, Ben Mahfoudh K, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fish JA, et al. FunGene: the functional gene pipeline and repository. Front. Microbiol. 2013;4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 73.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaw AK, et al. It’s all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- 75.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stewart CN, Excoffier L. Assessing population genetic structure and variability with RAPD data: Application to Vaccinium macrocarpon (American Cranberry) J. Evolution. Biol. 1996;9:153–171. doi: 10.1046/j.1420-9101.1996.9020153.x. [DOI] [Google Scholar]
- 78.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


