Abstract
In this article, we summarize the knowledge and best practices learned from bulk and single molecule measurements to address some of the frequently experienced difficulties in single molecule Förster Resonance Energy Transfer (smFRET) measurements on G-quadruplex (GQ) structures. The number of studies that use smFRET to investigate the structure, function, dynamics, and interactions of GQ structures has grown significantly in the last few years, with new applications already in sight. However, a number of challenges need to be overcome before reliable and reproducible smFRET data can be obtained in measurements that include GQ. The annealing and storage conditions, the location of fluorophores on the DNA construct, and the ionic conditions of the experiment are some of the factors that are of critical importance for the outcome of measurements, and many of these manifest themselves in unique ways in smFRET assays. By reviewing these aspects and providing a summary of best practices, we aim to provide a practical guide that will help in successfully designing and performing smFRET studies on GQ structures.
Introduction
As early as 1910, it was known that guanine rich nucleic acids self-associate to form gels, which are now known to be due to G-quadruplex (Bang 1910). G-quadruplex structures (GQs) are non-canonical nucleic acid secondary structures that form in guanine-rich regions of the genome (Gellert et al. 1962), most prominently at the telomeres (Sundquist and Klug 1989; Blackburn 1991) and promoters (Todd et al. 2005; Huppert and Balasubramanian 2007). In the telomeric context, GQs are implicated in maintenance of telomeres and are known to inhibit telomerase activity (Fletcher et al. 1998; Blackburn et al. 2006). When formed at the promoter regions, they are proposed to regulate gene expression at the transcription level (Du et al. 2008; Qin and Hurley 2008). Furthermore, GQs can form at the untranslated regions (UTR) of mRNA and influence translation level gene expression (Kumari et al. 2007; Huppert et al. 2008; Morris et al. 2010). GQs are formed of multiple G-tetrad layers in which a guanine occupies each corner of the tetrad. These guanines are connected to each of their nearest neighbors within the tetrad by two hydrogen bonds, resulting in a total of eight hydrogen bonds per tetrad. The GQ structure is further stabilized by various factors including stacking of the tetrad layers, hydration, and monovalent cations that intercalate within or between the tetrad layers (Lane et al. 2008; Stegle et al. 2009; Chaires 2010). This distinct structure endows GQ a number of unique characteristics and has made it an attractive system for biotechnological and biomedical applications. GQs have found use as a molecular switch (Johnson et al. 2013), a biosensor (Wang et al. 2011; Zhou et al. 2012; Taylor et al. 2013), a scaffold for small molecule binding (Rahman et al. 2012), and recently as an effector in a high throughput sequencing assay (Chambers et al. 2015). There has recently been a flurry of activity on designing and synthesizing GQ binding small molecules due to their potential as anti-cancer drugs (Balasubramanian et al. 2011; Husby et al. 2013; Iida et al. 2013; McLuckie et al. 2013; Ohnmacht et al. 2014; Neidle 2015). Direct imaging of GQ structures within human cells, both at telomeric and non-telomeric sites, and demonstration of their modulation during the cell cycle have highlighted the significance of protein-GQ interactions (Biffi et al. 2013). These observations have also fortified earlier work that showed GQ destabilization by protein activity to be required for normal progression of various cellular pathways, including DNA replication and repair (Johnson et al. 2010; Paeschke et al. 2011; Paeschke et al. 2013).
The thermodynamic and kinetic properties of GQs strongly depend on a number of variables (Lane et al. 2008; Phan 2010; Tran et al. 2011), including the layers and loops that form the structure (Guedin et al. 2010) and the neighboring ssDNA overhangs (Viglasky et al. 2010), the ionic conditions (Tran et al. 2011), and the type of crowding agents that are present in the environment (Miller et al. 2010; Zhou et al. 2016). However, these complexities have made GQs attractive systems to study with single molecule Förster Resonance Energy Transfer (smFRET) due to its high spatial and temporal resolution and sensitivity to heterogeneities in the system. smFRET enables monitoring structural changes at the sub-nanometer length scale with tens of millisecond time resolution (Ha et al. 1996; Roy et al. 2008). Force spectroscopy measurements have shown that the end to end distance change upon unfolding of a GQ is in the ~ 1 nm range (You et al. 2015). Therefore, it is possible to monitor GQ dynamics by smFRET in isolation (Ying et al. 2003; Lee et al. 2005; Shirude and Balasubramanian 2008; Okumus and Ha 2010; Kruger and Birkedal 2013; Long and Stone 2013; Tippana et al. 2014) or while it interacts with single stranded DNA binding proteins (Kruger et al. 2010; Hwang et al. 2012; Qureshi et al. 2012; Ray et al. 2013; Hwang et al. 2014; Ray et al. 2014; Zhou et al. 2014), helicases (Budhathoki et al. 2014; Chatterjee et al. 2014; Zhou et al. 2014; Budhathoki et al. 2015), or GQ-stabilizing small molecules (Jena et al. 2009; Kreig et al. 2015). Furthermore, monitoring more subtle features, such as different conformations of GQ, is also possible with carefully designed DNA constructs (Long and Stone 2013; Budhathoki et al. 2015).
Despite the large variety of GQ related studies that utilize smFRET, the number of groups that has contributed to the field has remained relatively small. Understanding the complications of GQ in different settings and attaining consistent results in smFRET measurements often require significant amount of time and effort even for experienced users of smFRET. We believe this is an important factor that limits the number of groups that have productively utilized smFRET for investigating GQ and its interactions. This article highlights some of the frequently encountered issues and presents strategies to address them. We have particularly focused on the GQ formed by human telomeric sequence (hGQ) as it is widely used and is one of the most polymorphic GQ structures, hence fairly challenging to work with. In this context, we should mention that unless required for biologically compelling reasons hGQ is not an ideal model system to work with due to complications associated with its polymorphism. For example, if a GQ is desired to be used as a molecular switch with just an “on” (folded) and an “off” (unfolded) state, then using a GQ that has a single conformation at ionic conditions of interest would be a practical approach. On the other hand, hGQ might have different folded conformations, hence multiple 'on states', depending on the details of how and when it was annealed and the ionic conditions of the measurement. In order to provide a broad overview, different aspects of smFRET assay are discussed in this article, including designing the DNA construct, the annealing protocol, and salt titrations. In addition, how different aspects of protein-GQ interactions are manifested in smFRET data are briefly reviewed. Better practices that would increase the likelihood of reproducible and reliable results are summarized. Even though we review and reference a large set of published data, most of the data presented in this article (except Figure 1) are original and not published before. In particular, utilizing TMAA to obtain a weakly folded GQ and demonstration of low GQ stability via a protein-mediated GQ unfolding assay, and using LiOH as an effective GQ destabilizer are new for both bulk FRET and smFRET studies and the data presented in this article are first examples of their kind. In these respects, this article deviates from a traditional review article.
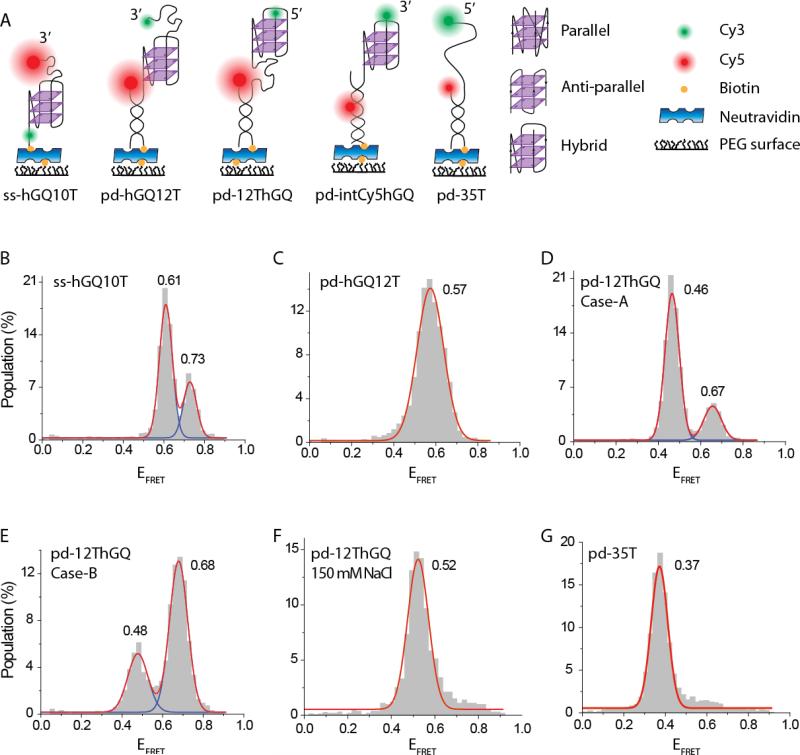
Fig. 1.
(A) Schematics of DNA constructs and the three different folding conformations of hGQ. (B) smFRET histogram showing the folding pattern for ss-hGQ10T construct at 150 mM KCl. The high FRET peak represents the parallel conformation and low FRET peak represents the anti-parallel (or hydrid) conformation. (C) smFRET histogram showing the folding pattern for pd-hGQ12T construct at 150 mM KCl. A single broad peak representing a combination of multiple conformations is observed. (D) smFRET histogram showing the folding pattern for pd-12ThGQ construct at 150 mM KCl. The sample was folded at elevated temperatures in 150 mM KCl, resulting in the anti-parallel (or hybrid) conformation to be dominant. (E) pd-12ThGQ was annealed in the absence of salt and folded at room temperature, resulting in the parallel conformation to be dominant. (F) pd-12ThGQ in 150 mM NaCl. A single folded peak representing the anti-parallel conformation is observed. (G) pd-35T in 150 mM KCl. A single peak representing coiled DNA, which should be similar to unfolded GQ state, is observed.
Materials and Methods
The relevant aspects of materials and methods are specified in respective sections, so only an overview will be provided here. Prism-type total internal reflection fluorescence microscopy was used to acquire smFRET data. The setup was built around an Olympus IX-71 microscrope that utilized an Olympus 60x 1.20 NA water objective to collect the fluorescence signal. An Andor Ixon EMCCD camera (iXon DV 887-BI EMCCD, Andor Technology, CT) was used for imaging. A SpectraPhysics Excelsior laser with λ=532 nm was used to excite the donor fluorophores. Also, a JDS Uniphase He-Ne laser with λ=632.8 nm was used to directly excite the acceptor fluorophores, when necessary. The imaging solution used in all of the measurements contained Tris base (50mM, pH 7.5), 2 mM Trolox, 0.8 mg/ml glucose, 0.1 mg/ml bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, 2 mM MgCl2, and a certain concentration of KCl, NaCl, LiCl, or Trimethylammonium acetate (TMAA), as specified in respective section. All buffers were filter sterilized before use. All DNA oligonucleotides were purchased from Integrated DNA Technologies (Iowa, USA) as HPLC or PAGE purified. The replication protein A (RPA) was purchased from Enzymax LLC (Kentucky, USA).
Unless specified otherwise, all measurements were performed on a surface coated with polyethylene-glycol (PEG). PEG and biotin-PEG were mixed in the ratio of ~100:1 (biotin-PEG-5000 and m-PEG-5000 from Laysan Bio Inc.). A detailed description of the surface passivation similar to what we utilized in our assays is reported in reference (Joo and Ha 2012). An improved version of this protocol has also been made available, along with guiding videos, in reference (Chandradoss et al. 2014). We have also tested a bovine serum albumin (BSA) coated surface for comparison purposes. The slide cleaning protocol for both BSA and PEG surfaces was kept the same. Quartz slides (Technical Glass Products, Inc., OH) and glass cover slips were sonicated for 20 minutes in acetone and then 20 minutes in KOH. After a thorough washing with distilled and deionized water, the slide and slip were dried and sample chamber was formed (Selvin et al. 2007). For a BSA surface, 1 mg/ml biotinylated-BSA (Sigma-Aldrich, A8549) was then incubated in the chamber for 20 minutes. For both PEG and BSA surfaces, we either added 0.1 mg/ml neutravidin and incubated for 1 min or added 0.01 mg/ml neutravidin and incubated for 15 minutes before introducing 15-30 pM DNA. The DNA incubation time showed some variation depending on how much DNA was added. We typically incubated 15 pM DNA for 5 minutes and checked the number of molecules on the surface. We aimed to have around 300 FRET-pairs in our imaging area so we added more DNA if the number of DNA molecules on surface was significantly lower than this number. In order to ensure proper folding and avoid possible complications due to buffer flow (possible induced surface interactions or poor mixing due to manual pipetting), GQ-forming DNA constructs were incubated in the indicated salt concentration for at least 15 minutes before data were acquired. Both long movies (1000-4000 frames) and short movies (20 frames) were recorded for performing different types of analysis. Each movie contained 250-350 molecules, and smFRET traces which contain time dependent donor and acceptor intensity and FRET efficiency were generated for each molecule using a software generously provided by Prof. Taekjip Ha at the University of Illinois-Center for Physics of the Living Cell (available for download at https://cplc.illinois.edu/software/). The long movies are generally used to study dynamics of the system, which are particularly significant if GQ is not stable enough or interacts with destabilizing proteins (Budhathoki et al. 2015). All acquired movies were included in the histograms, and each histogram was constructed from several thousand molecules, each imaged for 2 seconds (20 frames at an integration time of 100 ms/frame).
The FRET efficiency was calculated by EFRET=IA/(γ ID+IA), where IA is the acceptor intensity and ID is the donor intensity. The gamma parameter (γ) is defined as: , where ϕA and ϕD are the donor and acceptor quantum yields, respectively, and ηA and ηD are the sensitivities of sensor to the donor and acceptor emissions, respectively (Deniz et al. 2001). For Cy3 and Cy5 and the camera we use γ ≈ 1. About 5-20% of all molecules had only a donor fluorophore and formed the donor only (DO) FRET peak at EDO=0.12±0.03. The DO correction was made by moving the DO peak to EFRET=0.0, subtracting it from the histogram, and renormalizing the FRET scale. The following formula is used to move the DO peak and renormalizing the scale: EFRET =(EF-before-EDO)/(1-EDO), where EFRET is the FRET efficiency after DO correction (all FRET efficiencies reported in this manuscript are after DO correction has been made), EF-before is the FRET efficiency before DO correction. The position of the DO peak depends on a number of factors including the emission spectra of donor and acceptor fluorophores, the transmission-reflection characteristics and the exact angle of the dichroic used to separate (and possibly recombine depending on the experimental design) the donor/acceptor emissions. So it would naturally show variations in different setups. It is also possible to avoid this DO peak by selecting molecules based only on their intensity in the acceptor channel. We prefer to select molecules based on their intensities in either channel, i.e. if they are brighter than a certain threshold intensity in either the donor or acceptor channel. This ensures that low FRET populations would not be missed out because of this screening process.
Bulk FRET measurements were performed in a SpectraMax M4 Multi-Mode Microplate Reader using a 96-well plate. The DNA samples (both ss-hGQ4nt and pd-12T) were annealed at 1 μM concentration in 10 mM Tris (pH 7.5) and 150 mM KCl before they were slowly cooled down to room temperature. ss-hGQ4nt folds into a stable GQ at the end of this process while pd-12T forms a partial duplex DNA construct. Immediately before the measurement, the DNA samples were diluted to 50 nM in 10 mM Tris (pH 7.5) and proper concentration of LiOH. All bulk FRET measurements were performed after incubating in this solution (10 mM Tris, 50 nM DNA, 7.5 mM KCl, and proper concentration of LiOH) for 10 minutes. The samples were excited with 500 nm light and fluorescence emission was collected between 530 nm-750 nm in 5 nm steps.
Results
Designing the DNA Construct
In total internal reflection based smFRET assay, the DNA constructs are typically immobilized to a passivated surface via biotin-neutravidin or a similar linker. This immobilization is typically achieved by using a partial duplex DNA (pdDNA) construct that has a double stranded DNA (dsDNA) stem and a ssDNA overhang that contains the GQ-forming sequence. In this arrangement, biotin and one of the fluorophores, typically the acceptor, are placed on one DNA strand and the other fluorophore is placed on the other strand. Where on the DNA construct the donor-acceptor fluorophores are placed varies across different studies. We would like to highlight two designs that have provided high enough resolution capable of clearly distinguishing between the different conformations of hGQ. The end to end separations of hGQ for the parallel and anti-parallel conformations were estimated to be ≈0.9 nm and ≈1.7 nm, respectively (Long and Stone 2013). Hence, in order to distinguish between the two conformations, a significant FRET change should result when the end to end separation changes by approximately ≈0.8 nm. This can be achieved if the fluorophores are placed such that the separation (R) between them is at the most sensitive range of FRET, i.e. R≈0.58 R0, where R0 is the Förster radius for the donor/acceptor fluorophores. Due to the compact size of most GQ structures, placing the donor-acceptor fluorophores directly across the GQ would result in EFRET>0.85 for most commonly used FRET fluorophore pairs, such as Cy3-Cy5 or TAMRA-Cy5. However, this is a range where FRET is not very sensitive to distance changes, and the fluorophores need to be separated from each other in order to reduce the FRET efficiency to its more sensitive range. This can be achieved by placing one of the fluorophores internally within the duplex stem, 7-8 bp away from duplex end, while keeping the other fluorophore at the far end of hGQ (see pd-intCy5hGQ construct in Figure 1A for a schematic and references (Ying et al. 2003; Lee et al. 2005; Long and Stone 2013) for sequences of such constructs). Another way to achieve this is to keep 8-20 nucleotide (nt) of ssDNA overhang between the duplex stem and the hGQ (Budhathoki et al. 2015; Budhathoki et al. 2016). In this case one fluorophore can be placed at the end of duplex stem and the other fluorophore at the far end of hGQ. Figure 1A shows a schematic of this construct (pd-12ThGQ) and Figure 1D shows the smFRET distribution with two clearly identifiable peaks at EFRET=0.46±0.03 and EFRET=0.67±0.04 at 150 mM KCl. Interestingly, keeping an overhang of similar length at the other end of GQ, not between the GQ and duplex, did not result in as clear a separation of peaks for hGQ. Figure 1A shows a schematic of this construct (pd-hGQ12T) and Figure 1C shows the smFRET distribution with a single peak at EFRET=0.57±0.06 at 150 mM KCl. This peak is mid-way between the two peaks observed for pd-12ThGQ and is broader than either of them, suggesting that it represents a combination of multiple conformations. As the same fluorophores, linkers, and attachment chemistries are used in both cases, we do not think the observed variation is due to fluorophore properties or photophysics. Even though the two constructs have different polarities, i.e. pd-12ThGQ has a 5’ free end while pd-hGQ12T has a 3’ free end, this does not influence the folding pattern. We have reported folding data on pd-hGQ12T construct with either free 5’-end or 3’-end in an earlier study, and demonstrated that the folding pattern for these constructs is very similar even though the polarities are switched (Budhathoki et al. 2014).
Whether one or the other type of DNA construct, pd-12ThGQ or pd-intCy5hGQ, should be preferred depends on the specifics of the measurement. For example, if hGQ-protein interactions are studied and the protein needs an ssDNA longer than a few nt to bind, then pd-12ThGQ type construct could be preferred (Budhathoki et al. 2015). If on the other hand, having the ssDNA/dsDNA junction free of fluorophores is important or not having a large gap between duplex and GQ is desired, then placing the fluorophore internally as in pd-intCy5-hGQ would be advantageous. If not having a duplex stem is preferred, it is also possible to obtain a clear separation of the FRET peaks representing the two conformations of hGQ when both fluorophores are placed on the same strand, which also has a biotin at one end. Figure 1A shows a schematic of this construct (ss-hGQ10T) and Figure 1B shows the smFRET distribution with two clearly identifiable peaks at EFRET=0.61±0.03 and EFRET=0.73±0.03 at 150 mM KCl. In ss-hGQ10T the Cy3 was separated by 2-nt on the 5'-side of GQ and Cy5 was placed at the 3'-end, 10 nt away from GQ. The disadvantage of such a design is that it is typically costly to place all three modifications into a one strand and the yield of the final product is low. There are also some limitations in terms of the GQ structures that can be designed with all three modifications in one strand.
In both DNA designs that showed two well separated peaks, pd-12ThGQ and ss-hGQ10T, the low FRET peak is considered to represent the antiparallel (or hybrid) conformation while the high FRET peak represents the parallel conformation. This can be demonstrated by performing similar measurements in the presence of Na+ in which only the anti-parallel conformation is known to form (Tran et al. 2011). Figure 1F demonstrates smFRET data on pd-12ThGQ in 150 mM NaCl where a single peak at EFRET=0.52±0.05 is observed, identifying this peak as the peak for anti-parallel conformation. This construct was annealed and maintained in 150 mM NaCl throughout the measurement. A similar observation was also reported in reference (Long and Stone 2013), where the hybrid conformation was suggested to have a very similar FRET peak to the antiparallel conformation. Noer et al. recently reported multiple antiparallel conformations in 100 mM NaCl for a hGQ construct. In this study relatively broad histograms and dynamic smFRET traces (62% of traces showed multiple transitions) were observed at 100 mM NaCl. The authors identified folded populations at EFRET=0.50, EFRET=0.60, and EFRET=0.85 in addition to a significant unfolded population at EFRET=0.20. They interpreted the EFRET=0.60 peak to represent basket type anti-parallel conformation and the EFRET=0.85 peak the chair type anti-parallel conformation. Clearly, these results are significantly different from what we report in this manuscript and those reported by Long et al. (Long and Stone 2013), which also show a single FRET peak and no dynamics in their smFRET traces at 100 mM NaCl. Before we discuss the possible reasons for the differences, we should discuss whether the high FRET peak that was proposed to represent the chair-type antiparallel conformation has any implications for the high FRET peak we observed in 150 mM KCl and interpreted to represent the parallel conformation. First of all, CD and NMR data on hGQ in KCl show the existence of multiple conformations, not just different versions of the anti-parallel conformation (Ambrus et al. 2006; Phan et al. 2006; Maleki et al. 2016). Therefore, interpreting both peaks observed in KCl as different versions of antiparallel conformation would contradict these measurements. Secondly, the FRET peak that is proposed to represent the chair type antiparallel conformation (EFRET=0.85) is significantly higher than the peak we propose to represent the parallel conformation (EFRET=0.70), even though the low FRET peaks which are considered to represent basket-type antiparallel conformation are consistent with each other (EFRET=0.50-0.60). Finally, under our assay and sample preparation conditions we always observed only a single, relatively sharp peak in 150 mM NaCl and did not observe an additional FRET peak that could be representing another conformation. Therefore, we do think the EFRET=0.70 peak in our data represents the chair-type antiparallel conformation.
The question of why reports on hGQ in NaCl from different groups seem to be inconsistent with each other is an example worth discussing as it illustrates possible complications that could arise when working with GQ constructs. Noer et al. and Long et al. use DNA constructs with very similar linkers and fluorophore positions. Partial duplex DNA constructs were used in both studies. One fluorophore was internally labeled within the duplex handle while the other one was attached to the 3’-end immediately after the GQ forming sequence, without any spacer nucleotides in between. The donor-acceptor fluorophores were the same although their positions on DNA were switched (Cy3 internally labeled in one case vs. Cy5 in the other). Both studies followed very similar annealing and sample preparation protocols, where the sample was annealed and maintained in 100 mM Na+ at every step of sample preparation and imaging. In addition, similar image integration times (100 ms vs. 150 ms) were used in both studies. Despite these extensive similarities, all molecules were folded into a relatively sharp distribution and no significant dynamics was observed in one study (Long and Stone 2013), while in the other about a third of the molecules did not fold into GQ, and the remaining molecules showed a much broader distribution with rich dynamics (Noer et al. 2016). Therefore, there must be a feature in the Noer et al. construct that makes it significantly less stable than Long et al. construct and also our construct, which also folds into a sharp single peak (Figure 1F). A potentially significant difference we identified between these two studies is that a 2-nt spacer is kept between the duplex DNA handle in Long et al. construct while a 1-nt spacer is utilized in Noer et al. construct. We also keep at least 2-nt or longer spacers between the duplex handle and the GQ in our constructs. Whether a minimum spacer length is required to reduce potentially destabilizing interactions between the GQ and the duplex DNA handle can be answered only after further systematic studies however, it is a possibility that should be taken into account while designing the DNA construct. Another potentially significant difference between the two constructs is that the internally labeled fluorophore was placed 5-nt from the 5’-end in Noer et al. study while it was 7-nt from the 5’-end in the Long et al. study. The 5-nt between the 5’-end and the labeling position were complementary to hGQ sequence (AACCC). So unwinding of 5-bp of between the fluorophore and the junction would make this sequence (AACCC) available to bind to the G-rich sequence and potentially destabilize the GQ. Again it is not clear if this is a cause for the weak stability of the GQ, but other groups utilizing an internal label within the duplex DNA handle have placed the fluorophore 7 or 8 bp away from the DNA end (Ying et al. 2003; Lee et al. 2005).
Another important DNA design issue is how close the fluorophores can be placed to the GQ before they significantly interact with it and enhance its stability. It was shown that coupling a Cy5 fluorophore in the immediate vicinity of hGQ, to the 5'-end of DNA via phosphoramidite methodology, raised the thermal melting temperature of hGQ from Tm=59±3 °C (without Cy5) to Tm=67±3 °C (with Cy5) in 100 mM KCl (Ying et al. 2003). Interestingly, this study did not show a significant change in the melting temperature of hGQ in 100 mM NaCl with or without Cy5 at the end of GQ. More recently, molecular dynamics simulations have shown that a Cy3 molecule that is attached to the 3' end of hGQ with a six-carbon N-hydroxysuccinimide (NHS) linker attached to a primary amine, significantly interacts with the GQ (Sondergaard et al. 2015). Fluorescence lifetime measurements on this construct have shown that these interactions result in a significant shift in fluorescence lifetime compared to a free dye not attached to DNA (Sondergaard et al. 2015). In these measurements, the free dye exhibited a mono-exponential decay with a lifetime of 207±2 ps, while the dye attached to the end of GQ showed a multi-exponential decay with an additional ≈2 ns decay component. This long lived component is comparable to that observed when Cy3 is attached at the end of duplex DNA. Similar strong interactions were observed for a shorter linker, with the interactions confined to a smaller volume in the immediate vicinity of the attachment point of Cy3. These studies also showed that the generally assumed approximation of freely rotating fluorophore dipoles, i.e. κ2=2/3, is in general not valid for neither short nor long linkers, with long linkers being a better approximation to this limit. There are several different commercially available options in terms of the lengths of linkers connecting the fluorophores to the DNA. The 6 carbon long linkers (e.g. amino modifier C6, amino modifier C6 dT, or thiol modifier C6 S-S) are typically considered the standard length while 3 carbon long linkers are considered short (e.g. thiol modifier C3 S-S) and 10-12 carbon long linkers are considered long (e.g. amino modifier C12). The nomenclature given in parenthesis might show variations among different vendors. Unfortunately, a systematic study of how long the linkers or spacer sequences should be in order to reduce such interactions to insignificant levels has not be performed yet. Therefore, we prefer to leave at least a 2-nt gap (typically TT sequence) between the GQ and fluorophore and use standard length linkers in order reduce the impact on GQ stability and photophysical properties of the fluorophore.
How and where the fluorophores are coupled at the DNA are particularly significant in studies involving protein-mediated GQ unfolding. In particular, we observed significant differences in BLM-mediated GQ unfolding activity when the Cy3 fluorophore was placed internally vs. at the 5’-end (Budhathoki et al. 2015). The internal labeling of the fluorophore via the DNA backbone, which is the standard internal labeling method provided by some companies, gave rise to significant inhibition of BLM helicase activity and resulted in poor GQ unfolding. In cases where a helicase tracks the DNA backbone during translocation, either avoiding the internal labeling or performing the labeling via a modified base, typically via amino modifier dT, should be preferred (Yodh et al. 2009).
Annealing the DNA
Annealing conditions can have profound effects on GQ structures, particularly those that can fold in different conformations. A careful study of how different annealing, storage, and sample preparation protocols result in different folding patterns was performed by Long and Stone (Long and Stone 2013). It was demonstrated that folding hGQ in 100 mM KCl at high temperature vs. folding it at room temperature resulted significantly different distribution of conformations. In that study, the DNA was annealed at 95 °C for four minutes in 100 mM KCl and was slowly cooled down to room temperature. In one case, this DNA was immobilized on surface and studied via smFRET while the salt concentration was maintained at 100 mM KCl throughout the assay (Case I). In the second case, the annealed DNA was incubated on surface in the absence of salt and allowed to unfold at room temperature before adding 100 mM KCl (Case II). In both cases, the parallel and anti-parallel conformations were present however, the antiparallel conformation was dominant in Case I while the parallel conformation was dominant in Case II. The authors concluded that the antiparallel conformation has lower free energy as the hGQ primarily folds into that conformation at high temperatures. On the other hand, it was suggested that folding the hGQ at room temperature causes it to be kinetically trapped in the parallel conformation. Such hGQ molecules eventually transition to the antiparallel conformation in a slow process that takes many hours at room temperature.
We have performed similar measurements on hGQ with slight variations in the assay and have obtained results consistent with the study of Long and Stone (Long and Stone 2013). In our case, we annealed hGQ at 90 °C for three minutes either in 150 mM KCl (Case A) or in the absence of KCl (Case B), and allowed the DNA to slowly cool down to room temperature. The salt conditions were maintained at 150 mM KCl for Case A throughout the smFRET measurements. On the other hand, DNA prepared as in Case B was immobilized on a PEG surface in the absence of KCl, and then was folded into GQ by introducing 150 mM KCl to the chamber at room temperature. While the antiparallel (or hybrid) conformation was dominant in Case A (Figure 1D), the parallel conformation was primarily populated in Case B (Figure 1E). However, it should be mentioned that we noticed the distribution of folding conformations to be very sensitive to relatively minor variations in annealing and preparation conditions. Which of these conformations is physiologically more relevant for GQs that are unfolded and folded at 37 °C under relatively significant crowding conditions is not clear at this point (Buscaglia et al. 2013; Zhou et al. 2016). The practical solution we have pursued in our studies is to ensure that the initial distribution of folding conformations remains consistent among the different conditions that are tested for a particular system, e.g. when comparing hGQ unfolding efficiency of different proteins, we have ensured that the initial distributions of folding conformations are consistent for all proteins under study. For reproducible initial folding state, it is very important to not only maintain consistent annealing and cooling conditions but also perform the measurements after relatively similar times after cooling is completed. If a dominant parallel conformation is desired, it is important to anneal the sample in the absence of salt or fold it at room temperature and perform the measurements within a few hours after annealing is completed. If the measurements are performed significantly longer times after annealing, e.g. after an overnight incubation in KCl, then it is likely that a significant population of GQ molecules would transition into the anti-parallel state. If on the other hand a dominant anti-parallel conformation is desired, then it is important to anneal the DNA in K+. In this case, the waiting period after annealing is less significant as folding pattern is less likely to change over time. To our knowledge, a systematic study of how the annealing temperature, annealing time, cooling rate, and incubation time after annealing affect the folding distribution of hGQ has not been performed yet.
To conclude this section, we demonstrate smFRET data on a DNA construct that has a 35 thymine long ssDNA overhang (pd-35T). This overhang is as long as the overhang of GQ forming constructs pd-12ThGQ and pd-hGQ12T, both 35 nt. We use this construct as a reference point for the unfolded GQ state. pd-35T construct demonstrates a peak at EFRET=0.37±0.03 at 150 mM KCl (Figure 1G), which is lower than the peaks observed in pd-12ThGQ and pd-hGQ12T constructs, further supporting the proposal that both observed peaks in 150 mM KCl represent folded conformations.
Imaging Surface
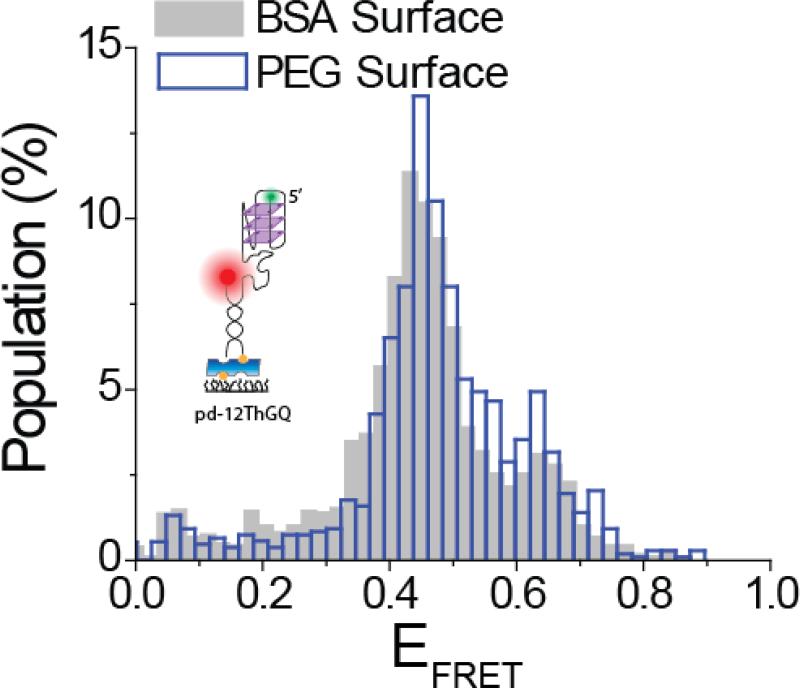
Most smFRET studies on GQ have been performed on polyethylene glycol (PEG) surface (Budhathoki et al. 2014; Tippana et al. 2014) or a bovine serum albumin (BSA) surface (Lee et al. 2005; Long and Stone 2013). In order to perform a direct comparison between these two surfaces, we imaged hGQ samples prepared under identical conditions on either a BSA or a PEG surface. The protocol for preparing these surfaces are given in Materials and Methods section. As shown in Figure 2, we obtained very similar folding patterns on the two surfaces. The minor variations between the two histograms are within the variation we obtain among different sample chambers that have the same surface. Therefore, a BSA surface, which is significantly easier and less time consuming to prepare compared to a PEG surface, should be adequate in cases where only GQ structure is probed (Long and Stone 2013; Noer et al. 2016). If proteins or small molecules are also used in these measurements, then the decision about what type of surface to use should be made on a case by case basis, as proteins and small molecules might show large variations in terms of their propensity to non-specific binding to these surfaces. In this context, a recent surface passivation method utilizing BSA is reported to be at least as effective as PEG at minimizing non-specific interactions of DNA and proteins with the surface (Hua et al. 2014).
Fig. 2.
pd-12ThGQ (schematics shown in inset) construct imaged in 150 mM KCl in either PEG (empty blue bins) or BSA (filled gray bins) surface. The folding patterns are very similar for the two surfaces and the minor variations are within the variation we would get when different sample channels having the same surface are used.
On a related note, it was shown that hGQ can be encapsulated within lipid vesicles, typically dimyristoylphosphatidylcholine (DMPC), that are immobilized on a supported lipid bilayer (such as EggPC), PEG, or BSA surface (Ishitsuka et al. ; Okumus et al. 2004; Cisse et al. 2007; Okumus et al. 2009; Okumus and Ha 2010). Comparative studies of hGQ that is encapsulated within a vesicle or immobilized on a BSA surface yielded consistent results in terms of the observed FRET levels (Lee et al. 2005). Vesicle encapsulation provides an alternative medium to study GQ interactions with various ligands, such as proteins or small molecules, which require high ligand concentration. A single molecule encapsulated within a vesicle of 100 nm diameter yields an effective concentration of approximately 3 μM. It is typically not challenging to access such high concentrations with non-fluorescent ligands. However, if interactions of GQ with fluorescent small molecules or fluorescently-labeled proteins are under investigation, then vesicle encapsulation enables accessing much higher concentrations than otherwise possible, while maintaining single molecule detection capability. In the absence of vesicle encapsulation, it is typically not possible to increase the concentration of fluorescent ligand beyond tens of nanomolar as the high fluorescent background would interfere with single molecule detection.
Ionic Conditions
Despite a recent smFRET study reporting multiple conformations (Noer et al. 2016), it is generally accepted that folding hGQ in physiologically relevant (100-150 mM) NaCl results in a single antiparallel conformation, which manifests as a single peak in smFRET histograms (Long and Stone 2013). On the other hand, at least two different conformations are observed for hGQ in similar KCl concentrations, hence two peaks may be observed in smFRET histograms if these conformations result in different end-to-end distances. With a properly designed DNA construct, the distinct folding patterns can be used to study conformation dependence of protein-mediated destabilization or small-molecule induced stabilization, among other topics. However, GQ stability is in general significantly higher in KCl compared to NaCl (Tran et al. 2011), with rare exceptions (Saintome et al. 2016). Therefore, the observed variations between NaCl and KCl may not be purely due to different conformations. For example, it was demonstrated that performing the same competition assay between RPA and POT1 to access a hGQ forming telomeric sequence in NaCl or KCl resulted in contrasting conclusions for the impact of hGQ in this competition, primarily due to the difference in the stability of hGQ in these salts (Ray et al. 2014).
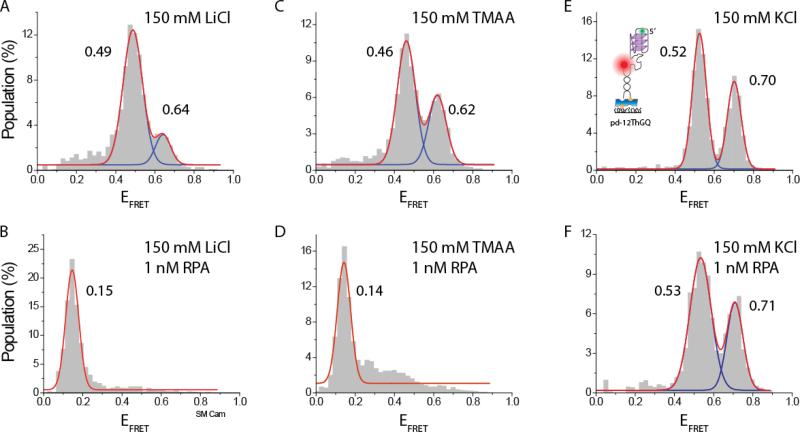
Occasionally, it might be necessary to identify the FRET level representing the unfolded GQ state when GQ interacts with an external destabilizing agent, such as a protein. However, in protein-mediated GQ unfolding, it may not be possible to distinguish the unfolded GQ state from 'protein-bound unfolded GQ state' or 'protein-bound folded GQ state' as they might all have similar FRET levels. In this case, it is practical to use DNA constructs that manifest only one of these states for a reliable identification. In order to identify the FRET level for the unfolded GQ state it is possible to use a DNA construct with a polythymine (poly-T) overhang that has a similar length to that of the GQ forming construct (see the schematic of pd-35T for an example) or a mutated version of the GQ construct in which several G's are replaced with other nucleotides such that GQ or a possible G-triplex (GT) formation is prevented. As such sequences would not form secondary structures, they should mimic the FRET level of the unfolded GQ state. Similarly, binding of a protein to the poly-thymine of mutated GQ constructs can be used to identify the FRET level for the 'protein-bound unfolded state'. The advantage of this approach is that the assay conditions can be kept identical to those used for the GQ forming construct. However, these measurements are indirect and often require purchasing multiple fluorescently labeled and purified DNA constructs which could be costly. It would be ideal if folding of the GQ could be prevented under ionic conditions that mimic those of interest. This would eliminate the need for using additional DNA constructs and could potentially improve reproducibility and accelerate the data acquisition process as the measurements can be performed within the same sample chamber. LiCl is often used in bulk studies to either prevent GQ formation or obtain a less stable GQ compared to those that form in KCl or NaCl. However, we observe a compact structure for the pd-12ThGQ construct in 150 mM LiCl with similar FRET peaks to the folded GQ peaks observed in KCl or NaCl (Figure 3A). In this case, the sample was folded at room temperature (annealed in the absence of salt) and two peaks at EFRET=0.49±0.05 and EFRET=0.64±0.05 were observed. The folding pattern was very similar for the sample that was annealed in 150 mM LiCl and maintained at that salt throughout the measurement. In bulk assays such as circular dichroism (CD), hGQ typically does not demonstrate GQ signature spectra in LiCl (see Supplementary Figure S2 of reference (Ray et al. 2014)). In the absence of supporting evidence from other methods, it is not possible to a claim that the observed peaks are due to GQ rather than another secondary structure that forms because of the electrostatic shielding provided by Li+. However, the similarity of the peak positions to those observed in KCl and NaCl suggests that these peaks most likely represent folded GQs. This interesting contrast suggests that smFRET may be more sensitive in detecting GQs with low stability compared to bulk methods such as CD.
Fig. 3.
smFRET studies on pd-12ThGQ at 150 mM LiCl in (A) the absence of RPA and (B) in the presence of 1 nM RPA. Similar studies at 150 mM TMAA in (C) the absence of RPA and (D) in the presence of 1 nM RPA. Similar studies at 150 mM KCl in (E) the absence of RPA and (F) in the presence of 1 nM RPA. While 1 nM RPA is adequate to unfold all of GQ constructs in LiCl, and most in TMAA, it does not unfold any of the structures in KCl. The schematics of pd-12ThGQ is shown in (E) since GQ formation in KCl is demonstrated by other methods as well, which is not the case for LiCl or TMAA.
TMAA has been identified as a promising volatile electrolyte in electrospray mass spectrometry that does not intercalate within the GQ structure and significantly reduces non-specific binding of K+ adducts to the external sites in the GQ. The trimethylammonium ion is too large to coordinate between G-quartets and has a high enough affinity to external sites that can drive K+ adducts away from these sites. When the TMAA/KCl ratio is high, e.g. 1 mM KCl and 100 mM TMAA, almost all non-specific binding of K+ adducts can be prevented, while K+ ions that specifically intercalate between the G-quartets are not disturbed and clearly identified (Marchand and Gabelica 2014). The implication of these for smFRET measurements is that TMAA might enable maintaining ionic strength comparable to that of physiological conditions while not stabilizing the GQ, which could be significant in various contexts. However, our smFRET measurements showed that 150 mM TMAA also results in FRET peaks similar to those of other salts: EFRET=0.46±0.05 and EFRET=0.62±0.05. Therefore, it may also not be possible to use TMAA to identify the peak position representing the unfolded GQ state in smFRET measurements.
Whether LiCl or TMAA can be used to identify the unfolded GQ state depends on the stability of the folded GQ structure. In our case, they were not useful however, 100 mM LiCl was reported to result in a completely unfolded GQ state for a weak GQ structure in another smFRET assay (Noer et al. 2016). However, regardless of the specifics of the GQ construct, LiCl and TMAA are expected to result in a significantly weaker GQ compared to KCl. In order to confirm this, we tested the stability of pd-12ThGQ construct against RPA-mediated GQ unfolding in 150 mM of LiCl, TMAA or KCl (Figure 3). These data show that while 1 nM RPA almost completely unfolds pd-12hGQ in TMAA and LiCl, it does not result in any unfolding in KCl. In these data, the unfolding of the GQ and binding of RPA to the unfolded DNA results in a low FRET peak at EFRET=0.15±0.03 in LiCl and EFRET=0.14±0.03 in TMAA. Therefore, even though LiCl or TMAA cannot be used to obtain the unfolded GQ state, they might be useful in determining the protein-bound unfolded state as they would require much less protein concentrations, compared to KCl, to unfold the GQ.
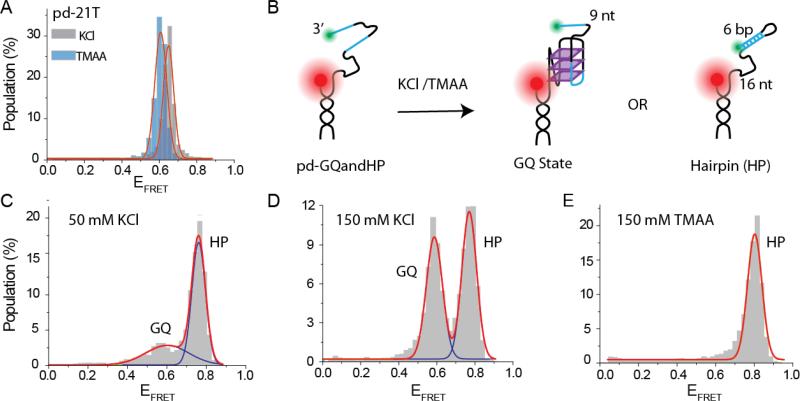
We also performed several measurements in TMAA to investigate its potential use in smFRET assays. In the first measurement we studied whether a DNA construct with a poly-T overhang results in similar FRET peaks in KCl and TMAA. We used pd-21T construct which has a 18 base pair (bp) duplex stem and 21 thymine overhang, and performed smFRET measurements in 150 mM KCl or 150 mM TMAA. As shown in Figure 4, the FRET peaks are very similar for the two cases suggesting the compaction due to electrostatic shielding is similar in the two cases. This essentially confirms that in the unfolded GQ state, similar FRET peaks are expected in TMAA and KCl. We then performed a competition assay between hGQ formation and hairpin formation using a construct shown in Figure 4B, which was adapted from reference (Liu et al. 2010). The ssDNA segment of this construct is capable of either forming a hGQ or a hairpin. When kept in 50 mM KCl, the construct results in two FRET peaks, which we attribute to the hairpin and hGQ but do not know which peak represents which structure. Repeating the same competition in 150 mM KCl resulted in a significant increase in the lower FRET peak population, suggesting that the low FRET peak represents the hGQ while the high FRET peak represents the hairpin (Figure 4D). Therefore, performing this competition in 150 mM TMAA is expected to result in a significantly higher population in the high FRET peak since the TMAA is expected to stabilize the hairpin at a much greater level compared to GQ. As shown in Figure 3E, the FRET distribution in 150 mM TMAA shows a complete domination of the higher FRET peak with almost no population at the lower FRET peak. In our experience, this is an unusually clean result within the context of GQ competition assays, and we believe demonstrates the potential of TMAA for these assays.
Fig. 4.
(A) The FRET peaks for pd-21T construct in 150 mM TMAA and 150 mM KCl. The similarity of the peak positions suggests the compaction of the coiled DNA due to electrostatic shielding is similar for KCl and TMAA. (B) A schematic of a competition assay between GQ and hairpin formation. The cyan colored segments in the overhang are complementary to each other and form the 6-bp stem of the hairpin when hybridized (see Table 1 for sequence). Hairpin formation would prevent GQ folding therefore, the system should either form a GQ or a hairpin. (C) The smFRET distribution at 50 mM KCl. (D) The smFRET distribution at 150 mM KCl. The population of lower FRET peak has increased compared to 50 mM KCl data suggesting that this peak represents the GQ state and the higher FRET peak represents the hairpin state. (E) The smFRET distribution at 150 mM TMAA. A single peak consistent with the hairpin state is observed, which agrees with significantly reduced stabilization of the GQ state in TMAA.
Unfolding the GQ
Bulk measurements have shown that incubating a folded GQ in 50 mM LiOH for about 10 minutes is an effective and possibly universal way of unfolding GQ structures (Ribeyre et al. 2009). Being able to quickly and effectively unfold the GQ would provide a significant convenience in smFRET measurements, particularly when an ion exchange is necessary or when the ion concentration needs to be reduced. Under such conditions, unfolding the GQ essentially resets the system and enables testing new conditions without the influence of earlier measurements. The standard method to unfold GQ is to perform a buffer exchange and incubate the sample in the absence of any salt. However, depending on the GQ this could be a slow process that might take many hours. A quick and more efficient method would have significant practical advantages, making LiOH a potentially attractive alternative.
Motivated by these, we attempted to unfold the pd-12ThGQ construct which was attached to a biotin-PEG surface and has a 18-bp duplex stem (see Figure 1A for a schematic). We tried 50 mM LiOH and 10-minute incubation as implemented in bulk studies. However, this resulted in unwinding of the 18 bp duplex stem and release of the donor fluorophore that was attached to the non-biotinylated strand. DNA is known to denature at high pH due to deprotonation of bases (Ageno et al. 1969), which limits the LiOH concentration that could be used in these assays. As most smFRET studies are performed with pdDNA constructs that have a duplex stem, we sought to test whether it is possible to unfold the GQ before unwinding the duplex stem at a particular LiOH concentration.
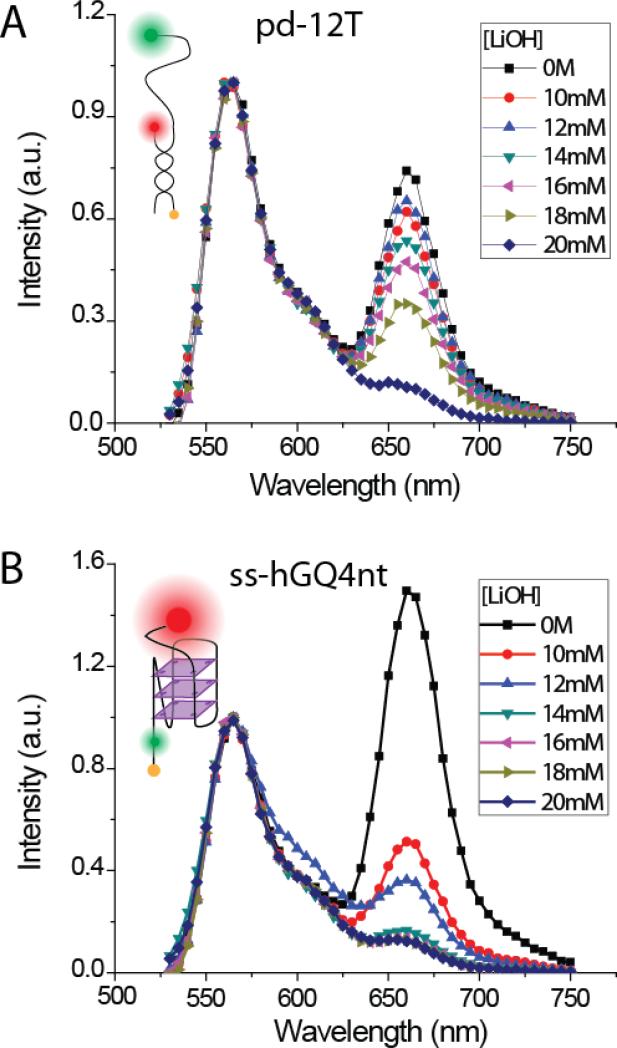
In order to determine an approximate LiOH concentration range, we performed bulk FRET measurements on two DNA constructs: one for probing GQ unfolding and the other for unwinding the duplex stem. ss-hGQ4nt was used to probe GQ unfolding, which is a single stranded DNA, without a duplex stem, that contains a hGQ-forming sequence and donor-acceptor fluorophores (Cy3-Cy5). Unfolding of hGQ would increase the separation between Cy3-Cy5 and result in a drop in the FRET efficiency. In bulk FRET this corresponds to a decrease in the acceptor peak and an increase in the donor peak. pd-12T construct was used for probing duplex unwinding since it contains an 18 bp duplex stem and just a 12T poly-thymine overhang. The donor is on one strand and acceptor on the other strand. Unwinding the duplex stem separates the strands and results in FRET decrease. Our initial measurements suggested 10-20 mM LiOH range to be potentially promising at which significant GQ unfolding is not accompanied by duplex unwinding. Therefore, we performed more detailed measurements in this concentration range as shown in Figure 5. The peak at 550 nm represents the donor emission and the peak at 670 nm represents the acceptor emission (Figure 5). The data are normalized with respect to the maximum of the donor peaks, and the magnitude of the acceptor peak is a measure of FRET. As Figure 5A demonstrates, the duplex unwinding is gradual in this concentration range, with only 12% reduction in acceptor peak amplitude between 0 M and 12 mM LiOH. On the other hand, GQ unfolds at significant levels at 12 mm LiOH, with 76% reduction in acceptor peak amplitude between 0 and 12 mM LiOH. Therefore, we identified 12 mM LiOH as a promising concentration at which significant GQ unfolding could be obtained with minimal duplex unwinding, and proceeded with performing smFRET measurements on pd-12ThGQ construct which contains a 18-bp duplex stem and a hGQ forming overhang (see Figure 1A for a schematic).
Fig. 5.
Bulk FRET measurements to identify a range of LiOH concentrations at which GQ is unfolded without significant unwinding of the 18-bp duplex DNA stem. (A) Bulk FRET measurements on pd-12T (see inset for construct schematics) construct showing gradual decrease in the acceptor signal as LiOH concentration is increased. The decrease in acceptor signal represents unwinding of the duplex stem. (B) Bulk FRET measurements on ss-hGQ4nt construct (see inset for construct schematics). A sharp decrease in the acceptor signal even at 10 mM LiOH indicates significant GQ unfolding for this construct. 12 mM LiOH was identified as a potentially promising concentration at which most GQ molecules are unfolded while only a small fraction of duplex stems is unwound.
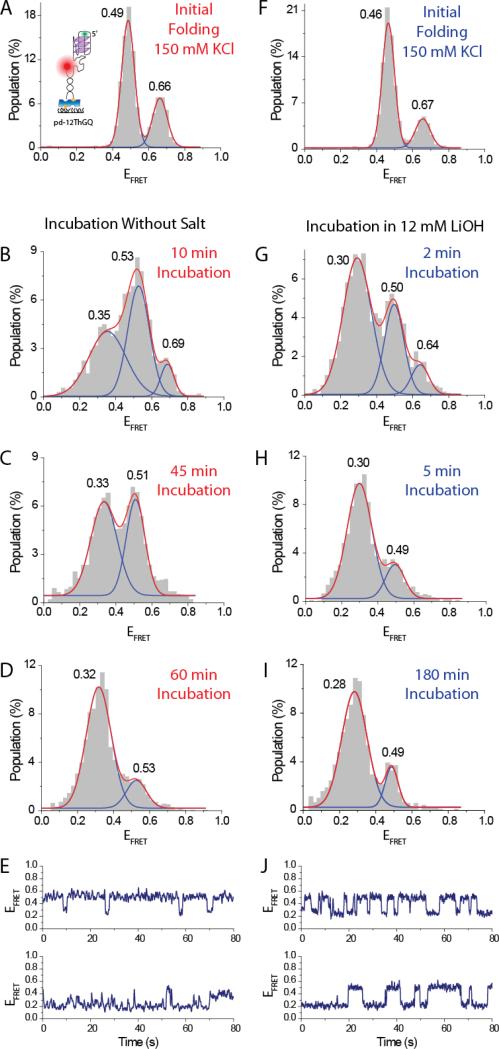
Figure 6A shows the initial folding of pd-12ThGQ at 150 mM KCl, which shows the clearly identifiable two peaks at EFRET=0.49±0.03 and EFRET=0.66±0.04, corresponding to the two folding conformations. We then performed a buffer exchange and incubated the sample in 50 mM Tris, in the absence of salt. We accumulated data at different times as shown in Figure 6B-D. Similarly GQ unfolding results in FRET reduction, resulting in a peak at EFRET≈0.33. The unfolding is gradual with most of the GQ constructs unfolded 60 minutes after the buffer exchange. In order to investigate the nature of the small folded population, we examined the smFRET time traces. Figure 6E shows two such traces where we observed transitions between folded and unfolded states even after 60 min incubation. Such transitions would explain the persistence of this small folded peak. We believe the high FRET peak (EFRET≈0.50) observed in these traces and in histogram of Fig. 6D (also observed in histogram of Fig. 6I and traces of Fig. 6J in LiOH case) represents the transiently folded GQ state in the anti-parallel conformation due to several reasons including the similarity of the FRET levels. The assignment of FRET levels to particular GQ folding conformations was discussed in Fig. 1, and we refer to those arguments for this assignment. In addition, the anti-parallel conformation of GQ was demonstrated to be the thermodynamically more stable conformation, further supporting this interpretation. Finally, the alternative interpretation of the EFRET≈0.50 level to be due to some intermediate GQ folding state (such as G-triplex) is not likely as the FRET level representing such an intermediate folding state is expected to be lower than the FRET level representing folded GQ, as some part of the structure would be unfolded in the intermediate folded state. But this is not the case and the FRET peaks are very similar. In principle, it is possible that this FRET peak could be representing some other secondary structure that is as compact as GQ, but we are not aware of the existence of such a structure.
Fig. 6.
A comparison of GQ unfolding by incubating the sample in the absence of salt or in 12 mM LiOH. (A) Initial folding of pd-12ThGQ construct in 150 mM KCl. (B), (C), and (D) show smFRET distributions after incubating the sample in the absence of salt for 10 min, 45 min, and 60 min, respectively. A low FRET peak representing the unfolded state is gradually populated in time. (E) smFRET time traces showing the dynamics between the two peaks in the histogram after 60 min incubation. The spontaneous transitions to the high FRET state are observed. (F) The DNA is refolded by introducing 150 mM KCl into the chamber. (G), (H), and (I) show smFRET distributions after incubating the sample in 12 mM LiOH for 2 min, 5 min, and 180 min, respectively. The 5 min and 180 min incubation in LiOH are very similar suggesting that the system has reached steady state in terms of the distribution of folded and unfolded states after 5 min incubation. The 5 min incubation in 12 mM LiOH results in a similar distribution as 60 min incubation in the absence of salt. (J) smFRET time traces showing transitions between the folded and unfolded states even after 180 min incubation in 12 mM LiOH. The transitions to the high FRET state explain the persistence of the folded peak in the histogram even after 180 min incubation.
We then introduced 150 mM KCl to the chamber and refolded the GQ as shown in Figure 6F. A folding pattern very similar to the initial folding is obtained with FRET peaks at EFRET=0.46±0.03 and EFRET=0.67±0.04. We then introduced 12 mM LiOH in 50 mM Tris and incubated the sample for different time intervals. After this incubation period we introduced imaging buffer at 50 mM Tris in the absence of salt. Figure 6G-I show the data after 2, 5, and 180 min incubation in 12 mM LiOH. These data demonstrate an unfolded peak at EFRET≈0.30. At 5 min incubation in 12 mM LiOH (Figure 6H), we reach a similar unfolding level as obtained at 60-min incubation in the absence of salt (Figure 6D), suggesting a significant acceleration in the GQ unfolding process. We observed negligible difference between 5-min and 180 min incubation periods, suggesting the sample has reached steady state after 5-min incubation. The persistence of the high FRET peak even after 180 min incubation can again be explained by the spontaneous transitions to the high FRET state, i.e. the folded state (Figure 6J). In both Fig. 6H and Fig. 6J, the single molecule traces demonstrate significantly different kinetics from each other, which illustrates the heterogeneity of the system. However, these heterogeneities are lost at the ensemble level as the population of folded and unfolded states essentially reaches a steady state after a certain time. To illustrate, the histograms of 5 min and 180 min incubation in LiOH look very similar, despite the large variations in the kinetics of the single molecules.
We also checked the number of donor molecules that dissociated from the surface upon incubation at 12 mM LiOH. We observed 97% and 89% of all molecules remained on surface after 2 min and 5 min incubation, respectively. Taking into account the small fraction of molecules that might have photobleached, despite the very short imaging time to minimize photobleaching, it is expected that the duplex stem of <10% of molecules were unwound after incubating at 12 mM LiOH for 5 min. A similar measurement at 14 mM LiOH showed 57% of all molecules to remain on surface after only 0.5 min incubation, illustrating the narrow range of LiOH concentration that could be used in this assay. Finally, we tested whether the GQ could be unfolded and refolded multiple times with 12 mM LiOH and 150 mM KCl exchange. We performed three unfolding and refolding treatments and obtained a similar folding pattern in each of them.
Discussion
smFRET has proved to be a versatile and potent tool in studying GQ structures and their interacting partners. It has been shown that smFRET data provide high resolution and high content information on both steady state and the dynamics of GQ structures and their interactions. However, these structures, particularly the most commonly used hGQ, have their complications and particular attention has to be paid in the way the sample is prepared, stored, and studied to attain reproducible and reliable results. With this article we aimed to illustrate some of these experimental complications and provide practical insights to address them.
Table 1.
Sequences of the DNA constructs. The sequences that form the stem are shown in bold fonts. The sequences forming GQ are underlined. In construct pd-GQandHP, the hairpin forming sequences are shown in italic fonts.
| Construct | Sequence (5′ to 3′) | Stem Complement |
|---|---|---|
| pd-hGQ12T | TGGCGACGGCAGCGAGGC TT GGGTTAGGGTTAGGGTTAGGG T12-Cy3 | Strand 1 |
| pd-12ThGQ | Cy3-TT GGGTTAGGGTTAGGGTTAGGG T12 TGGCGACGGCAGCGAGGC | Strand 2 |
| ss-hGQ10T | Biotin-TTT-Cy3-TT GGGTTAGGGTTAGGGTTAGGG T10-Cy5 | |
| ss-hGQ4nt | Biotin-TTT/iCy3/ TT GGGTTAGGGTTAGGGTTAGGG TTAG-Cy5 | |
| pd-35T | Cy3-T35 TGGCGACGGCAGCGAGGC | Strand 2 |
| pd-21T | TGGCGACGGCAGCGAGGC T21-Cy3 | Strand 1 |
| pd-12T | Cy3-T12 TGGCGACGGCAGCGAGGC | Strand 2 |
| pd-GQandHP | TGGCGACGGCAGCGAGGC TT GGGTTAGGGTTAGGGTTAGGG TTTCCTAAC-Cy3 | Strand 1 |
| Strand 1 | Cy5-GCC TCG CTG CCG TCG CCA-Biotin | |
| Strand 2 | Biotin- GCC TCG CTG CCG TCG CCA-Cy5 |
Acknowledgements
We thank Prof. Victoria Birkedal and Prof. Michael Stone for sharing extensive information regarding their experimental protocols with us. This study was funded by U.S. National Institutes of Health [grant number 1R15GM109386 to H.B.].
Footnotes
Compliance with ethical standards
Conflict of interest All the authors declare no conflict of interests.
Ethical approval This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Ageno M, Dore E, Frontali C. The alkaline denaturation of DNA. Biophys J. 1969;9:1281–1311. doi: 10.1016/S0006-3495(69)86452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang I. Untersuchungen über die Guanylsäure. Biochem. Ztschr. 1910;26:293–311. [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- Budhathoki Jagat B, Maleki P, Roy William A, Janscak P, Yodh Jaya G, Balci H. A Comparative Study of G-Quadruplex Unfolding and DNA Reeling Activities of Human RECQ5 Helicase. Biophys J. 2016;110:2585–2596. doi: 10.1016/j.bpj.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhathoki JB, Ray S, Urban V, Janscak P, Yodh JG, Balci H. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014;42:11528–11545. doi: 10.1093/nar/gku856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhathoki JB, Stafford EJ, Yodh JG, Balci H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015;43:5961–5970. doi: 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia R, Miller MC, Dean WL, Gray RD, Lane AN, Trent JO, Chaires JB. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires JB. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- Chandradoss SD, Haagsma AC, Lee YK, Hwang JH, Nam JM, Joo C. Surface passivation for single-molecule protein studies. J Vis Exp. 2014 doi: 10.3791/50549. 10.3791/50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Zagelbaum J, Savitsky P, Sturzenegger A, Huttner D, Janscak P, Hickson ID, Gileadi O, Rothenberg E. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun. 2014;5:5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci U S A. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu Rev Phys Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Sun D, Salazar M, Hurley LH. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua B, Han KY, Zhou R, Kim H, Shi X, Abeysirigunawardena SC, Jain A, Singh D, Aggarwal V, Woodson SA, Ha T. An improved surface passivation method for single-molecule studies. Nat Methods. 2014;11:1233–1236. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husby J, Todd AK, Platts JA, Neidle S. Small-molecule G-quadruplex interactions: Systematic exploration of conformational space using multiple molecular dynamics. Biopolymers. 2013;99:989–1005. doi: 10.1002/bip.22340. [DOI] [PubMed] [Google Scholar]
- Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Kreig A, Calvert J, Lormand J, Kwon Y, Daley JM, Sung P, Opresko PL, Myong S. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Majima S, Nakamura T, Seimiya H, Nagasawa K. Evaluation of the interaction between long telomeric DNA and macrocyclic hexaoxazole (6OTD) dimer of a G-quadruplex ligand. Molecules. 2013;18:4328–4341. doi: 10.3390/molecules18044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitsuka Y, Okumus B, Arslan S, Chen KH, Ha T. Temperature-independent porous nanocontainers for single-molecule fluorescence studies. Anal Chem. 82:9694–9701. doi: 10.1021/ac101714u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena PV, Shirude PS, Okumus B, Laxmi-Reddy K, Godde F, Huc I, Balasubramanian S, Ha T. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J Am Chem Soc. 2009;131:12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Okyere R, Joseph A, Musier-Forsyth K, Kankia B. Quadruplex formation as a molecular switch to turn on intrinsically fluorescent nucleotide analogs. Nucleic Acids Res. 2013;41:220–228. doi: 10.1093/nar/gks975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harb Protoc 2012. 2012 doi: 10.1101/pdb.top072058. [DOI] [PubMed] [Google Scholar]
- Kreig A, Calvert J, Sanoica J, Cullum E, Tipanna R, Myong S. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv749. 10.1093/nar/gkv749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger AC, Birkedal V. Single molecule FRET data analysis procedures for FRET efficiency determination: probing the conformations of nucleic acid structures. Methods. 2013;64:36–42. doi: 10.1016/j.ymeth.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Kruger AC, Raarup MK, Nielsen MM, Kristensen M, Besenbacher F, Kjems J, Birkedal V. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur Biophys J. 2010;39:1343–1350. doi: 10.1007/s00249-010-0587-x. [DOI] [PubMed] [Google Scholar]
- Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JQ, Chen CY, Xue Y, Hao YH, Tan Z. G-quadruplex hinders translocation of BLM helicase on DNA: a real-time fluorescence spectroscopic unwinding study and comparison with duplex substrates. J Am Chem Soc. 2010;132:10521–10527. doi: 10.1021/ja1038165. [DOI] [PubMed] [Google Scholar]
- Long X, Stone MD. Kinetic partitioning modulates human telomere DNA G-quadruplex structural polymorphism. PLoS One. 2013;8:e83420. doi: 10.1371/journal.pone.0083420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki P, Ma Y, Iida K, Nagasawa K, Balci H. A single molecule study of a fluorescently labeled telomestatin derivative and G-quadruplex interactions. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1090. 10.1093/nar/gkw1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand A, Gabelica V. Native electrospray mass spectrometry of DNA G-quadruplexes in potassium solution. J Am Soc Mass Spectrom. 2014;25:1146–1154. doi: 10.1007/s13361-014-0890-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLuckie KI, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, Tannahill D, Lowe C, Balasubramanian S. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J Am Chem Soc. 2013;135:9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3' sequence of d(AG3(TTAG3)3). J Am Chem Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- Neidle S. A Personal History of Quadruplex-Small Molecule Targeting. Chem Rec. 2015 doi: 10.1002/tcr.201500011. 10.1002/tcr.201500011. [DOI] [PubMed] [Google Scholar]
- Noer SL, Preus S, Gudnason D, Aznauryan M, Mergny JL, Birkedal V. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016;44:464–471. doi: 10.1093/nar/gkv1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht SA, Varavipour E, Nanjunda R, Pazitna I, Di Vita G, Gunaratnam M, Kumar A, Ismail MA, Boykin DW, Wilson WD, Neidle S. Discovery of new G-quadruplex binding chemotypes. Chem Commun (Camb) 2014;50:960–963. doi: 10.1039/c3cc48616h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B, Arslan S, Fengler SM, Myong S, Ha T. Single molecule nanocontainers made porous using a bacterial toxin. J Am Chem Soc. 2009;131:14844–14849. doi: 10.1021/ja9042356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B, Ha T. Real-time observation of G-quadruplex dynamics using single-molecule FRET microscopy. Methods Mol Biol. 2010;608:81–96. doi: 10.1007/978-1-59745-363-9_6. [DOI] [PubMed] [Google Scholar]
- Okumus B, Wilson TJ, Lilley DM, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MH, Ray S, Sewell AL, Basu S, Balci H. Replication Protein A Unfolds G-Quadruplex Structures with Varying Degrees of Efficiency. J Phys Chem B. 2012 doi: 10.1021/jp300546u. 10.1021/jp300546u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman KM, Tizkova K, Reszka AP, Neidle S, Thurston DE. Identification of novel telomeric G-quadruplex-targeting chemical scaffolds through screening of three NCI libraries. Bioorg Med Chem Lett. 2012;22:3006–3010. doi: 10.1016/j.bmcl.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc Natl Acad Sci U S A. 2014;111:2990–2995. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Celik U, Balci H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J. 2013;104:2235–2245. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintome C, Amrane S, Mergny JL, Alberti P. The exception that confirms the rule: a higher-order telomeric G-quadruplex structure more stable in sodium than in potassium. Nucleic Acids Res. 2016;44:2926–2935. doi: 10.1093/nar/gkw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin PR, Lougheed T, Tonks Hoffman M, Park H, Balci H, Blehm BH, Toprak E. Constructing Sample Chambers for Fluorescence Imaging with One-Nanometer Accuracy (FIONA). CSH Protoc 2007. 2007 doi: 10.1101/pdb.prot4867. pdb prot4867. [DOI] [PubMed] [Google Scholar]
- Shirude PS, Balasubramanian S. Single molecule conformational analysis of DNA G-quadruplexes. Biochimie. 2008;90:1197–1206. doi: 10.1016/j.biochi.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard S, Aznauryan M, Haustrup EK, Schiott B, Birkedal V, Corry B. Dynamics of fluorescent dyes attached to G-quadruplex DNA and their effect on FRET experiments. Chemphyschem. 2015;16:2562–2570. doi: 10.1002/cphc.201500271. [DOI] [PubMed] [Google Scholar]
- Stegle O, Payet L, Mergny JL, MacKay DJ, Leon JH. Predicting and understanding the stability of G-quadruplexes. Bioinformatics. 2009;25:i374–382. doi: 10.1093/bioinformatics/btp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Taylor A, Joseph A, Okyere R, Gogichaishvili S, Musier-Forsyth K, Kankia B. Isothermal quadruplex priming amplification for DNA-based diagnostics. Biophys Chem. 2013;171:1–8. doi: 10.1016/j.bpc.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Tippana R, Xiao W, Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42:8106–8114. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PL, Mergny JL, Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglasky V, Bauer L, Tluckova K, Javorsky P. Evaluation of human telomeric g-quadruplexes: the influence of overhanging sequences on quadruplex stability and folding. J Nucleic Acids 2010. 2010 doi: 10.4061/2010/820356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu L, Ren J, Qu X. Visualizing Human Telomerase Activity with Primer-Modified Au Nanoparticles. Small. 2011 doi: 10.1002/smll.201101938. 10.1002/smll.201101938. [DOI] [PubMed] [Google Scholar]
- Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H, Wu J, Shao F, Yan J. Stability and kinetics of c-MYC promoter G-quadruplexes studied by single-molecule manipulation. J Am Chem Soc. 2015;137:2424–2427. doi: 10.1021/ja511680u. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tateishi-Karimata H, Mergny JL, Cheng M, Feng Z, Miyoshi D, Sugimoto N, Li C. Reevaluation of the stability of G-quadruplex structures under crowding conditions. Biochimie. 2016;121:204–208. doi: 10.1016/j.biochi.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Du Y, Zhang L, Dong S. A label-free, G-quadruplex DNAzyme-based fluorescent probe for signal-amplified DNA detection and turn-on assay of endonuclease. Biosens Bioelectron. 2012;34:100–105. doi: 10.1016/j.bios.2012.01.024. [DOI] [PubMed] [Google Scholar]