Abstract
Antibiotic resistant Staphylococcus aureus is a major public health concern effecting millions of people annually. Medical science has documented completely untreatable S. aureus infections. These strains are appearing in the community with increasing frequency. New diagnostic and therapeutic options are needed to combat this deadly infection. Interestingly, around 50% of the proteins in S. aureus are annotated as hypothetical. Methods to select hypothetical proteins related to antibiotic resistance have been inadequate. This study uses differential gene expression to identify hypothetical proteins related to antibiotic resistant phenotype strain variations. We apply computational tools to predict physiochemical properties, cellular location, sequence-based homologs, domains, 3D modeling, active site features, and binding partners. Nine of 23 hypothetical proteins were <100 residues, unlikely to be functional proteins based on size. Of the 14 differentially expressed hypothetical proteins examined, confident predictions on function could not be made. Most identified domains had unknown functions. Six hypothetical protein models had >50% confidence over >20% residues. These findings indicate the method of hypothetical protein identification is sufficient; however, current scientific knowledge is inadequate to properly annotate these proteins. This process should be repeated regularly until entire genomes are clearly and accurately annotated.
Keywords: Annotations, Hypothetical proteins, Methicillin, S.aureus
Background
Antibiotic therapy has been the marvel of modern medicine since the advent of Penicillin in the 1920s. Over seventy billion doses of antibiotics are consumed globally each year [1]. Antibiotics are a low-cost resource to treat food-borne and other sanitation-related infections that commonly affect poor people. Among wealthier countries, antibiotics play a pivotal role as a prophylactic, controlling infections associated with medical practices such as surgery [1,2]. Unfortunately, this usage exposes normal microbial flora to anti-bacterial drugs, allowing them to develop resistances so the drugs lose effectiveness. Medical science has been unable to cultivate new antibiotics as fast as resistances to current therapies are rising [2,3]. Infectious organisms that are resistant to every antibiotic developed have been reported. This antibiotic resistance crisis is a critical challenge for humanity’s medical future.
Staphylococcus aureus, an opportunistic pathogen that was originally associated with hospital-acquired infections, was the first organism to show resistance to Penicillin and its synthetic offspring like Methicillin. Though hospital-acquired Methicillinresistant S. aureus (MRSA) cases proliferated through the late 20th century, recent years have seen decreases in the number of hospital-acquired MRSA infections due to improvements in sanitation procedures and increases in Vancomycin use despite its potential side effects [4]. Unfortunately, community-acquired MRSA infections have dominated recently since over 100 million people harbor MRSA strains as part of normal skin flora according to Dutch and United States prevalence data [5]. Therefore, the United States Center for Disease Control lists MRSA and Vancomycin-resistant S. aureus (VRSA) strains as serious and concerning public health threats, respectively, estimating over 80,000 invasive MRSA infections with 20,000 related deaths annually, many in immuno-compromised patients including children [5]. While there are no S. aureus strains currently resistant to all antibiotics, completely resistant strains of other infectious organisms have emerged so the same outcome will likely befall S. aureus soon.
A challenge to developing new antibiotic therapies is genome annotation. Around 50% of proteins identified in the S. aureus genome are annotated as hypothetical [6,7]. At annotation, hypothetical proteins are predicted by sequence only and lack homology to known proteins. Researchers further define hypothetical proteins by their larger than 100 amino acids size, since smaller sequences likely represent other macromolecular structures such as short interfering RNA (siRNA) rather than functional proteins [8]. True hypothetical proteins have similar features to other hypothetical proteins due to lack of experimental evidence to predict function for the protein family, though frequently hypothetical proteins found in databases represent old genome annotations in need of update. Several studies have used various methods to identify hypothetical proteins related to antibiotic resistance in S. aureus. Early studies randomly selected hypothetical proteins for characterization [6,7, 9,10]. While this approach developed and demonstrated computational procedures that contribute to hypothetical protein characterization, it is limited in its ability to identify hypothetical proteins specifically connected to antibiotic resistance. To improve the selection process, we formerly developed crossspecies approach that used proteins with experimentally established structures from the major facilitator superfamily; a large, highly conserved protein family associated with antibiotic resistance [7]. This approach worked because of the large percentage of hypothetical proteins in the S. aureus genome, but it becomes inadequate if a hypothetical protein related to resistance has no well-characterized homolog in another species, a common challenge for hypothetical proteins. Better methods for identifying antibiotic resistant-related hypothetical proteins are needed.
Microarray and other forms of publicly accessible gene expression data can provide an excellent repository for targeted identification of resistance linked hypothetical proteins in S. aureus. For example, Ham and colleagues examined mRNA expression between antibiotic resistant (MRSA; ATCC 33591, shown to be susceptible only to Vancomycin and Kanamycin) and sensitive (MSSA; ATCC 25923) strains using Affymetrix GeneChip® technology [11]. They statistically compared mRNA expression levels between the strains to uncover potential mechanisms of resistance, but did not consider hypothetical proteins. This presents an opportunity to characterize hypothetical proteins whose differential expression constitutes a drug-resistant genomic background.
This study uses computational procedures to characterize statistically significant differentially expressed hypothetical proteins from the microarray data generated by Ham and associates. By comparing natural gene expression between antibiotic sensitive and resistant strains, new insight into strain background differences is gained. These variations could uncover new resistance mechanisms, further developing into a useful diagnostic tool or potential antibiotic therapeutic target. This would improve outcomes for patients infected with MRSA strains through faster and more effective treatment options.
Methodology
Normalized mRNA expression data from Ham’s study is available at the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO; Dataset Record GDS4242; GEO accession GSE18289) [11]. Data consisted of 7774 entries, each with probe name and six samples representing triplicates of both MSSA (ATCC 25923) and MRSA (ATCC 33591) strains. Probe names were converted to gene names and descriptions per Affymetrix chip platform and non-hypothetical proteins were removed. Excel calculated T-scores and p-values based on Student’s T-test two-tailed, equal variance formulas. The study rejected hypothetical proteins with a p>0.05 as these were not differentially expressed. The National Center for Biotechnology Information (NCBI) and UniProt databases confirmed hypothetical protein annotation.
This study used numerous algorithms to characterize these hypothetical proteins and default program settings were used for all analyses. ExPASy’s Protparam server calculated physiochemical properties including number of amino acids, molecular weight, positively and negatively charged residues, theoretically isoelectric point (pI), extinction coefficient, aliphatic index (AI), instability index (II), and the grand average hydropathy (GRAVY) [12]. By hypothetical protein definition, those identified through differential expression yet smaller than 100 amino acids were excluded from further study.
PSortB and SOSUI servers predicted each hypothetical protein’s cellular location. PSortB predicted between cytoplasm, cytoplasmic membrane, cell wall, or extracellular locations [13]. SOSUI calculated transmembrane regions and solubility indices, a valuable confirmation of PSortB predictions [14]. These complementary algorithms provide confidence for cellular localization estimates.
Sequence similarity and domain identification projected functional features of hypothetical proteins. The Position-Specific Iterative (PSI) Basic Local Alignment Search Tool (BLAST) identified potential homologs from the NCBI database based on protein sequence similarities. Further, both Conserved Domain Database (CDD) BLAST and Pfam algorithms predicted potential domains within each hypothetical protein. CDD-BLAST uses a PSI-BLAST variation to identify domains by comparison of the protein sequence’s position specific scoring matrix to those in the NCBI database [15]. Alternatively, Pfam is a separately curated database of Hidden Markov Models and multiple sequence alignments representing protein domain families [16]. These complementary approaches provide a level of validation to this study’s findings.
For model development and characterization, we used the integrated Phyre2 and 3DLigandSite servers. Phyre2 produced a tertiary structure model, predicted ligand-binding sites, and analyzed the effect of amino acid variants through automatic homology detection methods [17]. Phyre2’s model advanced to 3DLigandSite for active site characterization and docking predictions. 3DLigandSite identifies homologous structures with bound ligands by searching a structural library then superimposing those ligands onto the Phyre2’s protein structure [18]. Together, Phyre2 and 3DLigandSite servers modeled the protein and characterized its binding site.
The Search Tool for Interactions of Chemicals (STITCH) database predicted potential ligand interactions for each hypothetical protein. STITCH draws upon scientific literature and several databases, including the formerly separate Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, which houses high-throughput experiment and conserved coexpression data, to calculate drug-target interactions, binding affinities, and biological pathways [19]. STITCH is a useful tool to predict protein and chemical binding partners.
Results
The mRNA expression dataset, GSE18289, was downloaded from GEO and Excel calculated the T-statistic and p-value for each protein. Twenty-seven proteins labeled as hypothetical in NCBI, 16 and 11 up- and down regulated in MRSA, respectively, had <0.05 p-values. Four of these proteins had predicted functions in UniProt, an endotoxin (SACOL0468, up regulated, T-score 9.00), exotoxin (SACOL1178, up regulated, T-score 10.17), phosphate dikinase regulatory protein (SACOL1620, down regulated, Tscore -9.80), and a lipoprotein (SACOL1531, up regulated, T-score 7.89). Since these proteins had predicted identities, they were excluded from further study. The remaining 23 proteins are listed by T-score in Table 1.
Table 1. Differential expression T-scores and physiochemical properties of 23 hypothetical proteins.
| Protein | T-score | # AA | MW | pI | # neg | # pos | EC | II | AI | GRAVY |
| SACOL0919 | 18.77 | 45 | 5270 | 9.03 | 4 | 6 | 2980 | 17.29 | 136.22 | 0.56 |
| SACOL1859 | 12.23 | 1016 | 120681 | 5.7 | 147 | 130 | 161360 | 37.5 | 95.75 | -0.415 |
| SACOL1346 | 10.83 | 64 | 7573 | 4.1 | 16 | 6 | 5960 | 21.26 | 92.81 | -0.42 |
| SACOL0356 | 9.29 | 78 | 8726 | 4.32 | 17 | 7 | 5960 | 41.83 | 86.28 | -0.529 |
| SACOL0326 | 7.48 | 74 | 8841 | 4.54 | 18 | 7 | 11460 | 52.27 | 77.7 | -0.938 |
| SACOL0323 | 7.32 | 102 | 11944 | 7.91 | 17 | 18 | 9970 | 33.41 | 89.8 | -0.762 |
| SACOL0109 | 6.83 | 135 | 15123 | 4.45 | 14 | 8 | 26930 | 39.15 | 132.89 | 0.757 |
| SACOL0087 | 6.62 | 35 | 4172 | 6 | 5 | 5 | 4470 | 25.62 | 94.57 | -0.149 |
| SACOL0075 | 6.04 | 200 | 22662 | 9.55 | 9 | 20 | 31860 | 42.51 | 121.35 | 0.665 |
| SACOL0644 | 5.35 | 208 | 24690 | 9.55 | 14 | 25 | 43890 | 29.04 | 125.48 | 0.448 |
| SACOL0350 | 3.8 | 118 | 13923 | 10.08 | 14 | 28 | 12950 | 26.44 | 76.02 | -0.804 |
| SACOL0362 | 3.77 | 66 | 7806 | 8.03 | 8 | 9 | 9970 | 37.99 | 125.45 | 0.185 |
| SACOL2481 | 3.23 | 121 | 14067 | 4.54 | 21 | 11 | 5960 | 35.52 | 118.43 | -0.098 |
| SACOL0835 | -2.56 | 209 | 24070 | 9.07 | 31 | 36 | 8940 | 64.82 | 34.16 | -1.974 |
| SACOL2241 | -6.45 | 129 | 14638 | 9.73 | 3 | 8 | 18450 | 26.53 | 155.74 | 1.209 |
| SACOL2123 | -6.59 | 223 | 25856 | 4.74 | 43 | 28 | 28550 | 41.8 | 93 | -0.289 |
| SACOL2491 | -8.97 | 63 | 7221 | 4.6 | 10 | 6 | 7450 | 25.52 | 97.46 | -0.146 |
| SACOL2571 | -9.8 | 63 | 7266 | 5.44 | 9 | 7 | 1490 | 17.06 | 103.65 | -0.233 |
| SACOL2076 | -10.78 | 45 | 5070 | 10.46 | 3 | 10 | 1 | 45.35 | 114.67 | -0.424 |
| SACOL1956 | -14.64 | 176 | 20513 | 9.25 | 10 | 15 | 21555 | 39.83 | 132.95 | 0.747 |
| SACOL0267 | -15.31 | 507 | 57978 | 8.02 | 93 | 95 | 36790 | 23.76 | 74.48 | -0.906 |
| SACOL0488 | -24.11 | 107 | 13458 | 5.23 | 26 | 21 | 15930 | 65.54 | 59.25 | -1.693 |
| SACOL0710 | -25.02 | 165 | 19009 | 5.08 | 27 | 17 | 10430 | 33.94 | 100.48 | -0.181 |
| # AA, number of amino acids; MW, molecular weight; pI, theoretical isoelectric point; # neg, total number of negatively charged residues (Asp + Glu); # pos, total number of positively charged residues (Arg + Lys); EC, extinction coefficient assuming all pairs of Cys residues form cystines; II, instability index; AI, aliphatic index; GRAVY, grand average hydropathy. 1As there are no Trp, Tyr, or Cys in the region considered, protein should not be visible by UV spectrophotometry. | ||||||||||
Few hypothetical proteins had well defined homologs in the NCBI database as identified by PSI-BLAST (Table 4). Most top homologs came from S. aureus and were vaguely annotated or had low sequence similarity to the hypothetical protein. Three proteins, SACOL0323, SACOL2481, and SACOL0710, had homologs from other species, Mucilaginibacter, Helicobacter mustelae, and Bacillus cereus, respectively. Interestingly, four hypothetical proteins had membrane protein for their top homolog. PSortB and SOSUI confirm that SACOL0109, SACOL0075, and SACOL2241 are likely membrane proteins too (Table 2 and 3, respectively). However, according to these algorithms, SACOL2481 is a soluble, cytoplasmic protein. Further, PSortB predicted SACOL0488 to reside in the cytoplasm, which PSI-BLAST’s top homolog confirmed, though PSortB was unable to confirm extracellular locations for SACOL0835 and SACOL0267 where top homologs are exported proteins. Interestingly, for SACOL2123, PSI-BLAST identified its top homolog as a PF11042 family member. This matched CDDBLAST domain identification of pfam11042, showing the interconnectivity of these computational tools, and was confirmed by Pfam itself.
Table 4. Top PSI-BLAST result for 14 hypothetical proteins.
| Protein | PSI-BLAST Match | Query Cover | E-value | Identity |
| SACOL1859 | NTPase | 100% | 0 | 100% |
| SACOL0323 | Metallophosphoesterase | 59% | 1.6 | 31% |
| SACOL0109 | Membrane protein | 100% | 3.00E-44 | 59% |
| SACOL0075 | Membrane spanning protein | 90% | 7.00E-124 | 98% |
| SACOL0644 | tandem five-TM protein | 100% | 1.00E-143 | 99% |
| SACOL0350 | Phage protein | 100% | 5.00E-80 | 98% |
| SACOL2481 | Outer membrane protein | 59% | 4.3 | 27% |
| SACOL0835 | Exported protein | 91% | 8.00E-128 | 100% |
| SACOL2241 | Membrane protein | 79% | 3.00E-64 | 100% |
| SACOL2123 | PF11042 family protein | 100% | 1.00E-91 | 65% |
| SACOL1956 | Permease | 100% | 3.00E-123 | 100% |
| SACOL0267 | Exported protein | 51% | 8.00E-170 | 98% |
| SACOL0488 | Cytosolic protein | 89% | 2.00E-59 | 100% |
| SACOL0710 | RHS repeat-associated core domain-containing protein | 87% | 1.00E-09 | 29% |
Table 2. PSortB cellular location of 14 hypothetical proteins.
| Protein | Location | Localization Score |
| SACOL1859 | Unknown | 2.501 |
| SACOL0323 | Cytoplasm | 7.5 |
| SACOL0109 | Cytoplasmic membrane | 10 |
| SACOL0075 | Cytoplasmic membrane | 10 |
| SACOL0644 | Cytoplasmic membrane | 10 |
| SACOL0350 | Unknown | 2.501 |
| SACOL2481 | Cytoplasm | 7.5 |
| SACOL0835 | Cytoplasmic membrane | 9.55 |
| SACOL2241 | Cytoplasmic membrane | 10 |
| SACOL2123 | Cytoplasm | 7.5 |
| SACOL1956 | Cytoplasmic membrane | 10 |
| SACOL0267 | Unknown | 3.332 |
| SACOL0488 | Cytoplasm | 7.5 |
| SACOL0710 | Cytoplasm | 7.5 |
| 1Equal probability of the protein being located in any cellular structure: cytoplasm, cytoplasmic membrane, cell wall, or extracellular. 2Equal probability of protein being located in cytoplasmic membrane, cell wall, or extracellular. | ||
Table 3. SOSUI results for 7 transmembrane hypothetical proteins.
| Protein | N-terminal | Transmembrane Region | C-terminal | Type | Length |
| SACOL0109 | 53 | IGKIAIWIGIVAQIYFSVVFVRM | 75 | PRIMARY | 23 |
| 89 | IFLLGLILALFTVLPTIFTAIYM | 111 | PRIMARY | 23 | |
| 123 | IVYAIIALCLYNFLSSILWLIGG | 145 | PRIMARY | 23 | |
| SACOL0075 | 7 | KIAIWIGIVAQIYFSVVFVRMIS | 29 | PRIMARY | 23 |
| 41 | IFLLGLILALFTVLPTIFTAIYM | 63 | PRIMARY | 23 | |
| 75 | IVYAIIALCLYNFLSSILWLIGG | 97 | PRIMARY | 23 | |
| SACOL0644 | 23 | YLLIDLVSTWLVYFFPFINWFIP | 45 | SECONDARY | 23 |
| 94 | QLDNKILISLCFIGFIGIAAFYI | 116 | PRIMARY | 23 | |
| 147 | SFIVFTYLLLGGCSILFLIWLMT | 169 | PRIMARY | 23 | |
| 174 | NLLVFIMWIIITIFFFLISMGSI | 196 | PRIMARY | 23 | |
| SACOL0835 | 23 | AKVVSIATVLLLLGGLVFAIFAY | 45 | PRIMARY | 23 |
| SACOL2241 | 10 | ALIGIFLILCEFFYGIPFLGATF | 32 | PRIMARY | 23 |
| 40 | PLLFNALLYLILTIILLVNRQNA | 62 | PRIMARY | 23 | |
| 65 | PIAIIPIFGIVGSFLAIIPFLGI | 87 | PRIMARY | 23 | |
| 90 | HWILFFLMILFVLVVLSAPTYIP | 112 | PRIMARY | 23 | |
| SACOL1956 | 16 | FIILQLVIALFVILFTYKWALGV | 38 | PRIMARY | 23 |
| 50 | LVYGFAGFIILLILHELIHRALF | 72 | PRIMARY | 23 | |
| 103 | QFSIIMLSPLILLSTGLLILIKV | 125 | PRIMARY | 23 | |
| 134 | MFSMHTAYCFIDILLVALTISSS | 156 | PRIMARY | 23 | |
| SACOL0267 | 6 | KIIIPIIIVLLLIGGIAWGVYAF | 28 | PRIMARY | 23 |
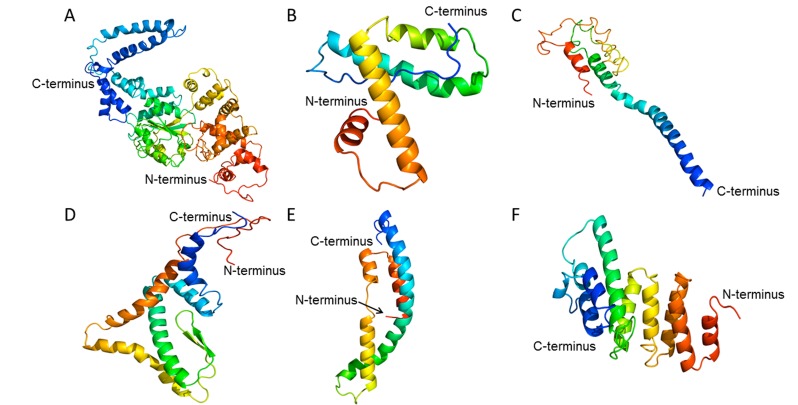
Phyre2 and 3DLigand servers performed hypothetical protein modeling and active site characterization. Similarity measurements of the hypothetical protein target to its experimental structure template are in Table 7. These findings represent Phyre2 running in normal mode. Hypothetical proteins with coverage >25% in normal mode were re-run under Phyre2’s intensive mode with the results show in Figure 2. Remarkably, under this mode, SACOL1859 and SACOL0710 models had 88% and 89% residues modeled with >90% confidence. No amino acids from the other four proteins could be modelled with that confidence. Unfortunately, 3DLigand was unable to make a prediction for any hypothetical protein examined in this study due to insufficient homologous structures with ligands bound.
Table 7. Phyre2 model data for 14 hypothetical proteins.
| Protein | Template | Template Description | Confidence | Coverage |
| SACOL1859 | c4kxfF | nlr family card domain-containing protein 4 | 99.70% | 30% |
| SACOL0323 | d1nu9c1 | immunoglobulin/albumin-binding domain-like | 37.80% | 25% |
| SACOL0109 | c3x29A | crystal structure of mouse claudin-19 | 73.20% | 45% |
| SACOL0075 | c4zxsD | virion egress protein ul31 | 55.60% | 20% |
| SACOL0644 | c4yjxB | ATP-dependent clp protease adapter protein | 30.70% | 7% |
| SACOL0350 | c2qdqA | talin-1 | 40.70% | 21% |
| SACOL2481 | c3daoB | putative phosphatse | 23.30% | 17% |
| SACOL0835 | c2ifmA | pf1 filamentous bacteriophage | 80.30% | 14% |
| SACOL2241 | c2ap8A | bombinin h4 | 43.20% | 10% |
| SACOL2123 | c1zctB | glycogenin-1 | 46.30% | 12% |
| SACOL1956 | c3b4rB | putative zinc metalloprotease mj0392 | 89.30% | 41% |
| SACOL0267 | c3jcuj | photosystem ii reaction center protein j | 50.40% | 5% |
| SACOL0488 | c4c46B | general control protein gcn4 | 80.60% | 29% |
| SACOL0710 | c1kt0A | lare fkbp-like protein, fkbp51, involved in steroid2 receptorcomplexes | 93.60% | 47% |
Figure 2.
Phyre2’s intensive mode models for hypothetical proteins SACOL1859 (A), SACOL0323 (B), SACOL0109 (C), SACOL1956 (D), SACOL0488 (E), and SACOL0710 (F). Image colored by rainbow N- to C-terminus.
STITCH predicted binding partners for hypothetical proteins. STITCH was unable to predict binding partners for the following hypothetical proteins: SACOL2481, SACOL0835, and SACOL2241. Most top binding partners were fellow hypothetical proteins with confidence scores listed in Table 8. This implies that more database annotation and/or wet bench work are needed to fully understand how these proteins work. SACOL0323, SACOL2123, and SACOL0710 had top matching binding partners that were not hypothetical proteins.
Table 8. Top STITCH predicted binding partners for 11 hypothetical proteins.
| Protein | Substrate | Score |
| SACOL1859 | SACOL1860 | 0.651 |
| SACOL0323 | SACOL0322 | 0.819 |
| SACOL0109 | SACOL0110 | 0.692 |
| SACOL0075 | SACOL0076 | 0.462 |
| SACOL0644 | SACOL0643 | 0.859 |
| SACOL0350 | SACOL0351 | 0.859 |
| SACOL2123 | SACOL2125 | 0.422 |
| SACOL2124 | 0.422 | |
| SACOL1956 | SACOL2519 | 0.685 |
| SACOL0267 | SACOL0266 | 0.694 |
| SACOL0488 | SACOL0487 | 0.859 |
| SACOL0486 | 0.859 | |
| SACOL0710 | SACOL0709 | 0.57 |
| SACOL0708 | 0.57 |
SACOL0323 matched a prophage L54a, Cro-like protein. SACOL2123 had equal scores to a M20/M25/M40 family peptidase (SACOL2125) and a hypothetical protein (SACOL2124). SACOL0710 equally matched a phosphotransferase mannose-specific family component IIA (SACOL0709) and a DAK2 domain-containing protein (SACOL0708). These results did not correlate with the findings from other programs used in this study.
Conclusion
Antibiotic resistance is a major global health crisis. Infections, like those caused by Methicillin-resistant S. aureus, are becoming untreatable, and increasing fatalities from these once curable diseases. Faster techniques to identify drug-resistant organisms and new therapeutics are needed to improve patient outcomes. Characterizing hypothetical proteins, particularly those contributing to resistance, may hold the key to unlock this health predicament. This work provides insight into hypothetical proteins related to antibiotic resistance, potentially leading to improved diagnostic tools and therapeutics against antibiotic resistant S. aureus. It characterized differentially expressed hypothetical proteins between Methicillin-sensitive and resistant strains whereas other studies have randomly selected or performed cross-species comparisons to identify hypothetical proteins of interest. Our approach to identify hypothetical proteins related to antibiotic resistance is an improvement over prior methods. However, computational algorithms were unable to confidently predict functions for any of the 14 differentially expressed hypothetical proteins examined. Most programs struggled to identify parameters, such as domains or binding partners. Those that were found usually had unknown functions or little sequence homolog. These results indicate that using statistically significant differential expression from a publically available antibiotic resistant strain comparison microarray study will identify proteins potentially related to antibiotic resistance for which more scientific knowledge is needed.
Table 5. CDD-BLAST domain data for 7 hypothetical proteins.
| Protein | Domains | Description | E-value |
| SACOL1859 | pfam13401 | AAA | 1.61E-04 |
| smart00382 | ATPase | 7.59E-03 | |
| SACOL0644 | pfam04276 | Protein of unknown function (DUF443) | 1.03E-37 |
| SACOL0350 | pfam07768 | PVL ORF-50-like family | 8.79E-47 |
| SACOL0835 | pfam16228 | Domain of unknown function (DUF4887) | 1.18E-12 |
| SACOL2123 | pfam11042 | Protein of unknown function (DUF2750) | 2.38E-20 |
| SACOL1956 | pfam11667 | Putative zincin peptidase | 4.49E-08 |
| SACOL0488 | pfam13654 | AAA | 5.17E-03 |
Table 6. Pfam domain data for 5 hypothetical proteins.
| Protein | Domain | Description | E-value |
| SACOL0644 | DUF443 | Unknown function | 9.80E-56 |
| SACOL0350 | PVL_ORF50 | Panton-Valentine leucocidin ORF-50- like family | 2.80E-45 |
| SACOL0835 | DUF4887 | Unknown function | 1.70E-50 |
| SACOL2123 | DUF2750 | Unknown function | 2.10E-21 |
| SACOL1956 | DUF3267 | Putative zincin peptidase | 1.60E-18 |
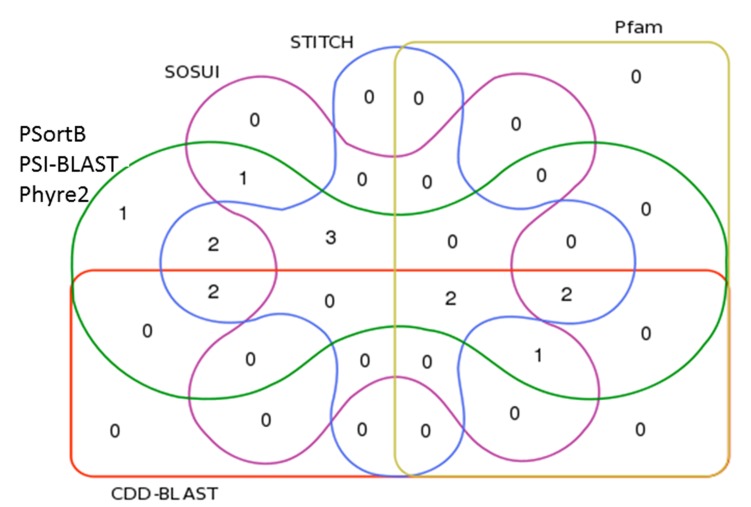
Figure 1.
Venn diagram illustrating overlap of study evalations. PSortB, PSI-BLAST, and Phyre2 (green line) characterized all 14 hypothetical proteins that passed Expasy’s size exclusion criteria. Only those algorithms found results for SACOL2481 (1). SACOL2241 also had a SOSUI (purple line) result (1). SACOL0710 and SACOL0323 had STITCH (blue line) results (2). SACOL0488 had both STITCH and CDD-BLAST (orange line) results (2). SACOL0267, SACOL0109, and SACOL0075 had SOSUI and STITCH results (3). SACOL2123 and SACOL0350 had CDD-BLAST, Pfam (yellow-line), and STITCH results (2). SACOL1956 and SACOL0644 had results from all programs (2) and SACOL0835 had results from all except STITCH (1).
Edited by P Kangueane
Citation: Marklevitz & Harris, Bioinformation 13(4): 104-110 (2017)
References
- 1.Woolhouse M,, et al. J of Glob Health. 2106;6(1) [Google Scholar]
- 2.Ventola CL. Pharmacy and Therapeutics. 2015;40(4)::277. [Google Scholar]
- 3.Nathan C, Cars O.N. Engl J Med. 2014;371(19)::1761. doi: 10.1056/NEJMp1408040. [DOI] [PubMed] [Google Scholar]
- 4.Rossolini GM,, et al. Curr Opin Pharmacol. 2014;18:56. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 5. http://www.cdc.gov/mrsa/tracking/
- 6.School K,, et al. Bioinformation. 2016;12(3)::209. doi: 10.6026/97320630012209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marklevitz J, Harris LK. Bioinformation. 2016;12(4)::254. doi: 10.6026/97320630012254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanagihara K,, et al. J Antimicrob Chemother. 2006;57(1)::122. doi: 10.1093/jac/dki416. [DOI] [PubMed] [Google Scholar]
- 9.Bharat Siva Varma P,, et al. J Infect Public Health. 2015;8(6)::526. doi: 10.1016/j.jiph.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Mohan R, Venugopal S. Bioinformation. 2012;8(15)::722. doi: 10.6026/97320630008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ham JS,, et al. Mol Cells. 2010;30(1)::71. doi: 10.1007/s10059-010-0090-3. [DOI] [PubMed] [Google Scholar]
- 12.Gasteiger E,, et al. Methods Mol Biol. 1999;112:531. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 13.Yu NY,, et al. Bioinformatics. 2010;26(13)::1608. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirokawa T,, et al. Bioinformatics. 1998;14:378. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A,, et al. NAR. 2015;43:D222. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnhammer EL,, et al. Proteins. 1997;28:405. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Kelley LA,, et al. Nat Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wass MN,, et al. NAR. 2010;38:W469. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczy D,, et al. NAR. 2016;44(D1)::D380. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://bioinformatics.psb.ugent.be/webtools/Venn/