Abstract
Invertebrate pests and parasites of humans, animals, and plants continue to cause serious diseases and remain as a high treat to agricultural productivity and storage. The rapid and accurate species identification of the pests and parasites are needed for understanding epidemiology, monitoring outbreaks, and designing control measures. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) profiling has emerged as a rapid, cost effective, and high throughput technique of microbial species identification in modern diagnostic laboratories. The development of soft ionization techniques and the release of commercial pattern matching software platforms has resulted in the exponential growth of applications in higher organisms including parasitology. The present review discusses the proof-of-principle experiments and various methods of MALDI MS profiling in rapid species identification of both laboratory and field isolates of pests, parasites and vectors.
Keywords: MALDI MS typing, MALDI TOF MS, intact protein profiling, pests, parasites, species, spectra reference databases
Introduction
Invertebrate pests and parasites of plants, mammals, birds, amphibians, and reptiles account for increased losses in the agricultural sector and continue to play a considerable role in the spread of infectious diseases (Paini et al., 2016; Poulin et al., 2016). Accurate and rapid species identification of pests and parasites is extremely important for initiating species-specific treatment procedures, understanding the epidemiology, monitoring of outbreaks, and designing control measures (Gibson, 2009; Furlong, 2015). Traditionally, trained taxonomists or entomologists visually examine or observe microscopically the morphological characteristics for species determination. However, in addition to being time-consuming, misidentification possibilities, distinguishing immature or development stages, damaged samples, cryptic species, and species differing by minor morphological characteristics make identification challenging and often impossible (McKeand, 1998). Molecular methods, whichever available, are accurate and applicable to any development stages. On the other hand, these methods are also labor intensive, expensive, time consuming, and difficult to apply for species for which sequences are not available (Wong et al., 2014). Furthermore, in certain cases such as Leishmania subtyping, which is crucial for treatment, identification generally requires several weeks for performing complex and expensive analyses (Roelfsema et al., 2011). In recent years, proteome based linear matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS profiling or MALDI MS typing), which is a well-established technique for microbial species identification, has also been successfully applied to a variety of parasites and their vectors (Seng et al., 2009; Bizzini and Greub, 2010; Patel, 2013; Nomura, 2015; Karger, 2016; Singhal et al., 2016; Yssouf et al., 2016). The popularity of the method is due to its rapidness, easy to use, high throughput analysis, accuracy, reliability equal to that of the molecular methods of species identification and cost-effectiveness despite the initial cost of the machine (Dhiman et al., 2011; Neville et al., 2011; Tran et al., 2015; Ge et al., 2016). The technique involves generation of MALDI MS spectra for a given species and pattern matching with that of the spectra database of the well-defined species to deduce the species information (Welker and Moore, 2011; Nagy et al., 2012; Suarez et al., 2013). The spectra pattern matching is carried out using automated commercial software suites, such as Bruker Biotyper (Bruker Daltonics, Bremen, Germany), VITEK MS (BioMérieux, Nuertingen, Germany: earlier Axima (Shimadzu)-SARAMIS (AnagnosTec) systems), Andromas (Andromas SAS), or MicrobeLynx (Waters) (Sogawa et al., 2011; Bille et al., 2012; Patel, 2013; Cassagne et al., 2016). The commercial software tools are usually integrated with their own spectra reference database, and utilize a unique algorithm for spectra processing, pattern matching, and result interpretation.
The manufacturer-provided database is limited only in terms of reference spectra for available microbial species, and currently the reference information for pests and parasites are not included. The software generally includes the possibility to create reference spectra for any organism to be integrated within the existing database. The database extension has been utilized to create additional reference spectra to enhance the identification confidence and to include reference spectra of the missing species, including higher organisms (Bohme et al., 2012; Murugaiyan et al., 2012, 2014; Hoppenheit et al., 2013). Several reviews have been dedicated to the recent developments of MALDI MS typing of plant nematodes and organisms related to parasitology (Ahmad and Babalola, 2014; Karger, 2016; Singhal et al., 2016; Yssouf et al., 2016). Therefore, the focus of this review is on the various approaches reported for MALDI MS typing based species identification of pests, parasites, and their vectors ranging from laboratory isolates to that of field samples.
MALDI MS basis of species identification
MALDI MS based species identification involves three steps:
Sample spotting onto a specially designed metal plate called the target plate,
MALDI TOF MS measurments and
Species deduction through pattern matching of the spectra with the database of spectra derived from known and/or well-defined species.
A small portion of biological substances e.g., microbial colonies or a drop of intact/crude protein extracted using simple procedures is directly added to the target plate and allowed to air dry. Then, the sample spot is overlaid with a drop of an excess concentration of UV-absorbing small organic compounds, referred to as a matrix. There are several choices of matrix such as α-cyano-4-hydroxycinnamic acid (HCCA/CHCA), sinapinic acid (3, 5- dimethoxy-4-hydroxycinnamic acid) (SA), and 2,5-dihydroxybenzoic acid (DHB). HCCA is most frequently reported matrix; however, there is no universally recommended matrix.
In the MALDI instrument, a small region of the crystalline sample-matrix spot (usually 0.05–0.2 mm in diameter) is irradiated using a pulsed beam of a laser, generally a nitrogen beam with a wavelength of 337 nm is used in most commercial machines. The matrix absorbs the laser energy, and rapidly heats up resulting in desorption (vaporization) or structural decomposition of the proteins and protonation to form a hot dense plume of ablated gases and ions (Clark et al., 2013). Using an electric field, the ions are accelerated into a vacuum tube that terminates in an ion detector. The ions are usually of single charge and the acceleration voltage results in the same kinetic energy applied to every single charged ions, which results in separation of ions based on mass/charge (m/z) ration in the drift or vacuum tube.
The time of flight “TOF” of ions is recorded as MALDI spectra where the x-axis represents m/z ratio and the y-axis represents the intensity (or number) of same/similar ions. MALDI MS ions are singly charged, representing the non-fragmented parent ion mass, the resulting spectra is simple, and therefore, the species of unknown samples can be easily deduced after data processing and direct pattern matching with that of the spectra established from well-defined species compiled as a database. The identity of spectral peaks or protein sequence information is not important, as the species deduced by matching the protein profiles usually at a range of 2–20 kDa (2,000–20,000 m/z) (Evason et al., 2001; Sauer and Kliem, 2010; Welker and Moore, 2011; Karlsson et al., 2015; Cassagne et al., 2016).
Advances in invertebrate pests, parasite, and vector profiling
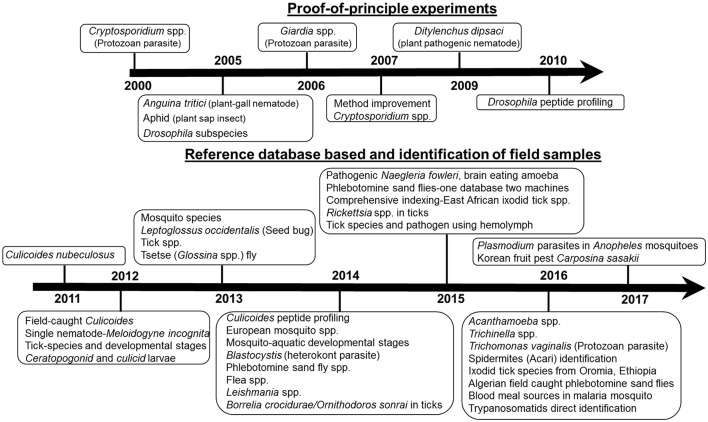
In the past 16 years (Figure 1), MALDI MS profiling has been successfully applied for species identification of different pests, parasites and vectors such as nematodes, protozoa, and arthropods.
Figure 1.
Time line on MALDI profiling application of pests, parasites, and vector species identification.
Proof of principle experiments
The early proof of principle experiments were focused on identification of biomarker peaks, standardization of sample preparation, matrixes and measurement optimization.
Protozoans and unicellular parasites
The first report of MALDI MS typing of protozoa was demonstrated using Cryptosporidium spp. associated with human infections. The species-specific spectra were reported from oocytes of C. parvum and C. muris isolated from feces of experimentally inoculated mice, lysed by freeze-thaw cycle, and spotted with HCCA as matrix (Magnuson et al., 2000). Later, it was shown that incubation of intact oocytes and purified sporozoites for 45 min with the matrix was critical for generating mass spectra with a large number of reproducible peaks for C. parvum oocysts (Glassmeyer et al., 2007). Subsequently, direct application of whole spores, spore shells, and soluble fractions of spore-forming unicellular parasites such as microsporidia, Encephalitozoon cuniculi, Encephalitozoon hellem, Encephalitozoon intestinalis, and Brachiola algerae isolated from humans and propagated on monolayers of Vero monkey kidney (E6) cells, displayed species-specific markers in the mass range of 2,000–8,000 Da (Moura et al., 2003). Later, species-specific peaks in the range of m/z 3,000–19,000 was reported for the water-borne protozoan parasite, Giardia spp., the causative agent of giardiasis. The cysts, cyst walls, and trophozoites of G. lamblia and G. muris isolated from feces of experimentally challenged mice, were washed, mixed with an equal volume of sinnapinic acid, incubated, and spotted for MALDI MS analysis (Villegas et al., 2006).
Insects and pests
Although not a pest or parasite, Drosophila has been used as a model for insect profiling possibilities. Protein extraction by simple grinding of adult whole insect in water was shown to generate species-specific spectra capable of distinguishing sibling species of Drosophila sub-species. The insect sex and matrix was not found to influence the spectra (Campbell, 2005). Likewise, species-specific peaks of varying intensities in a range of 3,000–25,000 m/z have been reported for three different aphids (plant phloem sap-feeding insects), green peach aphid Myzus persicae Sulzer, cowpea aphid Aphis craccivora Koch, and blue-green aphid (blue alfalfa aphid) Acyrthosiphon kondoi Shinji, independent of their dietary host plants (Perera et al., 2005).
Nematodes and developmental stage discrimination
Species-specific and diagnostic peaks have also been reported for simple extracts of three plant nematodes, Anguina tritici (wheat seed-gall nematode), its closely Anguina funesta (ryegrass nematode), and Meloidogyne javanica (root-knot nematode that infects horticultural and vegetable crops; Perera et al., 2005). Several years later, direct crushing of the root-knot nematode, Meloidogyne incognita was shown to be useful in rapid discrimination between the harmless and harmful J2 developmental stages and adult nematode (Ahmad et al., 2012).
Peptide profiling
In this approach, also referred as shotgun mass mapping or SMM, whole body protein extracts were subjected to trypsin digestion without purification or fractionation steps, and the resulting peptides were utilized for generation of MALDI MS spectra for insect vector species such as Drosophila (Feltens et al., 2010) and biting midges (Uhlmann et al., 2014). Feltens et al. had applied nano-high performance liquid chromatography coupled with electrospray ionization mass spectrometry for identification of some of the MALDI MS profiles and revealed that most of the proteins were of muscles and mitochondria. However, SMM is time-consuming and handling large set of samples is challenging.
Data independent analysis and clinical sample survey
The data independent species discrimination or grouping of microorganism is based on the visual examination for the presence or absence of peaks. This technique is very similar to those analyses performed before the days of the implementation of software with automated pattern matching algorithms; however, a different algorithm is used for rapid determination of the presence or absence of peaks for identification or discrimination. For example, this technique has been reported for the discrimination of the Leishmania subgenus Viannia or Leishmania exclusively based on the presence of 2 pairs of peaks (Mouri et al., 2014), as well as differentiation of protozoan parasitic Entamoeba histolytica and Entamoeba dispar (Azian et al., 2006).
Database-based enabled rapid species identification extended to field samples
MALDI MS typing based rapid species identification is usually achieved through pattern-matching of the unknown samples with that of a spectral reference library (database) created from known organisms. The main concerns at the proteome level are differences between the various developmental stages and complexity associated with various body parts. As listed in Table 1, vector related reference spectra were reported using commercial MALDI instrument-software suites. The proteins were extracted through homogenization and the parameters recommended for microbial species identification was followed. HCCA and SA were reported as the most utilized matrices for Bruker Biotyper and SARAMIS (Vitex MS), respectively. Despite the success of the procedures, these reference spectra remain in-house databases. Following the compilation of vector-specific reference spectra databases, the method proved to be rapid (~2–5 min/sample) as in the case of microbial species identification.
Table 1.
Reference database based species identification of parasite vectors.
| S. No | Organisms | Sample/database | Protein extraction † | Matrix* | Software and database ≠ | Comment | References | |
|---|---|---|---|---|---|---|---|---|
| Bruker BioTyper± | SARAMIS¥ | |||||||
| 1 | Culicoides biting midges | Whole insects or thoraxes | (1) Water and (2) FA (5-50%) | SA, DHB | + | Two laboratory reared -C. nubeculosus, better spectra with SA, recommended removal of abdomen, ethanol storage, and high concentration of formic acid influenced spectra | Kaufmann et al., 2011 | |
| 2 | Thoraxes with head, wings, and legs | 10% FA | SA | + | first field study, better results with fresh samples compared to ethanol stored | Kaufmann et al., 2012 | ||
| 3 | + | Ethanol preserved larvae, Spectra differed with development stage | Steinmann et al., 2013 | |||||
| 4 | HCCA | + | field survey at Senegal, Africa | Sambou et al., 2015 | ||||
| 5 | Mosquitoes | head and thorax | SA | + | Several months of ethanol preserved | Muller et al., 2013 | ||
| 6 | legs | 70% FA and 50% ACN | HCCA | + | Proteins extracted from legs is sufficient | Yssouf et al., 2013b | ||
| 7 | + | European mosquito species identification | Yssouf et al., 2014a | |||||
| 8 | + | Mosquitos' species and midgut microbial species identification after culturing | Tandina et al., 2016 | |||||
| 9 | Whole specimen | 70% FA | + | Culicidae juvenile database-aquatic developmental stages | Dieme et al., 2014 | |||
| 10 | Eggs | SA | + | Eggs as source for species surveillance | Schaffner et al., 2014 | |||
| 11 | Abdomen | 70% FA and 50% AC | HCCA | + | Blood mean source identification in malaria mosquito | Niare et al., 2016 | ||
| 12 | Head and thorax | Water | + | Plasmodium parasite identification in Anopheles | Laroche et al., 2017 | |||
| 13 | Ticks | Whole tick, nymphs, and larvae | Sonication with 6-M guanidinium chloride solution, acidified with TFA and eluted with 0.1% TFA/75% ACN | HCCA | + | Developmental stages for 7 tick species | Karger et al., 2012 | |
| 14 | Legs | 70% FA and 50% ACN | + | Proteins extracted from legs is sufficient | Yssouf et al., 2013a | |||
| 16 | 25% FA | SA | + | comprehensive indexing of East African ixodid tick species | Rothen et al., 2016 | |||
| 17 | 1 Spirochetes spotted directly 2 tick legs | 70% FA and 50% ACN | HCCA | + | Borrelia crocidurae /Ornithodoros sonrai in ticks. | Fotso et al., 2014 | ||
| 18 | 1 Hemolymph from leg 2 R. c. conorii strain extracte | 70% FA and 100% ACN | + | Rh. sanguineus specimens infected by R. c. conorii and A. variegatum specimens infected by R. africae | Yssouf et al., 2015a | |||
| 19 | 1 Tick legs 2 Cultured and purified Rickettsia species | + | Rickettsia spp in Ticks | Yssouf et al., 2015b | ||||
| 20 | Flea | Abdomen excised whole specimen | 70% FA and 50% ACN | + | Five flea species | Yssouf et al., 2014b | ||
| 21 | Glossina. spp Tsetse fly | Whole insect and dissected body parts | 70% FA and 100% ACN | + | Five lab reared Glossina. spp. Wings is sufficient | Hoppenheit et al., 2013 | ||
| 22 | Phlebotomine sand flies | Thoraxes | (1) water and (2) 25% FA | SA | + | Discrimination of five Mediterranean species, Water extract yielded superior result to that of formic acid extract | Dvorak et al., 2014 | |
| 23 | Thoraxes, wings, and legs | 70% FA and 50% ACN | HCCA | + | Six species caught from Algeria. | Lafri et al., 2016 | ||
| 24 | Thoraxes with wings and legs | 10% FA | SA, DHB | + | 1. Ultraflex III MALDI TOF mass spectrometer measurement and an in-house phyton script to transfer mzXML files in to SARAMIS software. 2. Spectra measurement using Axima Confidence to create a reference spectra database for 20 species | Mathis et al., 2015 | ||
Protein extraction by homogenization: FA, Formic acid; ACN, acetonitrile; TFA, Trifluroacetic acid;
Matrix: HCCA, α-Cyano-4-hydroxycinnamic acid; SA, Sinapinic acid; DHB, 2,5-Dihydroxybenzoic acid; Software and database:
Plus symbol indicates the commercial software in which the database was created and incorporated,
BioTyper™—software from Bruker Daltonics, Bremen, Germany.
-SARAMIS™ software (Spectral. ARchive And Microbial Identification System, originally developed by AnagnosTec, Potsdam-Golm, Germany and now acquired and redeveloped as Vitek-MS by bioMérieux, Marcy L'Etoile, France).
Parasites
In every reported case of parasite database approaches, Bruker BioTyper software tool and formic acid/acetonitrile extraction was applied. In the first such study, 56 clinical specimens belonging to 23 species of Leishmania were cultured, and promastigote pellets were utilized for database construction (Cassagne et al., 2014). Among 69 clinical isolates used for testing, only three samples were not identified. In a similar study, a database was constructed from four reference strains and the two clinical isolate were identified as L. infantum (Culha et al., 2014). In another study, using 19 enteric parasite Blastocystis isolates from 19 patients, a database for five subtypes was created and the remaining specimens were identified by matching (Martiny et al., 2014). Likewise, the differentiation of E. histolytica and E. dispar was demonstrated after establishment of the reference spectra and discriminating peaks were matched with the proteins identified through SDS-PAGE MALDI TOF MS based protein identification approach (Calderaro et al., 2015). Bruker BioTyper based database compilation were also reported for the food nematode Trichinella (Mayer-Scholl et al., 2016), Trichomonas vaginalis (Calderaro et al., 2016), Acanthamoeba spp. (Del Chierico et al., 2016) using a database for direct identification of trypanosomatids (Avila et al., 2016).
Toward “vector spectra reference database”
Kaufmann et al., were the first to report on utilization of SARAMIS premium software to create a reference database for two laboratory-reared C. nubeculosus biting midges (Kaufmann et al., 2011), and which was then extended to a larvae-specific database, screening of field collected samples, and applied for entomological surveys in Senegal, Africa. It was further reported as a means for discrimination of cryptic Anopheles, and demonstrated that the usefulness of mosquito eggs in species identification of field collected samples (Schaffner et al., 2014; Yssouf et al., 2014a). The Bruker Biotyper database has since been utilized to create a comprehensive “Vector specific reference spectra database” that includes spectra from leg proteins of 6 tick species, 30 mosquito species, one louse, one triatomine, one bed bug, and five flea species (Yssouf et al., 2013a,b, 2014b, 2015a). In addition, the package has been used to establish an aquatic developmental stage database starting from the larva stage to pupa of 6 mosquito species and detection of host blood meal and the presence of parasites (Niare et al., 2016; Laroche et al., 2017).
Pests
Despite the earlier proof of concept experiments for pests, the database approach was reported for only one plant pest in which Biotyper was utilized to distinguish evolutionary and morphologically close species of spider mites. Female adults of the Kanzawa (Tetranychus kanzawai), the two-spotted (T. urticae) spider mites and three other related species, namely T. phaselus Ehara (Tp), the bean red spider mite (T. ludeni Zacher) (Tl), and the tomato red spider mite (T. evansi Baker & Pritchard) (Te) were shown to be distinguishable. On the other hand, male adults and nymphs were reported to be non-distinguishable. Direct lysing of a single intact mite on target plates using double side carbon tape was also shown to generate spectra comparable to that of the extracts from 10 pooled individuals (Kajiwara et al., 2016). Recently, it was demonstrated that using MALDI MS, Tinkerbell LT, and its associated software MicroIDSys (ASTA Inc. Suwon, Korea), the larvae of the Korean apple pest, Carposina sasakii, could be effectively discriminated in about 15 min. Without such rapid identification methods, the export of these fruits might be hampered or rejected due to time-consuming pre-export inspection (Jeon et al., 2017).
Dual identification of vectors, parasites, and meal source
MALDI MS profiling has also been shown to be an effective method for simultaneous identification of vectors and parasite species, as in case of simultaneous identification of Borrelia crocidurae/Ornithodoros sonrai and Rickettsia spp. in ticks (Fotso et al., 2014; Yssouf et al., 2015b). Direct spotting of haemolymph from a dissected tick leg allows for the simultaneous and direct species identification of ticks and associated pathogens, and useful in species identification of parasites and vector while leaving the vector remains available for other laboratory investigations (Fotso et al., 2014). MALDI MS profiling has further been successfully applied to determine the feeding patterns of mosquitoes up to 24 h post-blood meal (Niare et al., 2016). A future goal for the application of MALDI MS profiling in pest/parasites will be the rapid species identification of vectors, parasites and blood meal either through individual analysis or through simultaneous monitoring processes. The recent demonstration of rapid distinguishing of the Plasmodium infection status among Anopheles stephensi mosquitoes underscores that MALDI MS typing could be useful in entomological surveys including species-specific infection status (Laroche et al., 2017).
Universal database and field samples
All the reported parasitology associated databases were developed as in-house databases and usually not accessable by other researchers, although several authors have agreed to supply the reference spectra upon request to other scientists possessing the same instrumentation and software tools. However, beyond a few initiations such as SpectraBank, there is no universal reference database such as protein database Swissprot/Uniprot for the purpose of species identification (Bohme et al., 2012). Recently, it was shown that the spectra generated from two different commercial MALDI MS instruments (Axima Confidence and Bruker Ultraflex III MS) could be analyzed in a single database (SARAMIS) in which a database was constructed for 20 species of phlebotomine sand flies based on measurements with Axima Confidence MS (Mathis et al., 2015). This underscores the possibilities of creation of online public reference databases that might be useful for any type of MALDI MS machines or spectral data formats. The online public database for parasitology is of great importance in terms of economy, time, and rapid analysis of samples collected from different geographical regions or hosts. The open source reference spectra database demands standardization of influencing parameters/processes affecting the sampling, such as developmental stage, specimen storage conditions, sample preparation/extraction methods, variations due to the spotting, instrumental, and post-measurement software settings.
Conclusion
Despite the achievements of MALDI TOF MS in microbial species identification, the application in parasitology remains limited to in-house databases integrated into commercial software. There is no commercial software tool or reference database available for parasitology, however, most of the successful reports were based on the principles, procedures, software, and databases best described for microorganisms. The crucial factors influencing parasitology MALDI MS typing includes: specimen associated parameters (age, developmental stage, sex, differing body parts, fed state), sample preparation procedures (sample storage, protein extraction solvents, and methods, sample spotting method, and matrix), and measurement parameters. MALDI MS typing of field specimens will be helpful in creating a distribution map and evaluation of the spread of parasites associated with disease. Few reports describe the spectral differences among the geographically different populations (Dieme et al., 2014; Dvorak et al., 2014; Hoppenheit et al., 2014). However, one should remain cautious, as the reliability of MALDI TOF MS in phylogenetic analysis is yet to be proven. The establishment of open source software and databases might be useful in future parasitological surveys and for rapid assessment of real time infection status.
Author contributions
JM conceived and wrote the manuscript. UR reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Karsten Tedin, Freie Universitaet Berlin, Germany and Vanessa Kowbel, University of Manitoba, Canada for their assistance of skillful grammatical corrections.
References
- Ahmad F., Babalola O. O. (2014). Application of mass spectrometry as rapid detection tool in plant nematology. Appl. Spectrosc. Rev. 49, 1–10. 10.1080/05704928.2013.798799 [DOI] [Google Scholar]
- Ahmad F., Gopal J., Wu H. F. (2012). Rapid and highly sensitive detection of single nematode via direct MALDI mass spectrometry. Talanta 93, 182–185. 10.1016/j.talanta.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Avila C. C., Almeida F. G., Palmisano G. (2016). Direct identification of trypanosomatids by matrix-assisted laser desorption ionization-time of flight mass spectrometry (DIT MALDI-TOF MS). J. Mass Spectrom. 51, 549–557. 10.1002/jms.3763 [DOI] [PubMed] [Google Scholar]
- Azian M. Y. N., Hakim S. L., Maslawaty M. N. (2006). Use of molecular tools to distinguish Entamoeba histolytica and Entamoeba dispar infection among the aborigines in Cameron Highlands. Trop. Biomed. 23, 31–36. [PubMed] [Google Scholar]
- Bille E., Dauphin B., Leto J., Bougnoux M. E., Beretti J. L., Lotz A., et al. (2012). MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. 18, 1117–1125. 10.1111/j.1469-0691.2011.03688.x [DOI] [PubMed] [Google Scholar]
- Bizzini A., Greub G. (2010). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16, 1614–1619. 10.1111/j.1469-0691.2010.03311.x [DOI] [PubMed] [Google Scholar]
- Bohme K., Fernandez-No I. C., Barros-Velazquez J., Gallardo J. M., Canas B., Calo-Mata P. (2012). Spectrabank: an open access tool for rapid microbial identification by MALDI-TOF MS fingerprinting. Electrophoresis 33, 2138–2142. 10.1002/elps.201200074 [DOI] [PubMed] [Google Scholar]
- Calderaro A., Piergianni M., Buttrini M., Montecchini S., Piccolo G., Gorrini C., et al. (2015). MALDI-TOF mass spectrometry for the detection and differentiation of Entamoeba histolytica and Entamoeba dispar. PLoS ONE 10:e0122448. 10.1371/journal.pone.0122448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderaro A., Piergianni M., Montecchini S., Buttrini M., Piccolo G., Rossi S., et al. (2016). MALDI-TOF mass spectrometry as a potential tool for Trichomonas vaginalis identification. BMC Infect. Dis. 16:261. 10.1186/s12879-016-1594-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P. M. (2005). Species differentiation of insects and other multicellular organisms using matrix-assisted laser desorption/ionization time of flight mass spectrometry protein profiling. Syst. Entomol. 30, 186–190. 10.1111/j.1365-3113.2004.00279.x [DOI] [Google Scholar]
- Cassagne C., Normand A. C., L'ollivier C., Ranque S., Piarroux R. (2016). Performance of MALDI-TOF MS platforms for fungal identification. Mycoses 59, 678–690. 10.1111/myc.12506 [DOI] [PubMed] [Google Scholar]
- Cassagne C., Pratlong F., Jeddi F., Benikhlef R., Aoun K., Normand A. C., et al. (2014). Identification of Leishmania at the species level with matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 20, 551–557. 10.1111/1469-0691.12387 [DOI] [PubMed] [Google Scholar]
- Clark A. E., Kaleta E. J., Arora A., Wolk D. M. (2013). Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 26, 547–603 10.1128/CMR.00072-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culha G., Akyar I., Yildiz Zeyrek F., Kurt O., Gunduz C., Ozensoy Toz S., et al. (2014). Leishmaniasis in Turkey: determination of Leishmania species by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Iran. J. Parasitol. 9, 239–248. [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F., Di Cave D., Accardi C., Santoro M., Masotti A., D'alfonso R., et al. (2016). Identification and typing of free-living Acanthamoeba spp. by MALDI-TOF MS Biotyper. Exp. Parasitol. 170, 82–89. 10.1016/j.exppara.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Dhiman N., Hall L., Wohlfiel S. L., Buckwalter S. P., Wengenack N. L. (2011). Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49, 1614–1616. 10.1128/JCM.02381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieme C., Yssouf A., Vega-Rua A., Berenger J. M., Failloux A. B., Raoult D., et al. (2014). Accurate identification of Culicidae at aquatic developmental stages by MALDI-TOF MS profiling. Parasit. Vectors 7:544. 10.1186/s13071-014-0544-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak V., Halada P., Hlavackova K., Dokianakis E., Antoniou M., Volf P. (2014). Identification of phlebotomine sand flies (Diptera: Psychodidae) by matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasit. Vectors 7:21. 10.1186/1756-3305-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evason D. J., Claydon M. A., Gordon D. B. (2001). Exploring the limits of bacterial identification by intact cell-mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 49–54. 10.1016/S1044-0305(00)00192-6 [DOI] [PubMed] [Google Scholar]
- Feltens R., Gorner R., Kalkhof S., Groger-Arndt H., Von Bergen M. (2010). Discrimination of different species from the genus Drosophila by intact protein profiling using matrix-assisted laser desorption ionization mass spectrometry. BMC Evol. Biol. 10:95. 10.1186/1471-2148-10-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotso A. F., Mediannikov O., Diatta G., Almeras L., Flaudrops C., Parola P., et al. (2014). MALDI-TOF mass spectrometry detection of pathogens in vectors: the Borrelia crocidurae/Ornithodoros sonrai Paradigm. PloS Negl. Trop. Dis. 8:e2984 10.1371/journal.pntd.0002984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong M. J. (2015). Knowing your enemies: integrating molecular and ecological methods to assess the impact of arthropod predators on crop pests. Insect Sci. 22, 6–19. 10.1111/1744-7917.12157 [DOI] [PubMed] [Google Scholar]
- Ge M. C., Kuo A. J., Liu K. L., Wen Y. H., Chia J. H., Chang P. Y., et al. (2016). Routine identification of microorganisms by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: success rate, economic analysis, and clinical outcome. J. Microbiol. Immunol. Infect. S1684–S1182, 30063–30069. 10.1016/j.jmii.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Gibson W. (2009). Species-specific probes for the identification of the African tsetse-transmitted trypanosomes. Parasitology 136, 1501–1507. 10.1017/S0031182009006179 [DOI] [PubMed] [Google Scholar]
- Glassmeyer S. T., Ware M. W., Schaefer F. W., III., Shoemaker J. A., Kryak D. D. (2007). An improved method for the analysis of Cryptosporidium parvum oocysts by matrix-assisted laser desorption/ionization time of flight mass spectrometry. J. Eukaryot. Microbiol. 54, 479–481. 10.1111/j.1550-7408.2007.00287.x [DOI] [PubMed] [Google Scholar]
- Hoppenheit A., Murugaiyan J., Bauer B., Clausen P. H., Roesler U. (2014). Analysis of Glossina palpalis gambiensis and Glossina tachinoides from two distant locations in Burkina Faso using MALDI TOF MS. Parasitol. Res. 113, 723–726 10.1007/s00436-013-3701-z [DOI] [PubMed] [Google Scholar]
- Hoppenheit A., Murugaiyan J., Bauer B., Steuber S., Clausen P. H., Roesler U. (2013). Identification of Tsetse (Glossina spp.) using matrix-assisted laser desorption/ionisation time of flight mass spectrometry. PLoS Negl. Trop. Dis. 7:e2305. 10.1371/journal.pntd.0002305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. H., Oh H., Yun C. S., Byun B., Park J., Park H. S., et al. (2017). Rapid and reliable species identification of Carposina sasakii from its morphological homologues, by MALDI-TOF mass spectrometry. J. Asia Pac. Entomol. 20, 411–415. 10.1016/j.aspen.2017.02.013 [DOI] [Google Scholar]
- Kajiwara H., Hinomoto N., Gotoh T. (2016). Mass fingerprint analysis of spider mites (Acari) by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for rapid discrimination. Rapid Commun. Mass Spectrom. 30, 1037–1042. 10.1002/rcm.7534 [DOI] [PubMed] [Google Scholar]
- Karger A. (2016). Current developments to use linear MALDI-TOF spectra for the identification and typing of bacteria and the characterization of other cells/organisms related to infectious diseases. Proteomics Clin. Appl. 10, 982–993. 10.1002/prca.201600038 [DOI] [PubMed] [Google Scholar]
- Karger A., Kampen H., Bettin B., Dautel H., Ziller M., Hoffmann B., et al. (2012). Species determination and characterization of developmental stages of ticks by whole-animal matrix-assisted laser desorption/ionization mass spectrometry. Ticks Tick Borne Dis. 3, 78–89. 10.1016/j.ttbdis.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Karlsson R., Gonzales-Siles L., Boulund F., Svensson-Stadler L., Skovbjerg S., Karlsson A., et al. (2015). Proteotyping: proteomic characterization, classification and identification of microorganisms - A prospectus. Syst. Appl. Microbiol. 38, 246–257. 10.1016/j.syapm.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Kaufmann C., Schaffner F., Ziegler D., Pfluger V., Mathis A. (2012). Identification of field-caught Culicoides biting midges using matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasitology 139, 248–258. 10.1017/S0031182011001764 [DOI] [PubMed] [Google Scholar]
- Kaufmann C., Ziegler D., Schaffner F., Carpenter S., Pfluger V., Mathis A. (2011). Evaluation of matrix-assisted laser desorption/ionization time of flight mass spectrometry for characterization of Culicoides nubeculosus biting midges. Med. Vet. Entomol. 25, 32–38. 10.1111/j.1365-2915.2010.00927.x [DOI] [PubMed] [Google Scholar]
- Lafri I., Almeras L., Bitam I., Caputo A., Yssouf A., Forestier C. L., et al. (2016). Identification of Algerian field-caught Phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl. Trop. Dis. 10:e0004351. 10.1371/journal.pntd.0004351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche M., Almeras L., Pecchi E., Bechah Y., Raoult D., Viola A., et al. (2017). MALDI-TOF MS as an innovative tool for detection of Plasmodium parasites in Anopheles mosquitoes. Malar. J. 16:5. 10.1186/s12936-016-1657-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M. L., Owens J. H., Kelty C. A. (2000). Characterization of Cryptosporidium parvum by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 66, 4720–4724. 10.1128/AEM.66.11.4720-4724.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny D., Bart A., Vandenberg O., Verhaar N., Wentink-Bonnema E., Moens C., et al. (2014). Subtype determination of Blastocystis isolates by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur. J. Clin. Microbiol. Infect. Dis. 33, 529–536. 10.1007/s10096-013-1980-z [DOI] [PubMed] [Google Scholar]
- Mathis A., Depaquit J., Dvorak V., Tuten H., Banuls A. L., Halada P., et al. (2015). Identification of phlebotomine sand flies using one MALDI-TOF MS reference database and two mass spectrometer systems. Parasit. Vectors 8:266. 10.1186/s13071-015-0878-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Scholl A., Murugaiyan J., Neumann J., Bahn P., Reckinger S., Nockler K. (2016). Rapid identification of the foodborne pathogen Trichinella spp. by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. PLoS ONE 11:e0152062. 10.1371/journal.pone.0152062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeand J. B. (1998). Molecular diagnosis of parasitic nematodes. Parasitology 117(Suppl.), S87–S96. [DOI] [PubMed] [Google Scholar]
- Moura H., Ospina M., Woolfitt A. R., Barr J. R., Visvesvara G. S. (2003). Analysis of four human microsporidian isolates by MALDI-TOF mass spectrometry. J. Eukaryot. Microbiol. 50, 156–163. 10.1111/j.1550-7408.2003.tb00110.x [DOI] [PubMed] [Google Scholar]
- Mouri O., Morizot G., Van Der Auwera G., Ravel C., Passet M., Chartrel N., et al. (2014). Easy identification of leishmania species by mass spectrometry. PLoS Negl. Trop. Dis. 8:e2841. 10.1371/journal.pntd.0002841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Pfluger V., Wittwer M., Ziegler D., Chandre F., Simard F., et al. (2013). Identification of cryptic Anopheles mosquito species by molecular protein profiling. PLoS ONE 8:e57486. 10.1371/journal.pone.0057486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan J., Ahrholdt J., Kowbel V., Roesler U. (2012). Establishment of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry database for rapid identification of infectious achlorophyllous green micro-algae of the genus Prototheca. Clin. Microbiol. Infect. 18, 461–467. 10.1111/j.1469-0691.2011.03593.x [DOI] [PubMed] [Google Scholar]
- Murugaiyan J., Walther B., Stamm I., Abou-Elnaga Y., Brueggemann-Schwarze S., Vincze S., et al. (2014). Species differentiation within the Staphylococcus intermedius group using a refined MALDI-TOF MS database. Clin. Microbiol. Infect. 20, 1007–1014. 10.1111/1469-0691.12662 [DOI] [PubMed] [Google Scholar]
- Nagy E., Becker S., Kostrzewa M., Barta N., Urban E. (2012). The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J. Med. Microbiol. 61, 1393–1400. 10.1099/jmm.0.043927-0 [DOI] [PubMed] [Google Scholar]
- Neville S. A., Lecordier A., Ziochos H., Chater M. J., Gosbell I. B., Maley M. W., et al. (2011). Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49, 2980–2984. 10.1128/JCM.00431-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niare S., Berenger J. M., Dieme C., Doumbo O., Raoult D., Parola P., et al. (2016). Identification of blood meal sources in the main African malaria mosquito vector by MALDI-TOF MS. Malar. J. 15:87 10.1186/s12936-016-1152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura F. (2015). Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): a revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta 1854, 528–537. 10.1016/j.bbapap.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Paini D. R., Sheppard A. W., Cook D. C., De Barro P. J., Worner S. P., Thomas M. B. (2016). Global threat to agriculture from invasive species. Proc. Natl. Acad. Sci. U.S.A. 113, 7575–7579. 10.1073/pnas.1602205113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. (2013). Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 57, 564–572. 10.1093/cid/cit247 [DOI] [PubMed] [Google Scholar]
- Perera M. R., Vanstone V. A., Jones M. G. K. (2005). A novel approach to identify plant parasitic nematodes using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 19, 1454–1460. 10.1002/rcm.1943 [DOI] [PubMed] [Google Scholar]
- Poulin R., Besson A. A., Morin M. B., Randhawa H. S. (2016). Missing links: testing the completeness of host-parasite checklists. Parasitology 143, 114–122. 10.1017/S0031182015001559 [DOI] [PubMed] [Google Scholar]
- Roelfsema J. H., Nozari N., Herremans T., Kortbeek L. M., Pinelli E. (2011). Evaluation and improvement of two PCR targets in molecular typing of clinical samples of Leishmania patients. Exp. Parasitol. 127, 36–41. 10.1016/j.exppara.2010.06.024 [DOI] [PubMed] [Google Scholar]
- Rothen J., Githaka N., Kanduma E. G., Olds C., Pfluger V., Mwaura S., et al. (2016). Matrix-assisted laser desorption/ionization time of flight mass spectrometry for comprehensive indexing of East African ixodid tick species. Parasit. Vectors 9:151. 10.1186/s13071-016-1424-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambou M., Aubadie-Ladrix M., Fenollar F., Fall B., Bassene H., Almeras L., et al. (2015). Comparison of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular biology techniques for identification of Culicoides (Diptera: ceratopogonidae) biting midges in senegal. J. Clin. Microbiol. 53, 410–418. 10.1128/JCM.01855-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S., Kliem M. (2010). Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8, 74–82. 10.1038/nrmicro2243 [DOI] [PubMed] [Google Scholar]
- Schaffner F., Kaufmann C., Pfluger V., Mathis A. (2014). Rapid protein profiling facilitates surveillance of invasive mosquito species. Parasit. Vectors 7:142. 10.1186/1756-3305-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P. E., Rolain J. M., et al. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551. 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- Singhal N., Kumar M., Virdi J. S. (2016). MALDI-TOF MS in clinical parasitology: applications, constraints and prospects. Parasitology 143, 1491–1500. 10.1017/S0031182016001189 [DOI] [PubMed] [Google Scholar]
- Sogawa K., Watanabe M., Sato K., Segawa S., Ishii C., Miyabe A., et al. (2011). Use of the MALDI BioTyper system with MALDI-TOF mass spectrometry for rapid identification of microorganisms. Anal. Bioanal. Chem. 400, 1905–1911. 10.1007/s00216-011-4877-7 [DOI] [PubMed] [Google Scholar]
- Steinmann I. C., Pfluger V., Schaffner F., Mathis A., Kaufmann C. (2013). Evaluation of matrix-assisted laser desorption/ionization time of flight mass spectrometry for the identification of ceratopogonid and culicid larvae. Parasitology 140, 318–327. 10.1017/S0031182012001618 [DOI] [PubMed] [Google Scholar]
- Suarez S., Ferroni A., Lotz A., Jolley K. A., Guerin P., Leto J., et al. (2013). Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods 94, 390–396. 10.1016/j.mimet.2013.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandina F., Almeras L., Kone A. K., Doumbo O. K., Raoult D., Parola P. (2016). Use of MALDI-TOF MS and culturomics to identify mosquitoes and their midgut microbiota. Parasit. Vectors 9:495. 10.1186/s13071-016-1776-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran A., Alby K., Kerr A., Jones M., Gilligan P. H. (2015). Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 53, 2473–2479. 10.1128/JCM.00833-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann K. R., Gibb S., Kalkhof S., Arroyo-Abad U., Schulz C., Hoffmann B., et al. (2014). Species determination of Culicoides biting midges via peptide profiling using matrix-assisted laser desorption ionization mass spectrometry. Parasit. Vectors 7:392. 10.1186/1756-3305-7-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas E. N., Glassmeyer S. T., Ware M. W., Hayes S. L., Schaefer F. W., III. (2006). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based analysis of Giardia lamblia and Giardia muris. J. Eukaryot. Microbiol. 53(Suppl. 1), S179–S181. 10.1111/j.1550-7408.2006.00223.x [DOI] [PubMed] [Google Scholar]
- Welker M., Moore E. R. (2011). Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst. Appl. Microbiol. 34, 2–11. 10.1016/j.syapm.2010.11.013 [DOI] [PubMed] [Google Scholar]
- Wong S. S., Fung K. S., Chau S., Poon R. W., Wong S. C., Yuen K. Y. (2014). Molecular diagnosis in clinical parasitology: when and why? Exp. Biol. Med. 239, 1443–1460. 10.1177/1535370214523880 [DOI] [PubMed] [Google Scholar]
- Yssouf A., Almeras L., Berenger J. M., Laroche M., Raoult D., Parola P. (2015a). Identification of tick species and disseminate pathogen using hemolymph by MALDI-TOF MS. Ticks Tick Borne Dis. 6, 579–586. 10.1016/j.ttbdis.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Yssouf A., Almeras L., Raoult D., Parola P. (2016). Emerging tools for identification of arthropod vectors. Future Microbiol. 11, 549–566. 10.2217/fmb.16.5 [DOI] [PubMed] [Google Scholar]
- Yssouf A., Almeras L., Terras J., Socolovschi C., Raoult D., Parola P. (2015b). Detection of Rickettsia spp in ticks by MALDI-TOF MS. PLoS Negl. Trop. Dis. 9:e0003473. 10.1371/journal.pntd.0003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yssouf A., Flaudrops C., Drali R., Kernif T., Socolovschi C., Berenger J. M., et al. (2013a). Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J. Clin. Microbiol. 51, 522–528. 10.1128/JCM.02665-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yssouf A., Parola P., Lindstrom A., Lilja T., L'ambert G., Bondesson U., et al. (2014a). Identification of European mosquito species by MALDI-TOF MS. Parasitol. Res. 113, 2375–2378. 10.1007/s00436-014-3876-y [DOI] [PubMed] [Google Scholar]
- Yssouf A., Socolovschi C., Flaudrops C., Ndiath M. O., Sougoufara S., Dehecq J. S., et al. (2013b). Matrix-assisted laser desorption ionization–time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PLoS ONE 8:e72380. 10.1371/journal.pone.0072380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yssouf A., Socolovschi C., Leulmi H., Kernif T., Bitam I., Audoly G., et al. (2014b). Identification of flea species using MALDI-TOF/MS. Comp. Immunol. Microbiol. Infect. Dis. 37, 153–157. 10.1016/j.cimid.2014.05.002 [DOI] [PubMed] [Google Scholar]