Abstract
During adult neurogenesis, newly formed olfactory bulb (OB) interneurons migrate radially to integrate into specific layers of the OB. Despite the importance of this process, the intracellular mechanisms that regulate radial migration remain poorly understood. Here, we find that microRNA (miRNA) let‐7 regulates radial migration by modulating autophagy in new‐born neurons. Using Argonaute2 immunoprecipitation, we performed global profiling of miRNAs in adult‐born OB neurons and identified let‐7 as a highly abundant miRNA family. Knockdown of let‐7 in migrating neuroblasts prevented radial migration and led to an immature morphology of newly formed interneurons. This phenotype was accompanied by a decrease in autophagic activity. Overexpression of Beclin‐1 or TFEB in new‐born neurons lacking let‐7 resulted in re‐activation of autophagy and restored radial migration. Thus, these results reveal a miRNA‐dependent link between autophagy and adult neurogenesis with implications for neurodegenerative diseases where these processes are impaired.
Keywords: adult neurogenesis, Argonaute, microRNA, olfactory bulb, RISC
Subject Categories: Development & Differentiation, Neuroscience
Introduction
In the adult rodent brain, neural stem cells (NSCs) reside in the subventricular zone (SVZ) where they continuously give rise to neuroblasts that migrate along the rostral migratory stream (RMS) into the olfactory bulb (OB). There they radially migrate into the OB where they differentiate into mature interneurons and integrate into the existing neuronal circuitry (Doetsch et al, 1999; Alvarez‐Buylla & Garcia‐Verdugo, 2002; Lledo et al, 2006). The function of adult‐born OB neurons has still not been fully elucidated, but they are thought to participate in mechanisms related to olfactory memory and odorant discrimination (Sultan et al, 2010; Lazarini & Lledo, 2011; Livneh et al, 2014). During adult neurogenesis, neuroblasts undergo rapid transitions in gene expression programs, which enable appropriate migration, morphological changes, and ultimately neuronal integration (Geschwind et al, 2001; Karsten et al, 2003; Gurok et al, 2004; Bonnert et al, 2006; Merkle et al, 2014). Alterations in these mechanisms contribute to dysregulation of NSCs, which has been implicated in neurodegenerative diseases (Lim & Alvarez‐Buylla, 2014; Bond et al, 2015). Thus, there is a great need to elucidate molecular mechanisms controlling NSC differentiation and maturation.
MicroRNAs (miRNAs), which are small non‐coding RNAs that mediate posttranscriptional gene regulation by destabilizing and inhibiting mRNA translation, have been suggested to play an important role in this process. Canonical miRNAs are processed by a conserved multistep process, where the primary‐miRNA transcript is first cleaved by the microprocessor complex, containing Dgcr8 and Drosha, into smaller hairpin precursor‐miRNAs. These are then exported into the cytoplasm where Dicer processes them into 19‐ to 22‐bp nucleotide duplexes. One of these strands is loaded into the RNA‐induced silencing complex (RISC) containing Argonaute (AGO) proteins. In the RISC, mature miRNAs bind target mRNAs through a 6‐ to 8‐nucleotide seed sequence. This small base‐pairing approach allows a single miRNA to target hundreds of mRNAs (Bartel, 2009; Krol et al, 2010). This, in combination with the posttranscriptional mode of regulation, enables miRNAs to provide a rapid and broad regulation of large gene networks. Interestingly, the different targets of miRNAs are often connected on a functional level, allowing the cooperative control of gene networks, which results in a broad effect on cell fate transitions. Several miRNAs have recently been implicated in the regulation of adult neurogenesis; for example, miR‐124 influences neuronal fate determination (Cheng et al, 2009; Akerblom et al, 2012), while miR‐7a controls subtype specification of adult‐born neurons (de Chevigny et al, 2012). However, many uncertainties remain about the role of miRNAs in adult neurogenesis. It is unknown whether new‐born neurons express a unique pool of miRNAs and the cellular mechanisms that are under the control of miRNAs in this process remain poorly understood.
In light of their potential broad regulation of adult neurogenesis, we decided to perform a large‐scale analysis of active miRNAs in new‐born OB neurons. By using this approach, which employed RNA immunoprecipitation (RIP) of the AGO2 protein in the RISC of new‐born neurons of the adult OB, we found that the let‐7 family is a highly expressed miRNA family in new‐born neurons. Knockdown of let‐7 resulted in impaired radial migration and maturation of these cells. Interestingly, we found that let‐7 provides a positive regulation of autophagy, an evolutionary conserved lysosomal degradation pathway that degrades macromolecules and organelles (Nikoletopoulou et al, 2015), in adult new‐born neurons. Let‐7 knockdown resulted in a reduction of autophagic activity while simultaneous overexpression of the autophagy‐inducing genes TFEB or Beclin‐1 rescued let‐7‐dependent phenotypes in adult‐born neurons, hereby providing a novel mechanistic link between miRNAs and autophagy in adult neurogenesis.
Results
The let‐7 family is highly expressed in new‐born OB neurons
To identify active miRNAs in new‐born OB neurons, we employed a modified version of miRNA tagging and affinity purification (miRAP), which was developed to profile active miRNAs in distinct neuronal populations in the mouse brain (He et al, 2012). We generated a lentiviral vector expressing a GFP‐AGO2 fusion protein under the control of the housekeeping PGK promoter (LV.GFP‐AGO2). The use of lentiviral vectors allows the targeting of dividing and non‐dividing cells in the RMS and results in transduction of a high number of new‐born neurons in the OB with a gradual increase over 12 weeks (Alonso et al, 2012; Akerblom et al, 2014). We therefore injected LV.GFP‐AGO2 into the RMS of adult mice, resulting in efficient, high‐level targeting of migrating neuroblasts (Fig 1A). Eight weeks after injection, we found numerous GFP‐AGO2‐expressing new‐born neurons in the OB as monitored by immunohistochemistry (Fig 1B). The GFP‐expressing cells integrated into the different layers of the OB in a characteristic pattern for new‐born interneurons (Lemasson et al, 2005) and co‐labeled with the neuronal marker NeuN (Fig 1B). The GFP expression was primarily localized to the cytoplasm, resembling endogenous AGO2 expression (Fig 1B′). Since AGO2 directly binds mature miRNAs, this experimental design allowed isolation of active miRNAs specifically from new‐born neurons. To validate this approach, the OB from mice was dissected 8 weeks after injection of the LV.GFP‐AGO2, when the majority of targeted neuroblasts have matured to neurons (Alonso et al, 2012), and RNA immunoprecipitation (RIP) was performed using an antibody against GFP. Using qRT–PCR, we found a high enrichment of the neuronal expressed miR‐124 and miR‐103 in LV.GFP‐AGO2‐injected animals compared to sham‐injected animals, confirming efficient RIP of miRNAs (Fig EV1A).
Figure 1. AGO2‐RIP‐seq on new‐born neurons of the olfactory bulb.
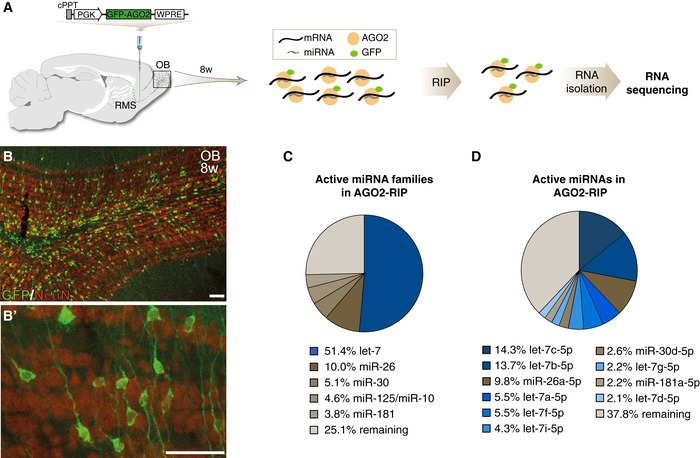
- Diagram of the LV.GFP‐AGO2 vector and the experimental workflow of the RIP‐seq approach.
- GFP‐AGO2 expression was detected in all layers of the OB. GFP‐AGO2‐expressing cells were co‐labeling with NeuN (B), and GFP‐AGO2 expression was cytoplasmic resembling endogenous AGO2 localization (B′). Scale bars: 40 μm.
- Pie chart of active miRNA families in new‐born neurons in the OB. Percentage of reads mapping to miRNA families are shown for the five most abundant miRNA families.
- Pie chart of active individual miRNAs in new‐born neurons in the OB. Percentage of reads mapping to miRNAs are shown for the ten most abundant miRNAs.
Figure EV1. Validation of RIP‐seq on new‐born neurons of the OB.
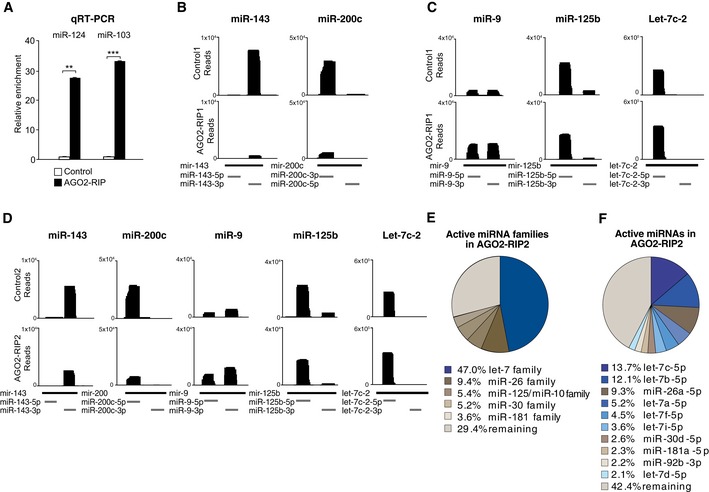
- qRT–PCR analysis showed that miRNAs are enriched in GFP‐AGO2 RIP samples compared to sham‐injected control samples; n = 4.
- Reads mapping to glia‐enriched miR‐143 and miR‐200c in control (upper panel) and GFP‐AGO2 RIP samples (lower panel).
- Reads mapping to miR‐9, miR‐125b, and let‐7c in control (upper panel)‐ and LV.GFP‐AGO2‐injected mice (lower panel).
- Reads mapping to miR‐143, miR‐200c, miR‐9, miR‐125b, let‐7c in an independent replicate of the GFP‐AGO2 RIP.
- Pie chart of active miRNA families in new‐born neurons in the AGO2‐RIP2 from an independent replicate. Percentage of reads mapping to miRNA families are shown for the five most abundant miRNA families.
- Pie chart of active individual miRNAs in new‐born neurons in the AGO2‐RIP2 from an independent replicate. Percentage of reads mapping to miRNAs are shown for the ten most abundant miRNAs.
In order to analyze miRNAs in new‐born neurons at a global level, we performed next‐generation small RNA sequencing on RIP samples of LV.GFP‐AGO2‐injected mice and of sham‐injected controls (see Materials and Methods for details on experimental design). The resulting reads were mapped to the mouse genome assembly (mm10), and miRNA expression was quantified using miRBase annotations (Kozomara & Griffiths‐Jones, 2014). To confirm that our analysis was restricted to new‐born neurons, we analyzed our data set for glia‐enriched miRNAs such as miR‐143 and miR‐200c (Jovicic et al, 2013) and found that these miRNAs had low read numbers in RIP samples of LV.GFP‐AGO2‐injected mice compared to control samples (Fig EV1B). We also found that miRNAs previously implicated in adult neurogenesis, including miR‐9 (Zhao et al, 2009) and miR‐125 (Akerblom et al, 2014), were highly abundant in RIP samples of LV.GFP‐AGO2‐injected mice, further validating the approach (Fig EV1C).
Recent studies suggest that one or a few very highly expressed miRNA families in each cell type are responsible for phenotypical maintenance or morphological switches (Busskamp et al, 2014; Jonsson et al, 2015; Shenoy et al, 2015). Therefore, we grouped miRNA families based on the miRbase classification (www.mirbase.org/) (Kozomara & Griffiths‐Jones, 2014) and analyzed their relative contribution to the total pool of active miRNAs in new‐born OB neurons (Fig 1C). We found that only a few miRNA families compose the vast majority in new‐born neurons. In particular, the let‐7 family was highly expressed in new‐born neurons, making up more than half of the sequencing reads, and may therefore be a key miRNA in regulating adult OB neurogenesis (Fig 1C).
In mice, the let‐7 family consists of 12 members that all share the same seed sequence and therefore are likely to have complementary targets (www.mirbase.org/) (Kozomara & Griffiths‐Jones, 2014). When we analyzed the relative contribution of individual let‐7 family members to the total pool of miRNAs in new‐born OB neurons, we found that seven members of the let‐7 family are among the ten highest expressed miRNAs, with let‐7c being most abundant (Fig 1D). To further confirm the robustness of the results, we performed an independent biological replicate, which yielded very similar results (Fig EV1D–F). Together these data demonstrate a high‐level expression of several let‐7 family members in new‐born OB neurons.
Knockdown of let‐7 in neuroblasts results in impaired radial migration and maturation of adult‐born neurons
In order to investigate a potential functional role for let‐7 we generated a lentiviral miRNA‐sponge vector (LV.let‐7.sp) (Fig 2A). miRNA sponges are imperfectly complementary transgenic transcripts that efficiently knock down miRNA activity in vivo (Gentner et al, 2009). The vector contained a GFP reporter allowing for simple detection of sponge‐expressing cells. We validated the functionality of the LV.let‐7.sp construct by demonstrating its ability to derepress a transcript known to be controlled by let‐7c (Rybak et al, 2008), using a luciferase reporter assay (Fig 2B).
Figure 2. Knockdown of let‐7 in migrating neuroblasts leads to an impairment in radial migration of new‐born neurons.
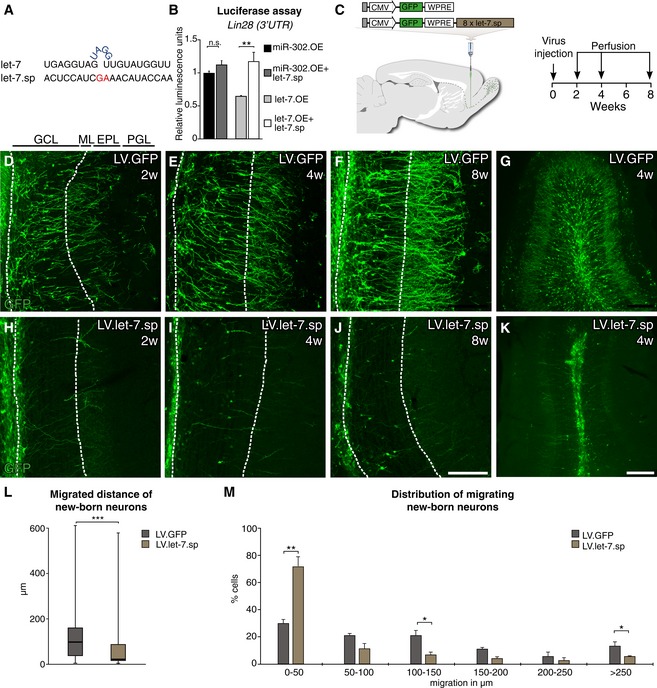
-
ASequences of the mature let‐7 and the let‐7.sp sequence.
-
BValidation of the let‐7.sp construct by luciferase assay. A miR‐302.OE construct, a miRNA not targeting LIN28, was used as a control; n = 3.
-
CVector design and experimental workflow of LV.let‐7.sp and LV.GFP virus injections into the RMS.
-
D–KImmunohistochemical analyses of the OB of injected animals 2, 4, and 8 weeks after injection. LV.GFP‐expressing cells migrated into the outer layers of the OB at all time points were examined. In contrast, LV.let‐7.sp‐expressing cells migrated into the center of the OB but failed to radially migrate into the outer layers. Layers of the OB are indicated (GCL, granule cell layer; ML, mitral cell layer; EPL, external plexiform layer; PGL, periglomerular layer). Scale bars: 100 μm (D–F and H–J) and 250 μm (G and K).
-
LBox plot of the migrated distances of all LV.let‐7.sp‐ and LV.GFP‐expressing cells from the center of the OB 4 weeks after LV injection. Boxes represent the 25th and 75th percentiles. Whiskers depict the minimum and maximum values. The median is represented by a horizontal line within the boxes. LV.let‐7.sp n = 97 cells (four animals); LV.GFP n = 100 cells (four animals).
-
MMigrated distance of new‐born neurons from the center of the OB 4 weeks after LV injection. Data are depicted in a histogram with distinct ranges in μm (x‐axis) plotted against the percentage of cells (y‐axis). LV.GFP n = 4 animals (100 cells); LV.let‐7.sp n = 4 animals (97 cells).
To be able to target migrating neuroblasts, we injected LV.let‐7.sp and LV.GFP control vectors into the RMS of adult mice and analyzed GFP expression in the OB 2, 4, and 8 weeks later (Fig 2C). In control‐injected mice, we found a gradual increase in neurons migrating into the different OB cell layers where they also matured in a characteristic pattern for adult‐born neurons (Fig 2D–G). In contrast, neuroblasts expressing the LV.let‐7.sp failed to migrate radially into the outer layers and instead remained in the center of the OB (Fig 2H–K).
We measured the distance that new‐born neurons had radially migrated from the center of the OB 4 weeks after injection and found a significant difference in the migration distances of LV.let‐7.sp‐ and LV.GFP‐expressing cells (Fig 2L and M). When counting the migrated distance in 50‐μm intervals, we found that most LV.let‐7.sp‐expressing neurons (70%) migrated distances within 50 μm, which is three times shorter than the distance most LV.GFP‐expressing cells were migrating, which was within 150 μm (Fig 2M).
To investigate whether an overall impairment of the generation of the neuroblasts or cell death was the reason for the observed phenotype, we quantified GFP‐positive cells in the OB of LV.let‐7.sp‐ and LV.GFP‐injected animals and found no significant difference in total numbers of GFP‐positive cells in the different groups (Fig EV2A). Caspase‐3 staining confirmed that cells lacking let‐7 displayed no evidence for increased apoptosis (Fig EV2B and C).
Figure EV2. Quantification of GFP‐positive cells in the OB and caspase‐3 staining.
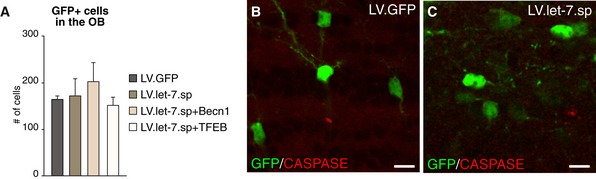
-
ATotal numbers of GFP‐positive new‐born neurons in the OB of LV.let‐7.sp (n = 4 animals)‐, LV.GFP (n = 4 animals)‐, LV.let‐7.sp + LV.Becn1 (n = 3 animals)‐, and LV.let‐7.sp + LV.TFEB (n = 3 animals)‐injected brains. Data are presented as mean ± SEM.
-
B, CCaspase‐3 staining on LV.let‐7.sp‐ and LV.GFP‐injected brains showed no differences between the two conditions. Scale bars: 10 μm.
let‐7 has previously been shown to be necessary for cell cycle exit of cells (Lee et al, 2011; Patterson et al, 2014). To test whether inhibition of let‐7 impairs differentiation of neuroblasts into neurons leading to ectopic proliferation in the OB, we injected BrdU into animals injected with LV.let‐7.sp or LV.GFP, 2 h before animals were sacrificed. We found no evidence for increased BrdU labeling in let‐7.sp‐expressing cells (Fig 3A and B), indicating that knockdown of let‐7 does not affect the cell cycle exit of neuroblasts.
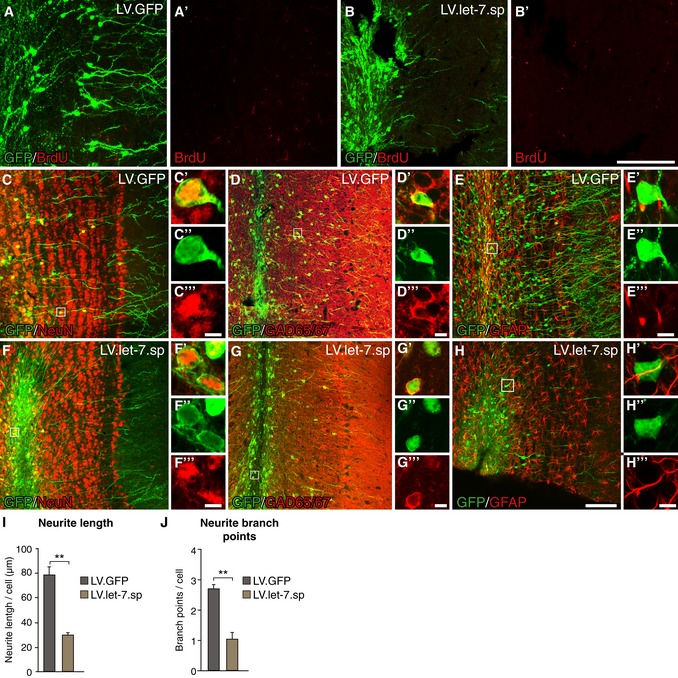
Figure 3. Cells lacking let‐7 express neuronal markers but fail to fully mature.
-
A, BBrdU staining of LV.GFP‐ and LV.let‐7.sp‐injected brains. A BrdU pulse was given 2 h before perfusion. Scale bars: 80 μm.
-
C–HLV.GFP‐ and LV.let‐7.sp‐expressing cells co‐labeled for NeuN (C, F), GAD65/67 (D, G), and the astrocyte‐marker GFAP (E, H). Scale bars: 80 μm (C‐H) and 5 μm (insets).
-
I, JTotal neurite length (I) and total number of branch points (J) of GFP‐positive cells in LV.GFP‐ and LV.let‐7.sp‐injected animals. LV.GFP n = 3 animals (357 neurons), LV.let‐7.sp n = 3 animals (235 neurons). Data are shown as mean ± SEM; unpaired two‐tailed t‐test, **P < 0.01.
On the contrary, we found that cells expressing LV.let‐7.sp co‐labeled for the pan‐neuronal marker NeuN and GAD65/67, a marker for inhibitory neurons, which was similar in LV.GFP control‐injected animals (Fig 3C, D, F and G). We found no evidence for co‐labeling with the astrocyte‐marker GFAP in LV.let‐7.sp‐injected animals (Fig 3E–H). This suggests that LV.let‐7.sp‐expressing cells, despite not migrating radially into the OB, obtain many of the characteristics normally found in adult‐born neurons, including the expression of markers associated with inhibitory neurons.
Although let‐7 knockdown does not affect the expression of mature neuronal markers, we noted that many LV.let‐7.sp‐expressing cells appeared to have an immature morphology. We therefore conducted an automated morphological profiling of GFP‐positive cells in LV.let‐7.sp‐ and LV.GFP‐injected animals. LV.GFP‐expressing cells had a complex morphology and an average total neurite length of 80 μm and on average 2.6 branch points per cell (Fig 3I and J). In contrast, the LV.let‐7.sp‐expressing cells just had an average total neurite length of 32 μm per cell and only 1.2 branch points per cell, demonstrating that let‐7 knockdown results in an immature neuronal morphology. Taken together, these results demonstrate that knockdown of let‐7 in adult‐born neuroblasts results in an impairment of radial migration into the OB and an impaired morphological maturation of new‐born neurons.
Let‐7 regulates autophagy in new‐born neurons
Computational predictions suggest that let‐7 targets hundreds of transcripts (TargetScan), hereby regulating multiple cellular mechanisms. However, we hypothesized that the impaired migration and maturation in new‐born neurons induced by loss of let‐7 may be influenced by altered autophagy. A number of recent studies implicate let‐7 in the control of intracellular metabolism and protein degradation pathways, including autophagy, by targeting genes in the amino acid‐sensing pathway (Zhu et al, 2011; Dubinsky et al, 2014). In addition, autophagy has been shown to be active at a basal level in most cells of the central nervous system (CNS) and recent studies have suggested that autophagy and autophagy‐related genes are important for neurogenesis (Yazdankhah et al, 2014; Li et al, 2016; Wu et al, 2016). However, a role for let‐7 in controlling autophagy in adult neurogenesis has not previously been investigated.
Dubinsky et al previously identified 14 genes in the amino acid‐sensing pathway as potentially being controlled by let‐7, herby providing a positive regulation of neuronal autophagic activation (Dubinsky et al, 2014) (Fig EV3A). Using qRT–PCR, we found that several of these genes were also expressed in the mouse OB (Figs 4A and EV3B). To test whether these genes were also miRNA targets and present in the RISC of new‐born neurons, we injected LV.GFP‐AGO2 and performed RIP 8 weeks after injection. We performed qRT–PCR on RIP samples and found that Slc7a5 and Slc3a2 were both enriched in AGO2‐RIP samples, demonstrating that they are miRNA targets in new‐born neurons (Fig 4B). We then performed luciferase assays and confirmed that Slc7a5 and Slc3a2 are direct let‐7 target genes (Fig 4C). Together, these data demonstrate that Slc7a5 and Slc3a2, which are both known amino acid transporters that play a crucial role in the regulation of autophagic activity (Nicklin et al, 2009), are direct let‐7 targets in adult‐born neurons.
Figure EV3. let‐7 positively regulates autophagy by targeting autophagy‐related genes.
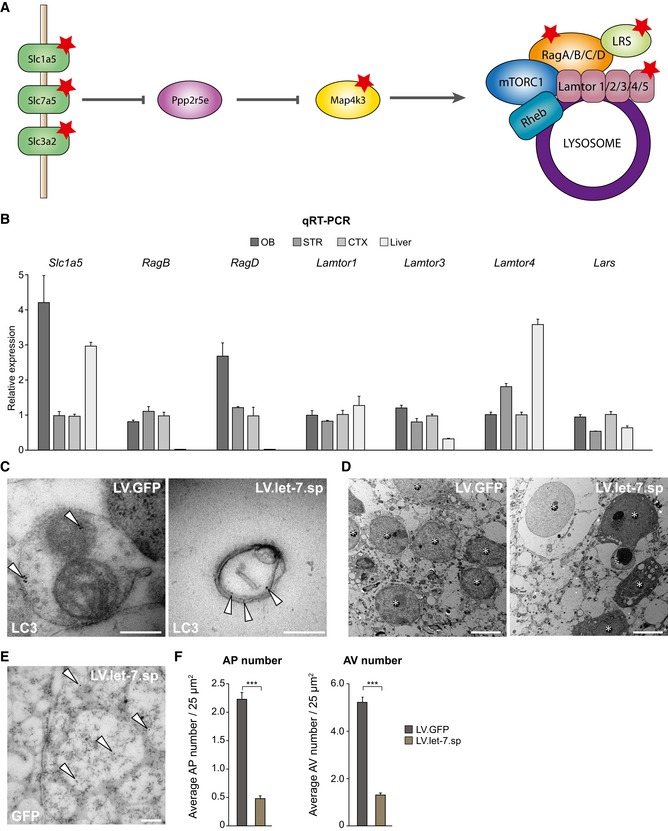
- Previously identified let‐7 targets in the amino acid‐sensing pathway are marked with a star. Graphic is adapted from Dubinsky et al (2014).
- Genes in the amino acid‐sensing pathway are expressed in the OB shown by qRT–PCR (n = 3).
- LC3 staining was conducted to identify and validate autophagic structures in the OB using electron microscopy. Gold particles are indicated with white arrows. Scale bars: 200 nm.
- Pictures for the quantification of autophagic structures were taken in close vicinity to the nuclei of GFP‐positive cells. Nuclei are indicated with a star. Scale bars: 5 μm.
- Electron microscopy picture showing gold particles in the nucleus of a let‐7.sp‐expressing new‐born neuron in the OB. Gold particles are highlighted with white arrows. Scale bar: 200 nm.
- Electron microscopy analysis and quantifications showed a decrease in the number of autophagosomes (APs) and late autophagic vesicles (AVs) in LV.let‐7.sp‐injected animals [n = 210 (two animals)] compared to LV.GFP control animals [n = 132 (three animals)].
Figure 4. let‐7 targets autophagy‐related genes and knockdown of let‐7 leads to an alteration in autophagy.
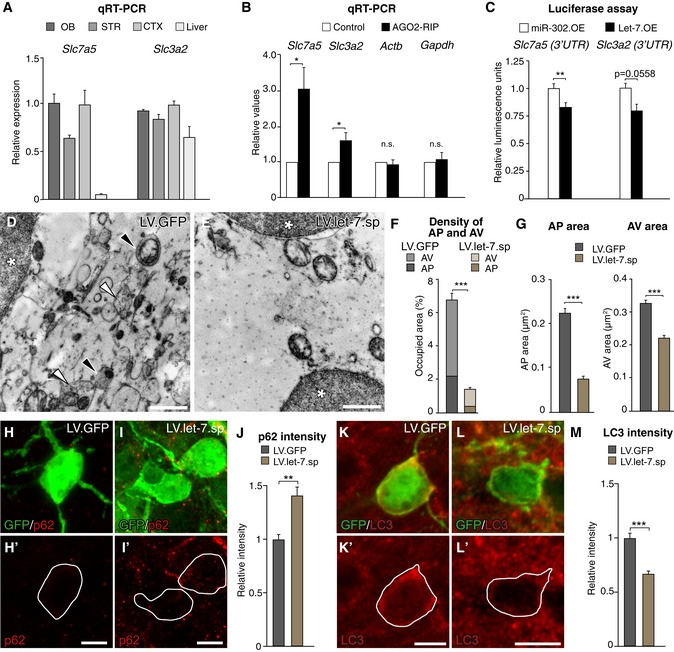
-
AmRNA levels of the amino acid transporter Slc7a5 and Slc3a2 in the brain and other tissues shown by qRT–PCR, n = 3 animals.
-
BSlc7a5 and Slc3a2 are significantly enriched in AGO2‐RIP samples compared to sham‐injected controls. Beta‐actin (Actb) and Gapdh were used as controls, n = 6.
-
C3′UTR luciferase assay was performed to determine functional let‐7 binding site in Slc7a5 and Slc3a2. Slc7a5 3′UTR n = 6 per group, Slc3a2 3′UTR n = 3 per group.
-
D, EElectron microscopy pictures taken from LV.GFP control‐ and LV.let‐7.sp‐expressing cells. * indicates nuclei, black arrows indicate autophagosomes (AP), white arrows indicate late autophagic vesicles (AVs). Scale bars: 1 μm.
-
FBar graph depicting the density of APs and AVs in LV.let‐7.sp‐ and LV.GFP‐expressing cells. LV.let‐7.sp n = 210 (two animals) and LV.GFP n = 132 (three animals).
-
GAverage area shown in μm2 of APs and AVs in LV.let‐7.sp‐ and LV.GFP‐expressing cells. LV.let‐7.sp n = 210 (two animals) and LV.GFP n = 132 (three animals).
-
H–Jlet‐7 knockdown leads to an increase in p62 levels in new‐born neurons compared to LV.GFP‐expressing cells. Scale bars: 10 μm (H, I). LV.GFP n = 3 animals (26 cells); LV.let‐7.sp n = 3 animals (35 cells).
-
K–MKnockdown of let‐7 leads to a significant decrease in LC3 levels in new‐born neurons compared to LV.GFP‐expressing cells. Scale bars: 10 μm (K, L); LV.GFP n = 3 animals (137 cells); LV.let‐7.sp n = 3 animals (234 cells).
To investigate if let‐7 inhibition impairs autophagy in new‐born neurons, we performed ultrastructural analysis using transmission electron microscopy (Figs 4D and E, and EV3C–E). We quantified the number and size of autophagic structures in GFP‐positive new‐born neurons in the OB of either LV.let‐7.sp‐ or LV.GFP‐injected animals (Fig EV3D and E). We found a lower content of autophagosomes (AP), as well as late autophagic structures (AVs), in LV.let‐7.sp‐ compared to LV.GFP‐injected animals (Figs 4F and EV3F). In addition, we found that both APs and AVs were smaller in let‐7.sp‐expressing cells (Fig 4G). To further analyze if autophagy is altered in the LV.let‐7.sp‐injected animals, we assessed the expression of Sequestosome‐1 (p62). p62 is a cargo transport protein that is selectively degraded by autophagy and is widely used as a marker to study autophagy; p62 accumulates when autophagy is inhibited while the levels of p62 decrease as a result of induced autophagy (Pircs et al, 2012; Klionsky et al, 2016). When performing immunohistochemistry of p62 on LV.let‐7.sp‐ and LV.GFP control‐injected mice, we found that p62 was localized primarily to the cytoplasm of new‐born neurons, with significantly more p62 in LV.let‐7.sp‐expressing cells compared to LV.GFP‐injected controls (Fig 4H–J). We additionally analyzed microtubule‐associated protein 1A/1B‐light chain 3 (LC3) (Klionsky et al, 2016). Levels of LC3 correlate with autophagy, and decrease of LC3 together with an increased p62 suggests impairment in autophagy. Immunohistochemistry of LC3 showed a higher level of LC3 in LV.GFP‐ compared to LV.let‐7.sp‐expressing cells, further confirming that knockdown of let‐7 leads to a decreased level of autophagy in new‐born neurons (Fig 4K–M).
Activation of autophagy rescues the let‐7‐dependent impaired radial migration
To provide a direct mechanistic link between the let‐7‐mediated impairment of migration and integration and the block of autophagy, we co‐expressed transcription factor EB (TFEB) or Beclin‐1 (Becn1), respectively, together with LV.let‐7.sp. TFEB is a master transcription factor that activates a large autophagy‐activating gene program (Settembre et al, 2011), and Becn1 is a positive regulator of autophagosome formation (He & Levine, 2010) (Fig EV4A–C). We co‐injected LV.let‐7.sp together with LV.TFEB or LV.Becn1 into the RMS and analyzed new‐born neurons 4 weeks after injection (Fig 5A). We found that co‐injection of either LV.Becn1 or LV.TFEB with the LV.let‐7.sp resulted in decreased p62 accumulation compared to when LV.let‐7.sp was injected alone, confirming a re‐activation of autophagy (Fig 5B–E).
Figure EV4. Validation of LV.TFEB‐ and LV.Becn1‐overexpressing constructs.
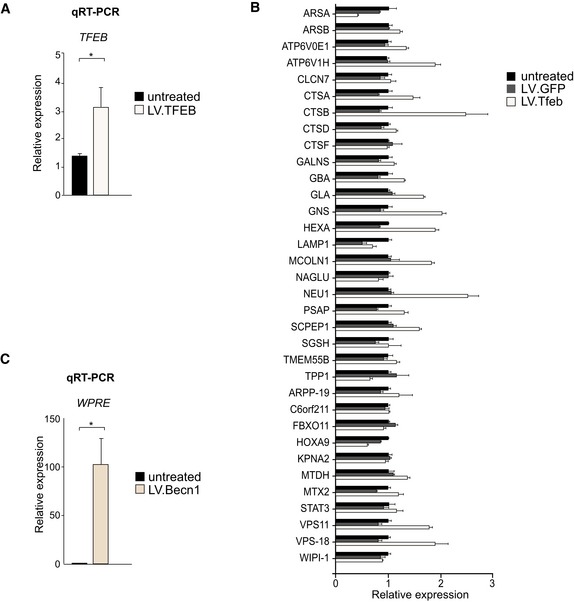
- Levels of TFEB in mouse neural progenitor cells upon LV.TFEB transduction shown by qRT–PCR; n = 2.
- Levels of lysosome‐ and autophagy‐related genes upon LV.TFEB transduction of 293T cells shown by qRT–PCR; n = 3.
- Levels of WPRE sequence upon LV.Becn1 transduction of mouse neural progenitor cells shown by qRT–PCR; n = 2.
Figure 5. Activation of autophagy rescues the impairment of radial migration.
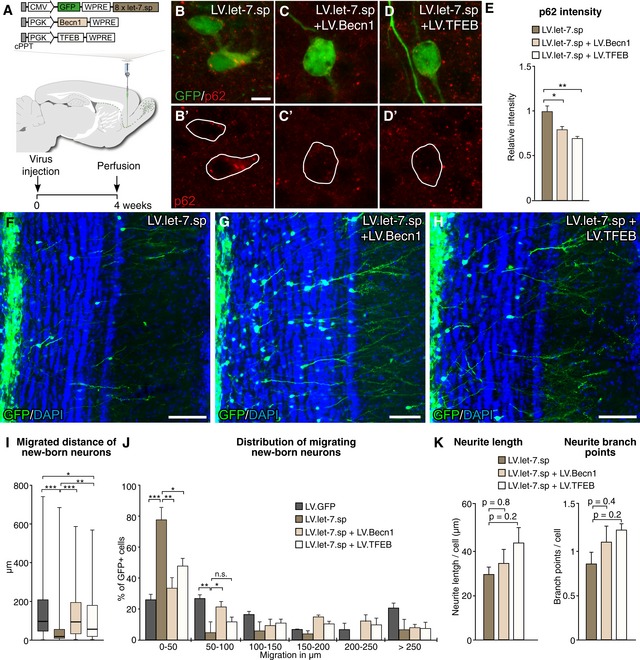
-
ALentiviral vector design and experimental workflow of LV injections into the RMS.
-
B–Ep62 levels upon LV.let‐7.sp injections (B), upon co‐expression of Becn1 (C) and upon co‐expression of TFEB (D). Scale bar: 10 μm. LV.let‐7.sp n = 3 animals (73 cells); LV.let‐7.sp + LV.Becn1 n = 3 animals (75 cells); LV.let‐7.sp + LV.TFEB n = 3 animals (98 cells).
-
F–HImmunohistochemical analysis of GFP‐positive new‐born neurons in the OB of LV.let‐7.sp (F)‐, LV.let‐7.sp + LV.Becn1 (G)‐, and LV.let‐7.sp + LV.TFEB (H)‐injected animals 4 weeks after injection. Scale bars: 80 μm.
-
IBox plot of the migrated distances from the center of the OB of all cells expressing either LV.GFP, LV.let‐7.sp alone, LV.let‐7.sp + LV.Becn1, or LV.let‐7.sp + LV.TFEB. Boxes represent the 25th and 75th percentiles. Whiskers depict the minimum and maximum values. The median is represented by a horizontal line within the boxes. LV.GFP n = 112 cells (three animals); LV.let‐7.sp n = 62 cells (three animals); LV.let‐7.sp + LV.Becn1 n = 88 cells (four animals), and LV.let‐7.sp + LV.TFEB n = 97 cells (five animals).
-
JMigrated distance of new‐born neurons from the center of the OB 4 weeks after LV injection. Data are depicted in a histogram with distinct ranges in μm (x‐axis) plotted against percentage of cells (y‐axis); LV.GFP n = 112 cells (three animals); LV.let‐7.sp n = 62 cells (four animals); LV.let‐7.sp + LV.Becn1 n = 88 cells (four animals); and LV.let‐7.sp + LV.TFEB n = 97 cells (five animals).
-
KTotal neurite length and total number of branch points of GFP‐positive new‐born neurons in LV.let‐7.sp‐, LV.let‐7.sp + LV.Becn1‐, and LV.let‐7.sp + LV.TFEB‐injected animals. LV.let‐7.sp n = 139 cells (three animals), LV.let‐7.sp + LV.Becn1 n = 110 cells (three animals), and LV.let‐7.sp + LV.TFEB n = 124 cells (three animals).
When analyzing the new‐born neurons, we found that co‐injection of either LV.TFEB or LV.Becn1 together with LV.let‐7.sp resulted in a rescue of the let‐7‐mediated impaired radial migration of new‐born neurons into the outer layers of the OB. The migration pattern of LV.TFEB‐ or LV.Becn1‐expressing neurons, co‐injected with LV.let‐7.sp, was similar to that of animals injected with control LV.GFP vectors (Fig 5F–H). We also measured the migrated distance of co‐injected neurons and found that the distance was significantly increased compared to that of cells expressing let‐7.sp‐expressing alone (Fig 5I and J). The number of total GFP‐positive cells in the OB upon LV.BECN‐1 and LV.TFEB overexpression was similar to the number of new‐born neurons upon LV.GFP and LV.let‐7.sp injections alone (Fig EV2A).
We then conducted automated morphological neuronal profiling to investigate whether the activation of autophagy could also rescue the maturation of new‐born neurons. Similar to our previous experiment, LV.let‐7.sp‐expressing cells showed a total neurite length of 30 μm with on average < 1 branch point per cell (0.83) (Fig 5K). We found no evidence for an increase in total neurite length or increase in total number of branch points upon co‐expression of Becn1 and TFEB, respectively (Fig 5K). Thus, although activation of autophagy significantly increases radial migration, it does not mediate a significant improvement of maturation of let‐7.sp‐expressing new‐born neurons in a 4‐week time frame suggesting that other let‐7‐regulated mechanisms underlie this phenotype.
Discussion
When migrating adult‐born neuroblasts reach the OB, they change their route of migration and move radially into the OB where they undergo a remarkable morphological transition and integration into the existing neuronal network (Gheusi et al, 2013). Radial migration of adult‐born neurons is tightly controlled since they integrate in a specific pattern into the OB, which differs from the integration pattern of developmentally born neurons (Lemasson et al, 2005). The placement of adult‐born neurons within the OB is thought to contribute to the unique physiological properties of these cells (Lledo et al, 2008; Malvaut & Saghatelyan, 2016). Despite the importance of appropriate radial migration in adult‐born neurons, the intracellular mechanism that regulates this process remains poorly understood. In this study, we find that let‐7 plays a key role in regulating this process.
Let‐7 has been extensively studied in various tissues and appears to play a key role in maintaining cellular homeostasis, and a number of recent studies implicate let‐7 in the control of metabolic and intracellular protein degradation pathways (Zhu et al, 2011; Dubinsky et al, 2014). For example, let‐7 was shown to regulate several genes in the amino acid‐sensing pathway to induce neuronal autophagy via the mTOR pathway (Dubinsky et al, 2014), and to regulate mTOR also in muscle and liver (Zhu et al, 2011). In the brain, let‐7 is highly expressed and its dysregulation has been implicated in neurodegenerative disorders (Lehmann et al, 2012). We describe herein a previously unknown role for let‐7 in the control of adult neurogenesis through the activation of autophagy.
An appropriate intracellular control of autophagy allows the cell to react and adapt to changing nutrient conditions by recycling cytoplasmic components and macromolecular building blocks. Since neurogenesis is a highly energy‐consuming process, it is likely that autophagy is crucial for providing neural progenitor cells with energy for differentiation, migration and integration. Reduction of autophagy in new‐born neurons due to loss of let‐7‐activity could therefore lead to a lack in energy necessary for radial migration into the OB. In humans, neuroblasts have also been found to be present in the SVZ but they do not migrate to the OB; rather they appear to directly radially migrate and integrate into the striatum (Ernst et al, 2014). Interestingly, adult striatal neurogenesis is impaired in patients with Huntington′s disease (HD) (Ernst et al, 2014), a neurodegenerative disorder characterized by an expansion of CAG repeats in the huntingtin gene resulting in misfolding and aggregation of the protein. It is well established that the formation of protein aggregates in the brain of patients with HD, and other neurodegenerative disorders such as Parkinson's disease and Alzheimer's disease, is linked to impairment in autophagy (Nixon, 2013; Martin et al, 2015). Our data raise the interesting hypothesis that the impairment in adult neurogenesis found in these disorders could be directly linked to alterations in autophagy. It will be very interesting to investigate if autophagy‐activating drugs, which have recently entered clinical testing as a treatment for HD (Frake et al, 2015), will impact on the impairment of adult neurogenesis.
miRNAs are emerging as key regulators of neurogenic processes highlighted by the growing number of miRNAs implicated in brain development and adult neurogenesis (Akerblom & Jakobsson, 2013; Petri et al, 2014). The major contribution of miRNAs to this process is also demonstrated by the ability of miR‐9 and miR‐124 to drive conversion of fibroblasts to neurons (Yoo et al, 2011). In this study, we find that the let‐7 family is a highly abundant miRNA in adult‐born neurons. Let‐7 is likely to target hundreds of mRNAs in new‐born neurons and hereby controls multiple intracellular mechanisms. This is in line with our data where activation of autophagy restored radial migration but not neuronal maturation following let‐7 inhibition. Other studies have found that let‐7 controls the nuclear receptor TLX to influence cell cycle progression of NSCs (Zhao et al, 2010). Let‐7 also directly targets the chromatin‐associated protein HMGA2, which in turn influences the Notch pathway that is important for regulating quiescence of NSCs (Patterson et al, 2014). Together with our findings, this starts to unravel a complex and important regulation of neurogenesis at multiple steps and levels through let‐7.
Cumulatively, this study extends the knowledge on miRNA regulation during adult neurogenesis and provides a novel and exciting finding on how let‐7 contributes to the migration of new‐born neurons through the control of autophagy. This provides a new mechanistic link between miRNAs, protein degradation and autophagy, and these findings open up for new possibilities to identify molecular targets for the treatment of neurodegenerative disorders.
Materials and Methods
Viral vectors
The GFP‐AGO2 sequence used in this study was designed according to (He et al, 2012), synthesized (GenScript), and cloned into a lentiviral backbone under the control of a PGK promoter. Since the AGO2‐RIP approach is dependent on a high number of transduced cells, we used lentiviral vectors for the delivery of the GFP‐AGO2 fusion protein. Lentiviral vectors allow for high transduction rates of both dividing and non‐dividing cells, leading to the transduction of high numbers of new‐born neurons (Alonso et al, 2012).
The let‐7.sp vectors contained eight repeats of an imperfectly complementary sequence forming a central bulge when binding to the respective miRNAs. The sponge sequence was cloned into a third generation lentiviral vector containing a strong promoter derived from cytomegalovirus (CMV), a requirement for successful miRNA inhibition.
Following sponge sequence was used: let‐7.sp, AACCATACAAAGCTACCTCA.
All cloning was performed using standard techniques. Lentiviral vectors were produced as previously described (Zufferey et al, 1997). Vectors were titrated using flow cytometry and qRT–PCR analysis as previously described (Georgievska et al, 2004). The titers of the vectors in this study were in the range of: 5 × 108 – 2 × 109 TU/ml.
A let‐7c overexpression vector was designed by cloning the sequence of the precursor miRNA and 200 nucleotides flanking either site between GFP and a WPRE under the control of an ubiquitin promoter.
The human TFEB and the mouse Becn1 overexpression sequences used in this study were designed according to Decressac et al (2013), synthesized (GenScript), and cloned into a lentiviral backbone under the control of a PGK promoter.
Viral vector injections
All animal‐related procedures were approved by and conducted in accordance with the committee for use of laboratory animals at Lund University. For vector injections into the rostral migratory stream (RMS), 1 μl of lentiviral vector was unilaterally injected into the right RMS of wild‐type 10‐week‐old C57BL/6 mice (females, Taconic). Coordinates from bregma were as follows: AP + 3.3, ML − 0.8, DV − 2.9. Sham injections were conducted the same way as lentiviral vector injections, but instead of lentiviral vectors 1 μl of PBS was injected into the RMS.
LV.let‐7.sp and LV.GFP viruses, respectively, were diluted 1:1 in phosphate‐buffered saline (PBS). For co‐injections in the rescue experiment, LV.let‐7.sp and LV.TFEB or LV. Becn1, lentiviral vectors were mixed in a ratio of 1:1 in a total volume of 1 μl.
A proportion of the animals were given an intraperitoneal pulse of 50 mg/kg BrdU (Sigma).
Immunofluorescence and microscopy
Mice were transcardially perfused with 4% paraformaldehyde (PFA) (Sigma), the brains post‐fixed for 2 h and transferred to PBS with 25% sucrose. Brains were coronally sectioned on a microtome (35 μm) and put in KPBS.
Standard immunohistochemistry was used, as published in detail elsewhere (Sachdeva et al, 2010). Primary antibodies were diluted as follows: chicken anti‐GFP 1:1,000 (Abcam, Cat# Ab13970), mouse anti‐NeuN 1:1,000 (Millipore, Cat# MAB377), mouse anti‐p62 1:500 (Abcam, Cat# ab91526), rabbit anti‐GAD67/65 1:5,000 (Chemicon, Cat# AB1511), rabbit anti‐GFAP 1:1,000 (DAKO, Cat# Z0334), rabbit anti‐LC3 1:500 (Sigma, Cat# L7543), and rabbit anti‐caspase‐3 (Cell Signaling Cat# 9661; 1:500). The dilution factor of the secondary antibodies was 1:500 (Molecular Probes, Cat# A11039) or 1:200 (Jackson ImmunoResearch Labs Cat# 715‐165‐151; Jackson ImmunoResearch Labs Cat# 711‐485‐152). All sections were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI, Sigma‐Aldrich, 1:1,000).
For the BrdU staining, the slices were fixed for 20 min in 4% PFA followed by incubation at 65°C in 1 M HCl before adding the primary antibody (1:500 rat anti‐BrdU, Serotec, Cat# OBT0030).
For imaging and analysis, a confocal microscope (Leica) or inverted microscope (Leica, DFC360 FX‐DMI 6000B) was used.
Migration analyses of GFP‐expressing cells in the OB
Two to three representative OB sections (35 μm) from four to six LV‐injected animals were used for quantification. Fluorescent images of GFP expression were taken with the same exposure using a 10× objective. A line was drawn in the middle of the OB sections to determine the center of the OB, and a square of the same size was set per section using Adobe Illustrator CS6. The migration of all GFP‐positive cells in this square was measured by determining the distance from the OB center to the center of the cell body. The distribution of migrated distances of all GFP‐positive cells in a specific group (LV.GFP, n = 100 cells (four animals); LV.let‐7.sp, n = 97 cells (four animals); LV.let‐7.sp + LV.TFEB, n = 97 cells (five animals); LV.let‐7.sp + LV.Becn1, n = 88 cells (four animals); let‐7.sp, n = 62 cells [three animals); LV.GFP, n = 112 (three animals)] was depicted in a box plot, and Mann–Whitney U‐test or Kruskal–Wallis test followed by a Dunn's multiple comparison test was conducted to test for statistical significance.
The percentage of cells per animal migrating distances within specific ranges were calculated, and an unpaired two‐tailed t‐test or an one‐way ANOVA followed by Tukey's post hoc test was conducted to test for statistical significance. Data are presented as mean ± SEM.
Quantification of total number of GFP‐positive cells
One to two representative OB sections (35 μm) from three to four LV‐injected animals were used for quantification of GFP‐positive cells. Fluorescent images of GFP expression were taken with the same exposure using a 5× objective. All GFP‐positive cells on an OB section were counted using CellCounter in ImageJ. Data are represented as mean ± SEM.
Automated neuronal profiling
The neuronal profiling was conducted with the software HCS Studio—Cellomics Scan 6.6.0 from Thermo Scientific. Pictures were taken with a 20× objective.
Representative OB pieces (35 μm) of similar size from three different animals that were sacrificed 4 weeks after vector injection were chosen per group, and neuronal profiling was conducted on all GFP‐positive neurons on these sections [LV.GFP n = 3 mice (357 analyzed neurons); LV.let‐7.sp n = 6 mice (374 analyzed neurons), LV.Becn1 + LV.let‐7.sp n = 3 mice (110 analyzed neurons), LV.TFEB + LV.let‐7.sp n = 3 mice (124 analyzed neurons)]. Cell bodies were detected by the software, and neurites were traced to determine the total length of neurites per cell and the total amount of branch points per cell. Crossings of neurites from different cells were not counted. An unpaired two‐tailed and an one‐way ANOVA followed by a Tukey's post hoc test were conducted to test for statistical significance. Data are presented as mean ± SEM.
Luciferase reporter assay
A 400‐bp sequence incorporating the let‐7 binding site in the 3′UTR of Slc7a5 and Slc3a2 and of Lin28a was cloned into the dual‐luciferase reporter vector pSICHECK‐2 (Promega). The luciferase reporter constructs were co‐transfected with either a let‐7 or miR‐302 non‐targeting overexpression construct or additionally with let‐7.sp construct into three independent replicates of 293T cells using Turbofect (Fermentas). Forty‐eight hours after transfection, cells were assayed for luminescence using a dual‐luciferase assay (Promega). One‐way ANOVA followed by a Tukey's multiple comparison post hoc test was performed in order to test for statistical significance. Data are presented as mean ± SEM.
Validation of TFEB and Becn1 overexpression constructs
Mouse neural progenitor cells were transduced with LV. Becn1 or LV. TFEB with an MOI of 10 to validate overexpression of the constructs. A total of 293T cells were transduced with the LV.TFEB or LV.GFP to test the upregulation of lysosome‐ and autophagy‐related genes. Cells were harvested 48 h after transduction for RNA extraction. Total RNA extraction was performed using the RNeasy Mini Kit (Qiagen).
Expression analyses of autophagy‐related genes
OB, striatum, cortex, and parts of the liver from three mice were dissected and the tissue was homogenized using TissueLyser LT (50 Hz, 2 min). Total RNA extraction was performed using the RNeasy Mini Kit (Qiagen).
For qRT–PCR, 1 ug of RNA was used for the reverse transcription performed with the Maxima First Strand cDNA Synthesis Kit for qRT–PCR according to supplier's recommendations. SYBR green qRT–PCR was performed and data were quantified using the ΔΔCt‐method and normalized to Gapdh expression.
All primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3/). Primer sequences are shown in Table EV1. Primer sequences for lysosome‐ and autophagy‐related genes were previously reported in Settembre et al (2011).
RNA‐interacting protein immunoprecipitation
For the qRT–PCR validation of the RIP samples and for sequencing experiments of RIP samples, we used PBS sham‐injected (n = 12) and LV.GFP‐AGO2‐injected (n = 18) mice.
LV vector‐injected mice were decapitated, and the OBs of three injected mice were quickly dissected, pooled, and homogenized (within 20 min) in ice‐cold lysis buffer (10 mM HEPES (pH = 7.3), 100 mM KCl, 0.5% NP40, 5 mM MgCl2, 0.5 mM dithiothreitol, protease inhibitors, recombinant RNase inhibitors, 1 mM PMSF) using TissueLyser LT (50 Hz, 2 min). The pooling of the tissue from different animals has the advantage of decreasing the variation within tissue differences due to vector injections. Homogenates were centrifuged for 15 min at 16,200 × g, 4°C to clear the lysate and separated into two individual samples, of which one was later used for qRT–PCR analyses and one for RNA sequencing. A 50‐μl sample served as INPUT control. The remaining RIP samples were incubated with anti‐GFP‐coated Dynabeads® Protein G beads (Life Technologies) at 4°C for 24 h with end‐over‐end rotation. The beads were coated with rabbit anti‐GFP polyclonal antibody (Abcam). After incubation, beads were collected on a Dynamagnet (1 min, on ice) and gently resuspended in low‐salt NT2 buffer (50 mM Tris–HCL (pH = 7.5), 1 mM MgCl2, 150 mM NaCl, 0.5% NP40, 0.5 mM dithiothreitol, 1 mM PMSF, protease inhibitors, and recombinant RNase inhibitors). The beads were transferred into a new collection tube and washed once with low‐salt NT2 buffer, followed by two washes with high‐salt NT2 buffer (50 mM Tris–HCl (pH = 7.5), 1 mM MgCl2, 600 mM NaCl, 0.5% NP40, 0.5 mM DTT, protease inhibitors, 1 mM PMSF, and recombinant RNase inhibitors). After the last washing step, the RNA fraction was resuspended in QIAzol buffer and RNA was isolated from RIP and INPUT samples according to the miRNeasy micro kit (Qiagen).
LNA‐Quantitative real‐time PCR
To synthesize cDNA from miRNA, we used the Universal cDNA synthesis kit (Exiqon) according to the supplier's recommendations. LNA™ PCR primer sets, hsa‐miR‐103a‐3p, and hsa‐miR‐124‐3p were purchased from Exiqon.
qRT–PCR analyses of RIP samples
cDNA was generated from five RIP samples using the Maxima First Strand cDNA Synthesis Kit for qRT–PCR according to supplier's recommendations. SYBR green qRT–PCR was performed. Data were normalized to sham‐injected control samples, and the relative enrichment of genes was calculated. Beta‐actin and Gapdh were used as controls. A one‐sample t‐test was performed to test for statistical significance.
Sequencing and data analysis of RIP samples
cDNA libraries of two independent GFP‐AGO2 RIP and two independent RIP control samples (each RIP sample contains tissue from three injected mice) were prepared using the NEB small RNA library prep kit for small RNA sequencing. Illumina high‐throughput sequencing (HiSeq2500 SR 1×50 run) was applied to the samples (total number of reads for small RNA sequencing: 71,276,024).
The 50‐bp single end reads were mapped to the mouse genome (mm10) and visualized in USCS genome browser. Reads mapping to miRNAs were quantified using miRbase (www.miRbase.org). Data were normalized to the total read number mapping to the genome.
For read distribution analyses, the percentage of reads mapping to individual miRNAs or to miRNA families was calculated.
The small RNA sequencing data from this publication have been submitted to the NCBI Gene Expression Omnibus database and assigned the GEO series accession number GSE83903.
Relative intensity measurements
p62 and LC3 relative intensity was measured in three representative sections from three animals in each case per group. Confocal pictures were taken with the same settings in each. The pictures were analyzed with ImageJ by circling GFP‐positive cells and measuring the integrated density. Average intensity was measured from LV.GFP n = 26, LV.let‐7.sp n = 35 cells in Fig 4H and I; LV.let‐7.sp n = 73, LV.let‐7.sp + LV.TFEB n = 75, LV.let‐7.sp + LV. Becn1 n = 98 cells in Fig 5B–D and LV.GFP n = 137, LV.let‐7.sp n = 234 cells in Fig 4K and L. Student's t‐test was performed in order to test for statistical significance. Data are presented as mean ± SEM.
Transmission electron microscopy
For transmission electron microscopy (TEM) analysis, LV.GFP‐ and LV.let‐7.sp‐injected mice were perfused 3 weeks post‐injection using 1.5% PFA, and a small (1–3 mm2) GFP fluorescent granule cell layer piece was dissected from the OB from three animals/group using a Leica MZ16 F microscope. The tissue was fixed with 1.5% PFA + 0.5% glutaraldehyde (0.1 M Sorensen phosphate buffer, pH = 7.2) and rinsed with 0.1 M Sorensen phosphate buffer. Leica AFS2 was used in progressive lowering of temperature (PLT) mode allowing for low temperature dehydrating in graded ethanol series and embedding in Lowicryl HM20. Ultra‐thin (around 50 nm) sections were cut with a Leica EMU C7 ultramicrotome and mounted on pioloform‐coated gold grids. Sections were incubated at 4°C overnight with chicken anti‐GFP 1:100 (Abcam, Cat# Ab13970) or rabbit anti‐LC3 1:100 (Sigma, Cat# L7543) diluted in blocking solution, BSA, or goat serum, respectively. Sections were rinsed with PBS and incubated with secondary antibodies for an hour at room temperature: rabbit anti‐chicken (gold 10 nm, TED PELLA, 1:20) or goat anti‐rabbit (gold 10 nm, TED PELLA, 1:20). The sections were examined in a FEI Tecnai Spirit BioTWIN 120kv electron microscope, and the late and early autophagic structures were quantified with the ImageJ software.
A LC3 staining was performed to validate and identify autophagic structures in the OB tissue (Fig EV3C). For statistical analysis, representative pictures were taken with the same magnification in all cases from two LV.let‐7.sp (n = 210)‐ and three LV.GFP (n = 132)‐injected animals. Autophagic structures were defined in GFP‐positive gold immunolabeled cytoplasmic areas based on size, cellular contents, and morphology (Eskelinen, 2008; Yla‐Anttila et al, 2009; Eskelinen & Kovacs, 2011; Klionsky et al, 2016). In each case, pictures were taken in close proximity to GFP‐positive nuclei. Unpaired, two‐tailed t‐test was performed in order to test for statistical significance. Data are presented as mean ± SEM.
Author contributions
RP, KP, MEJ, MÅ, PLB, TK, and JJ designed and performed research and analyzed data. RP and PLB performed Bioinformatics analysis. RP and JJ wrote the paper, and all authors reviewed the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We are grateful to all members of the Jakobsson laboratory. We also thank M. Persson Vejgården, U. Jarl, J. Johansson, A. Hammarberg, E. Ling, Christina Isaksson, B. Mattson, S. da Rocha Baez, M. Sparrenius, and I. Nilsson for technical assistance. We would also like to thank Lina Gefors and Lund University Bioimaging Center (LBIC) at Lund University for help with TEM. The work was supported by grants from the Swedish Research Council (# K2014‐62X‐22527‐01‐3), the Swedish Foundation for Strategic Research (# FFL12‐0074), the Swedish Brain Foundation, The Swedish Cancer Foundation, the Swedish excellence project Basal Ganglia Disorders Linnaeus Consortium (BAGADILICO), and the Swedish Government Initiative for Strategic Research Areas (MultiPark & StemTherapy).
The EMBO Journal (2017) 36: 1379–1391
References
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J (2012) MicroRNA‐124 is a subventricular zone neuronal fate determinant. J Neurosci 32: 8879–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerblom M, Jakobsson J (2013) MicroRNAs as neuronal fate determinants. Neuroscientist 20: 235–242 [DOI] [PubMed] [Google Scholar]
- Akerblom M, Petri R, Sachdeva R, Klussendorf T, Mattsson B, Gentner B, Jakobsson J (2014) microRNA‐125 distinguishes developmentally generated and adult‐born olfactory bulb interneurons. Development 141: 1580–1588 [DOI] [PubMed] [Google Scholar]
- Alonso M, Lepousez G, Sebastien W, Bardy C, Gabellec MM, Torquet N, Lledo PM (2012) Activation of adult‐born neurons facilitates learning and memory. Nat Neurosci 15: 897–904 [DOI] [PubMed] [Google Scholar]
- Alvarez‐Buylla A, Garcia‐Verdugo JM (2002) Neurogenesis in adult subventricular zone. J Neurosci 22: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Ming GL, Song H (2015) Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnert TP, Bilsland JG, Guest PC, Heavens R, McLaren D, Dale C, Thakur M, McAllister G, Munoz‐Sanjuan I (2006) Molecular characterization of adult mouse subventricular zone progenitor cells during the onset of differentiation. Eur J Neuorsci 24: 661–675 [DOI] [PubMed] [Google Scholar]
- Busskamp V, Krol J, Nelidova D, Daum J, Szikra T, Tsuda B, Juttner J, Farrow K, Scherf BG, Alvarez CP, Genoud C, Sothilingam V, Tanimoto N, Stadler M, Seeliger M, Stoffel M, Filipowicz W, Roska B (2014) miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 83: 586–600 [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F (2009) miR‐124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chevigny A, Core N, Follert P, Gaudin M, Barbry P, Beclin C, Cremer H (2012) miR‐7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci 15: 1120–1126 [DOI] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A (2013) TFEB‐mediated autophagy rescues midbrain dopamine neurons from alpha‐synuclein toxicity. Proc Natl Acad Sci USA 110: E1817–E1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia‐Verdugo JM, Alvarez‐Buylla A (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97: 703–716 [DOI] [PubMed] [Google Scholar]
- Dubinsky AN, Dastidar SG, Hsu CL, Zahra R, Djakovic SN, Duarte S, Esau CC, Spencer B, Ashe TD, Fischer KM, MacKenna DA, Sopher BL, Masliah E, Gaasterland T, Chau BN, Pereira de Almeida L, Morrison BE, La Spada AR (2014) Let‐7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab 20: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisen J (2014) Neurogenesis in the striatum of the adult human brain. Cell 156: 1072–1083 [DOI] [PubMed] [Google Scholar]
- Eskelinen EL (2008) To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 4: 257–260 [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Kovacs AL (2011) Double membranes vs. lipid bilayers, and their significance for correct identification of macroautophagic structures. Autophagy 7: 931–932 [DOI] [PubMed] [Google Scholar]
- Frake RA, Ricketts T, Menzies FM, Rubinsztein DC (2015) Autophagy and neurodegeneration. J Clin Invest 125: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L (2009) Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 6: 63–66 [DOI] [PubMed] [Google Scholar]
- Georgievska B, Jakobsson J, Persson E, Ericson C, Kirik D, Lundberg C (2004) Regulated delivery of glial cell line‐derived neurotrophic factor into rat striatum, using a tetracycline‐dependent lentiviral vector. Hum Gene Ther 15: 934–944 [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF, Kornblum HI (2001) A genetic analysis of neural progenitor differentiation. Neuron 29: 325–339 [DOI] [PubMed] [Google Scholar]
- Gheusi G, Lepousez G, Lledo PM (2013) Adult‐born neurons in the olfactory bulb: integration and functional consequences. Curr Topics Behav Neurosci 15: 49–72 [DOI] [PubMed] [Google Scholar]
- Gurok U, Steinhoff C, Lipkowitz B, Ropers HH, Scharff C, Nuber UA (2004) Gene expression changes in the course of neural progenitor cell differentiation. J Neurosci 24: 5982–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ (2012) Cell‐type‐based analysis of microRNA profiles in the mouse brain. Neuron 73: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ME, Nelander Wahlestedt J, Akerblom M, Kirkeby A, Malmevik J, Brattaas PL, Jakobsson J, Parmar M (2015) Comprehensive analysis of microRNA expression in regionalized human neural progenitor cells reveals microRNA‐10 as a caudalizing factor. Development 142: 3166–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi‐Carter R (2013) Comprehensive expression analyses of neural cell‐type‐specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci 33: 5127–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten SL, Kudo LC, Jackson R, Sabatti C, Kornblum HI, Geschwind DH (2003) Global analysis of gene expression in neural progenitors reveals specific cell‐cycle, signaling, and metabolic networks. Dev Biol 261: 165–182 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre‐Ghiso J, Airoldi EM et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths‐Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Lazarini F, Lledo PM (2011) Is adult neurogenesis essential for olfaction? Trends Neurosci 34: 20–30 [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim SK, Park CK, Jung KH, Lee SK, Kim M, Roh JK (2011) Let‐7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol 102: 19–24 [DOI] [PubMed] [Google Scholar]
- Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kalin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL et al (2012) An unconventional role for miRNA: let‐7 activates Toll‐like receptor 7 and causes neurodegeneration. Nat Neurosci 15: 827–835 [DOI] [PubMed] [Google Scholar]
- Lemasson M, Saghatelyan A, Olivo‐Marin JC, Lledo PM (2005) Neonatal and adult neurogenesis provide two distinct populations of new‐born neurons to the mouse olfactory bulb. J Neurosci 25: 6816–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lu G, Hu J, Shen X, Ju J, Gao Y, Qu L, Xia Y, Chen Y, Bai Y (2016) EVA1A/TMEM166 regulates embryonic neurogenesis by autophagy. Stem Cell Rep 6: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Alvarez‐Buylla A (2014) Adult neural stem cells stake their ground. Trends Neurosci 37: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Y, Adam Y, Mizrahi A (2014) Odor processing by adult‐born neurons. Neuron 81: 1097–1110 [DOI] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS (2006) Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci 7: 179–193 [DOI] [PubMed] [Google Scholar]
- Lledo PM, Merkle FT, Alvarez‐Buylla A (2008) Origin and function of olfactory bulb interneuron diversity. Trends Neurosci 31: 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaut S, Saghatelyan A (2016) The role of adult‐born neurons in the constantly changing olfactory bulb network. Neural Plast 2016: 1614329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR (2015) Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci 38: 26–35 [DOI] [PubMed] [Google Scholar]
- Merkle FT, Fuentealba LC, Sanders TA, Magno L, Kessaris N, Alvarez‐Buylla A (2014) Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat Neurosci 17: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Papandreou ME, Tavernarakis N (2015) Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ 22: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA (2013) The role of autophagy in neurodegenerative disease. Nat Med 19: 983–997 [DOI] [PubMed] [Google Scholar]
- Patterson M, Gaeta X, Loo K, Edwards M, Smale S, Cinkornpumin J, Xie Y, Listgarten J, Azghadi S, Douglass SM, Pellegrini M, Lowry WE (2014) let‐7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Rep 3: 758–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R, Malmevik J, Fasching L, Akerblom M, Jakobsson J (2014) miRNAs in brain development. Exp Cell Res 321: 84–89 [DOI] [PubMed] [Google Scholar]
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G (2012) Advantages and limitations of different p62‐based assays for estimating autophagic activity in Drosophila . PLoS ONE 7: e44214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG (2008) A feedback loop comprising lin‐28 and let‐7 controls pre‐let‐7 maturation during neural stem‐cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Sachdeva R, Jonsson ME, Nelander J, Kirkeby A, Guibentif C, Gentner B, Naldini L, Bjorklund A, Parmar M, Jakobsson J (2010) Tracking differentiating neural progenitors in pluripotent cultures using microRNA‐regulated lentiviral vectors. Proc Natl Acad Sci USA 107: 11602–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A, Danial M, Blelloch RH (2015) Let‐7 and miR‐125 cooperate to prime progenitors for astrogliogenesis. EMBO J 34: 1180–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S, Mandairon N, Kermen F, Garcia S, Sacquet J, Didier A (2010) Learning‐dependent neurogenesis in the olfactory bulb determines long‐term olfactory memory. FASEB J 24: 2355–2363 [DOI] [PubMed] [Google Scholar]
- Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM, Rubinsztein DC (2016) Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun 7: 10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdankhah M, Farioli‐Vecchioli S, Tonchev AB, Stoykova A, Cecconi F (2014) The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis 5: e1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla‐Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol 452: 143–164 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee‐Messer C, Dolmetsch RE, Tsien RW, Crabtree GR (2011) MicroRNA‐mediated conversion of human fibroblasts to neurons. Nature 476: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y (2009) A feedback regulatory loop involving microRNA‐9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y (2010) MicroRNA let‐7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA 107: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh‐Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M , Altshuler D, Daley GQ (2011) The Lin28/let‐7 axis regulates glucose metabolism. Cell 147: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo . Nat Biotechnol 15: 871–875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File


