Abstract
The screening of a metagenomic library of 250,000 clones generated from a hypersaline soil (Spain) allowed us to identify a single positive clone which confers the ability to degrade N-acyl homoserine lactones (AHLs). The sequencing of the fosmid revealed a 42,318 bp environmental insert characterized by 46 ORFs. The subcloning of these ORFs demonstrated that a single gene (hqiA) allowed AHL degradation. Enzymatic analysis using purified HqiA and HPLC/MS revealed that this protein has lactonase activity on a broad range of AHLs. The introduction of hqiA in the plant pathogen Pectobacterium carotovorum efficiently interfered with both the synthesis of AHLs and quorum-sensing regulated functions, such as swarming motility and the production of maceration enzymes. Bioinformatic analyses highlighted that HqiA showed no sequence homology with the known prototypic AHL lactonases or acylases, thus expanding the AHL-degrading enzymes with a new family related to the cysteine hydrolase (CHase) group. The complete sequence analysis of the fosmid showed that 31 ORFs out of the 46 identified were related to Deltaproteobacteria, whilst many intercalated ORFs presented high homology with other taxa. In this sense, hqiA appeared to be assigned to the Hyphomonas genus (Alphaproteobacteria), suggesting that horizontal gene transfer had occurred.
Introduction
Many microorganisms use a genetic regulatory mechanism to monitor their cell density and to establish a coordinated behaviour1–3. This regulation mechanism, initially identified in bacteria, has been termed quorum sensing (QS). It is based on the production of diffusible signal molecules (also called autoinducers) by bacteria, which accumulate in their surrounding environment and once they reach a threshold concentration allow the induction or repression of numerous genes2. To date, different types of signal molecules have been identified, but the most correspond to the class of the N-acylhomoserine lactones (AHLs)4, 5. AHL-based communication systems have been studied extensively and identified as playing a crucial role in the regulation of several functions in terrestrial and aquatic environments. Indeed, the production of exoenzymes in Pectobacterium carotovorum, motility of Aeromonas hydrophila, luminescence of Aliivibrio fischeri, antibiotic biosynthesis by Pseudomonas aeruginosa and conjugative transfer of plasmid Ti in Agrobacterium tumefaciens all involve an AHL-based regulation mechanism3, 6, 7. Nevertheless, little information is known about the QS systems of halophilic bacteria and even less on hitherto uncultivated microorganisms. A few studies based on a non-cultivation metagenomic approach have been carried out and new LuxR/LuxI-type QS systems have been identified from uncultivated microorganisms in activated sludge and forest soils8, 9. Regarding extreme environments, such as hypersaline soils, QS systems have only been described in cultivable bacteria belonging to the Halomonadaceae family10, 11. In this context, our understanding of QS signal communication between bacteria living in such extreme environments remains limited.
Besides the ability to produce and use AHL-based communication systems, the ability to inhibit QS-regulated functions or to degrade AHLs was also reported for very different organisms5. Such abilities have been described for bacteria, fungi, plants and animals, suggesting that all these organisms have developed various mechanisms to interfere with or disrupt these cellular communication systems5, 12–16. One of the first mechanisms historically described is related to the production of chemical compounds (quorum sensing inhibitors or QSI), acting as antagonists and interfering with the transcriptional regulator structure17–19. Another mechanism that strongly perturbs or even abolishes QS-regulated functions is related to the production of enzymes capable of degrading the AHL-signal molecules, known as quorum quenching (QQ)5. To date, 3 different groups of enzymes have been identified according to the enzymatic mechanism involved: the AHL lactonases (lactone hydrolysis), the AHL acylases (amide hydrolysis) and the AHL oxidases/oxidoreductases (oxidoreduction)16, 20–22. Indeed, very different AHL lactonase types and AHL acylase types have been identified, suggesting that convergent evolution events occurred and that other unrelated types may exist in the environment5. To date, these different types of AHL-degrading enzymes have been identified in very different lineages of bacteria, such as Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria, by using cultivation-dependent approaches and for some of them the related genes have been characterized22–27. Only some of the known genes encoding AHL-degrading enzymes have been characterized using a cultivation-independent approach based on the construction and screening of metagenomic libraries constructed from soil and marine environments28, 29. The construction of such libraries allows the screening of DNA from cultivated and hitherto uncultivated organisms from different environments, thus increasing the possibility to identify new genes of interest. Such an approach gives the opportunity to develop interfering processes based on the inhibition of AHL perception or AHL degradation, and it opens up new perspectives in industrial, medical and agronomic fields as QQ represents an environmentally friendly alternative for the use of chemicals and antibiotics. Notably, QQ has, among its main biotechnological applications, the ability to control biofouling and the regulation of bacterial pathogens.
In this study, we have constructed and screened a metagenomic library from a hypersaline soil to identify potential genes involved in QS or in QQ. To the best of our knowledge, AHL-producing halophilic bacteria have so far only been identified in hypersaline soils by cultivation-dependent methods and no AHL-degrading bacteria have been reported in these extremophilic environments10, 11. By using a fast screening method based on pooled clones rather than on the screening of individual clones, we were able to screen a fosmid library of 250,000 clones of E. coli for the ability to produce or degrade AHLs. This screening allowed the identification of one clone capable of degrading AHLs. Among the 42 kb of the fosmid insert, one ORF (hqiA) was identified and biochemically characterized by HPLC/MS as coding for an AHL lactonase. The analysis of the protein sequence revealed no homology with the well-known AHL-degrading enzymes, showing that HqiA represents a new type of enzyme in the AHL lactonase family. A detailed analysis revealed that it clustered into the cysteine hydrolase (CSHase) group and showed strong homology with isochorismatase-like and N-carbamoylsarcosine amidase-like enzymes. Moreover, characterization of the neighbouring ORFs allowed us to hypothesize the phylogenetic belonging of the fosmid DNA insert and to suggest that hqiA was inherited from a horizontal transfer.
Results
Screening of AHL-degrading genes from a metagenomic library
Environmental DNA extracted from a hypersaline soil located in the Finca La Salina (Rambla Salada, Murcia, Spain) was used to construct a metagenomic library. This library consists of 250,000 metagenomic fosmid clones each containing a DNA fragment of ~40 kb as determined by enzymatic restriction on randomly sub-sampled clones. The screening of the metagenomic library was done using a pool strategy, which consisted of the conditioning of the metagenomic library to obtain a theoretical number of 50 clones per well (4,800 clones per 96-well microplate). Using this strategy, a screening based on the appropriate AHL biosensor was done to identify AHL-producing or AHL-degrading clones. After a two-day incubation without AHL addition, none of the wells of the 52 microplates screened was positive for AHL production. On the contrary, after a two-day incubation with C6-HSL, one well did not activate the Chromobacterium violaceum CV026 biosensor. To identify the clone(s) involved, serial dilutions were made from the positive well to obtain individual colonies. The posterior screening of the QQ activity of these colonies allowed the identification of 17 positive clones, thus conferring to Escherichia coli the ability to degrade C6-HSL. The enzymatic restriction analyses revealed that the different clones recovered corresponded to a unique fosmid, which we named f10/17.1H (data not shown). This fosmid was purified and transferred into E. coli S17 λ pir to carry out additional AHL-degradation assays. These assays revealed that this fosmid conferred the ability to degrade a broad range of unsubstituted, oxo- and hydroxyl-substituted AHLs to E. coli (Fig. 1). To identify whether the phenotype observed was related to degradation or inhibition, TLC and HPLC/MS analyses were carried out. Based on AHL-degradation assays performed with short- and long-chain AHLs (C6-HSL and C12-HSL), both TLC and HPLC/MS analyses revealed that the activity corresponded to AHL degradation and not to an inhibition of the biosensor (Fig. 2). Notably, the HPLC/MS analyses confirmed that E. coli S17 λ pir carrying the fosmid f10/17.1H significantly degraded AHLs compared to the control E. coli S17 λ pir (P < 0.0001). Comparison of the C6-HSL and C12-HSL degradation assays also revealed that after 24 h of incubation, a significantly higher efficacy of degradation was obtained for C12-HSL (Fig. 2B, P < 0.0001).
Figure 1.
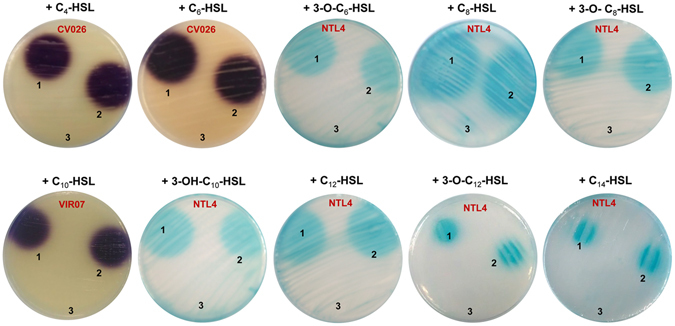
Diffusion agar-plate assay to detect AHL degradation using the biosensors Chromobacterium violaceum CV026, C. violaceum VIR07 and Agrobacterium tumefaciens NTL4 (pZLR4). Cell-free LB medium (1), E. coli S17 λ pir (2) and E. coli S17 λ pir harbouring the fosmid f10/17.1H (3) were supplemented with the same quantities of C4-HSL, C6-HSL, 3-O-C6-HSL, C8-HSL, 3-O-C8-HSL, C10-HSL, 3-OH-C10-HSL, C12-HSL, 3-O-C12-HSL and C14-HSL.
Figure 2.
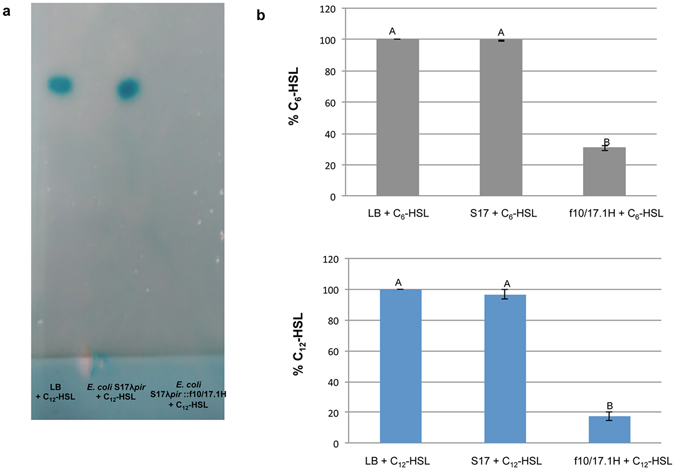
Confirmation of AHL-degradation activity. (a) TLC analysis using the biosensor Agrobacterium tumefaciens NTL4 (pZLR4). (b) HPLC/MS measurements of remaining C6-HSL and C12-HSL after 24 h incubation with E. coli S17 λ pir and E. coli S17 λ pir::f10/17.1H. LB medium was used as control. Initial AHL concentration was 25 µM. Error bars represent standard deviations. Different letters above the bars indicate that the values are significantly different according to a one-factor (treatment) ANOVA (P < 0.05) and the Tukey test.
In silico analysis of the f10/17.1H fosmid DNA sequence
The in silico analysis of the 42,318 bp environmental DNA revealed that it contained 46 open reading frames (ORF) in both orientations (Fig. 3). The analysis of the G + C content with GC-Profile highlighted a global G + C of 45.6 mol % (100 bp window), with three segments (1–8,963 bp: 49 mol %; 8,926–21,987 bp: 37.7% and 21,958–42,318 bp: 49.1 mol %) (Fig. 3). Besides these variations of the G + C content, a detailed analysis of the sequence revealed that the codon usage of most of the amino acids detected was equally distributed among their different forms.
Figure 3.
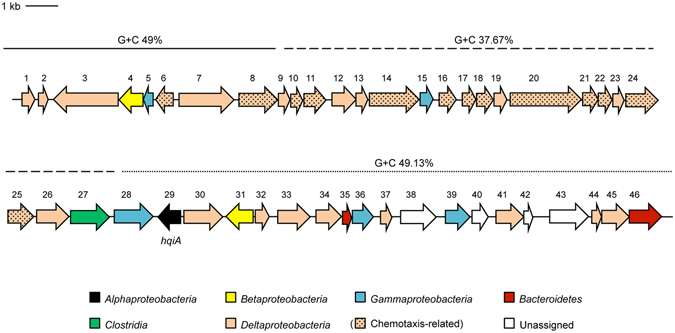
Genetic map of the DNA insert of the fosmid f10/17-1H. The 46 ORFs are numbered and coloured according to their phylogeny, ORF29 being (in black) hqiA. The chemotaxis related genes are also presented as square dots.
Comparison of the deduced protein sequences against the protein sequences available in the GenBank sequence database revealed that 31 out of 46 ORFs showed a high sequence homology with the class Deltaproteobacteria and among them, 25 ORFs equally distributed in the fosmid insert were related to sulphate-reducing bacteria (SRB) proteins (Table 1). The most represented genera related to the class Deltaproteobacteria were assigned to Desulfobacula (5 ORFs), Desulfatibacillum (3 ORFs), Desulfobacterium (4 ORFs), Desulfosarcina (2 ORFs), Desulfomonile (2 ORFs), Desulforegula (2 ORFs) and Desulfocapsa (2 ORFs). The other proteins identified on the fosmid were affiliated to Gammaproteobacteria (5 ORFs), Betaproteobacteria (2 ORFs), Alphaproteobacteria (1 ORF), Clostridia (1 ORF) and Bacteroidetes (2 ORFs) (Fig. 3). These affiliations were confirmed by phylogenetic analyses of the representative proteins of each genus based on maximum likelihood (phyML) (Fig. 4a). This approach confirmed the results obtained by BLAST, showing that the fosmid insert originated mainly from a Deltaproteobacteria genomic background. However, several ORFs (4, 5, 15, 27, 28, 29, 31, 35, 36, 38, 39, 40, 42, 43 and 46) related to other lineages, such as Clostridia, Bacteroidetes, Alpha-, Beta- or Gammaproteobacteria, were inserted in different locations between the Deltaproteobacteria related ORFs, suggesting that they were inherited by horizontal gene transfer.
Table 1.
Phylogenetic analysis of each ORF in the fosmid f10/17.1H.
| ORF | Length | Hypothetical function | Organism | Phylum/class |
|---|---|---|---|---|
| 1 | 416 | Hypothetical protein | Uncultured Deltaproteobacteria | Deltaproteobacteria |
| 2 | 326 | Thioredoxin | Uncultured Deltaproteobacteria | Deltaproteobacteria |
| 3 | 2072 | Two component system sensor histidine kinase, hybrid | Desulfobacula toluolica | Deltaproteobacteria |
| 4 | 737 | Hypothetical protein | Uliginosibacterium gangwonense | Betaproteobacteria |
| 5 | 311 | Nickel ABC transporter ATP-binding protein | Enterobacter lignolyticus | Gammaproteobacteria |
| 6 | 533 | Chemotaxis protein CheW | Desulfatibacillum alkenivorans | Deltaproteobacteria |
| 7 | 1730 | Fis family transcriptional regulator | Desulfobacter postgatei | Deltaproteobacteria |
| 8 | 1199 | Methyl-accepting chemotaxis sensory transducer | Desulfobacula toluolica | Deltaproteobacteria |
| 9 | 359 | Hypothetical protein | Desulfovibrio alaskensis | Deltaproteobacteria |
| 10 | 419 | Gliding motility protein | Desulfosarcina sp. | Deltaproteobacteria |
| 11 | 680 | CheY-like receiver, AAA-type ATPase and DNA-binding domain | Desulfocapsa sulfexigens | Deltaproteobacteria |
| 12 | 743 | Hypothetical protein | Desulfosarcina sp. | Deltaproteobacteria |
| 13 | 371 | Fis family transcriptional regulator | Desulfonatronospira thiodismutans | Deltaproteobacteria |
| 14 | 1541 | Chemotaxis protein CheA | Desulfonauticus sp. | Deltaproteobacteria |
| 15 | 422 | Fis family transcriptional regulator | Pseudomonas fuscovaginae | Gammaproteobacteria |
| 16 | 524 | CheY-like receiver, AAA-type ATPase and DNA-binding domain | Desulfocapsa sulfexigens | Deltaproteobacteria |
| 17 | 416 | Chemotaxis protein histidine kinase-like kinase | Lawsonia intracellularis | Deltaproteobacteria |
| 18 | 512 | Chemoreceptor glutamine deamidase CheD | Desulfobacula toluolica | Deltaproteobacteria |
| 19 | 398 | Histidine kinase | Desulforegula conservatrix | Deltaproteobacteria |
| 20 | 2216 | Chemotaxis protein A CheA | Desulfobacula toluolica | Deltaproteobacteria |
| 21 | 482 | Chemotaxis protein CheD | Desulforegula conservatrix | Deltaproteobacteria |
| 22 | 428 | Chemotaxis protein CheY | Uncultured Desulfobacterium sp. | Deltaproteobacteria |
| 23 | 353 | Hypothetical protein | Desulfobacterium autotrophicum | Deltaproteobacteria |
| 24 | 1046 | Chemotaxis response regulator protein-glutamate methylesterase | Uncultured Desulfobacterium sp. | Deltaproteobacteria |
| 25 | 824 | CheR-type MCP methyltransferase | Desulfatibacillum alkenivorans | Deltaproteobacteria |
| 26 | 1046 | L-threonine 3-dehydrogenase | Desulfomonile tiedjei | Deltaproteobacteria |
| 27 | 1184 | 2-amino-3-ketobutyrate coenzyme A ligase | Clostridium termitidis | Clostridia |
| 28 | 1232 | NhaP-type Na+ (K+)/H+ antiporter | Thioflavicoccus mobilis | Gammaproteobacteria |
| 29 | 725 | Isochorismatase | Hyphomonas jannaschiana | Alphaproteobacteria |
| 30 | 1271 | 4Fe-4S ferredoxin | Desulfomonile tiedjei | Deltaproteobacteria |
| 31 | 839 | GTP cyclohydrolase | Pandoraea sp. | Betaproteobacteria |
| 32 | 428 | Hypothetical protein | Uncultured Deltaproteobacteria | Deltaproteobacteria |
| 33 | 1019 | Phospholipid/glycerol acyltransferase | Desulfatibacillum alkenivorans | Deltaproteobacteria |
| 34 | 785 | Conserved hypothetical membrane protein | Uncultured Deltaproteobacteria | Deltaproteobacteria |
| 35 | 275 | Hypothetical protein | Polaribacter sp. | Bacteroidetes |
| 36 | 647 | Heme-binding protein | Methylomonas methanica | Gammaproteobacteria |
| 37 | 347 | Hypothetical protein | Uncultured Deltaproteobacteria | Deltaproteobacteria |
| 38 | 1124 | Uncharacterized ATPase | Uncultured bacteria | Unassigned |
| 39 | 761 | Hypothetical protein | Pseudoalteromonas luteoviolacea | Gammaproteobacteria |
| 40 | 506 | Hypothetical protein | Uncultured bacteria | Unassigned |
| 41 | 866 | Hypothetical protein | Thermodesulfobium narugense | Deltaproteobacteria |
| 42 | 245 | Unknown | — | Unassigned |
| 43 | 1127 | TonB-dependent siderophore receptor | — | Unassigned |
| 44 | 302 | Hypothetical protein | Desulfobacula sp. | Deltaproteobacteria |
| 45 | 845 | Unknown | Uncultured Desulfobacterium sp. | Deltaproteobacteria |
| 46 | 989 | Subtilisin-like serine protease | Ignavibacterium album | Bacteroidetes |
The table presents the ORF number, the length in bp, the hypothetical function and the phylogenetic affiliation at both genus and phylum/class levels.
Figure 4.
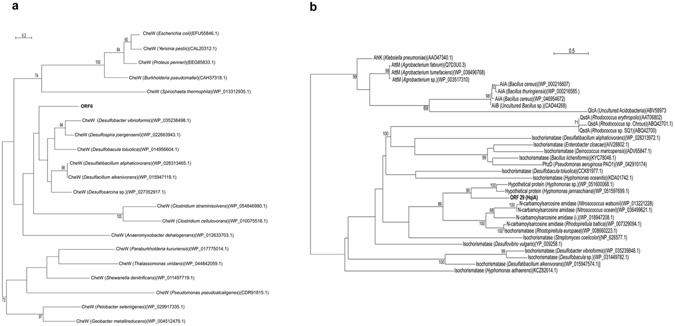
Phylogenetic analysis of the ORFs found in fosmid f10/17.1H based on protein sequences. (a) ORF6 as an example of affiliation of ORFs by phylogenetic analysis of representative proteins of each genus identified based on maximum likelihood (phyML). (b) Phylogenetic analysis of HqiA showed that it is not related to any known AHL lactonases, otherwise clustering in the cysteine hydrolase group with other hypothetical (HYPO), isochorimatase-like (ISO) and N-carbamoylsarcosine amidase (NCAR)-like enzymes.
In term of function, a detailed analysis of the fosmid sequence revealed a cluster of ORFs homologous to genes encoding chemotactic signalling systems. Indeed, from ORF6 to ORF25 up to 13 genes related to chemotaxis and motility are represented, such as homologues of CheA (cytoplasmic histidine kinase), CheD (receptor-kinase coupling protein), CheR (methyltransferase), CheW (receptor-kinase coupling protein) and CheY (response regulator). The functionality of these genes was analysed using E. coli RP437 and two mutants (∆cheA or ∆cheW) unable to swarm. No complementation of the swarming phenotype was obtained after transformation of the mutants with f10/17.1H compared to the wild-type strain (data not shown). From ORF26 to ORF31, five ORFs presented homologies with putative enzymes, such as L-threonine-3-dehydrogenase (ORF26), 2-amino-3-ketobutyrate coenzyme A ligase (ORF27), isochorismatase (ORF29), a GTP cyclohydrolase (ORF31) and a phospholipid acyltransferase (ORF33). From ORF32 to 46, most of the ORFs were assigned as hypothetical proteins, except some ORFs such as ORF36 and ORF43 presenting homologies with proteins related to iron access. Notably, no homologues of the known AHL-degrading enzymes such as lactonases (AttM, AiiA, QlcA, QsdA) or acylases (QuiP, PvdQ) have been detected among the 46 ORFs carried by the fosmid (Table 1 and Fig. 3).
Demonstration of the AHL degradation role conferred by the ORF29
A first step consisting in the cloning in pGEM-T of 5-kb fragments of the fosmid DNA insert was conducted to determine which region conferred AHL-degradation ability to E. coli. This first screening allowed the identification of a fragment containing from ORF26 to ORF31. Based on the sequence homology and the predicted enzymatic reaction associated to the ORF26, ORF27, ORF29, and ORF31, we selected and designed specific primers to clone the ORF29, which was predicted to encode an isochorismatase (237 aa). The isochorismatase corresponds to a large family of enzymes related to the cysteine hydrolases (CSHases) capable of cleaving ester and ether bonds. The ORF29 was first cloned in pGEM-T and then transferred to the low copy number plasmid pME6010 under the constitutive promoter pK. The AHL-degradation assays performed with pGEM-T or pME6010-based constructions revealed that ORF29 (named as hqiA gene) conferred to E. coli the ability to degrade all the AHLs tested (data not shown).
HqiA: a new AHL-degradation enzyme type
A detailed analysis of the amino acid sequence highlighted that HqiA has no significant homology with the known AHL lactonase or acylase enzyme types. Indeed, HqiA appeared not to be related at all to the known AHL acylases (i.e., PvdQ, QuiP) (data not shown) and very distant from the known AHL lactonases (i.e., AiiA, AttM, QlcA or QsdA) (Fig. 4b). It also confirmed that the HqiA protein sequence was more related to that of the Hyphomonas genus than to those of other Deltaproteobacteria genera, although less related homologues can be found in the genome of sequenced strains of Desulfobacterium, Desulfosarcina and Desulfocapsa. This phylogenetic analysis also highlighted that HqiA clustered into the CSHase group with other α/β-hydrolase enzymes, and more especially within isochorimatase-like and N-carbamoylsarcosine amidase-like enzymes. Moreover, HqiA shows the conserved catalytic domains (D, K and C) of the CSHase family and presents a very different three-dimensional structure than the other AHL lactonases based on a Phyre2 analysis30 (see Supplementary Fig. S1).
hqiA encodes for an AHL lactonase
To identify the function of the HqiA protein, hqiA was sub-cloned in the expression plasmid pMAL-c2TEV under the control of a Lac promotor to generate the fusion protein MBP-HqiA. The TEV protease-processed recombinant protein revealed a molecular weight of about 26 kD (see Supplementary Fig. S2). AHL-degrading activity of hqiA was confirmed against all the AHLs using a crude extract of induced E. coli carrying pMAL-c2TEV-ORF29, whilst no AHL degradation occurred with E. coli carrying the empty vector (data not shown). Furthermore, as revealed with biosensors, the degradation assays performed with the purified recombinant HqiA protein confirmed the AHL-degradation activity associated with the HqiA peptide (Fig. 5). To assign the enzymatic activity of the HqiA protein, the product of the enzymatic reaction between the purified recombinant protein and C12-HSL was identified by using both the lactone ring closure assay and HPLC/MS. Thus, after acidification of the supernatant with HCl and neutralisation of the pH, the C12-HSL-related signal was significantly recovered as evidenced using the A. tumefaciens biosensor (see Supplementary Fig. S3). Similar results were obtained with other AHLs (data not shown). The analysis by HPLC/MS of the main peak obtained after the C12-HSL degradation assay (HqiA) revealed a mass (M-) of 300, which corresponds to the open-ring form of the C12-HSL, while in the chromatogram corresponding to the control (RE) a molecular weight of 282, compatible with the closed form, was detected (Fig. 5).
Figure 5.
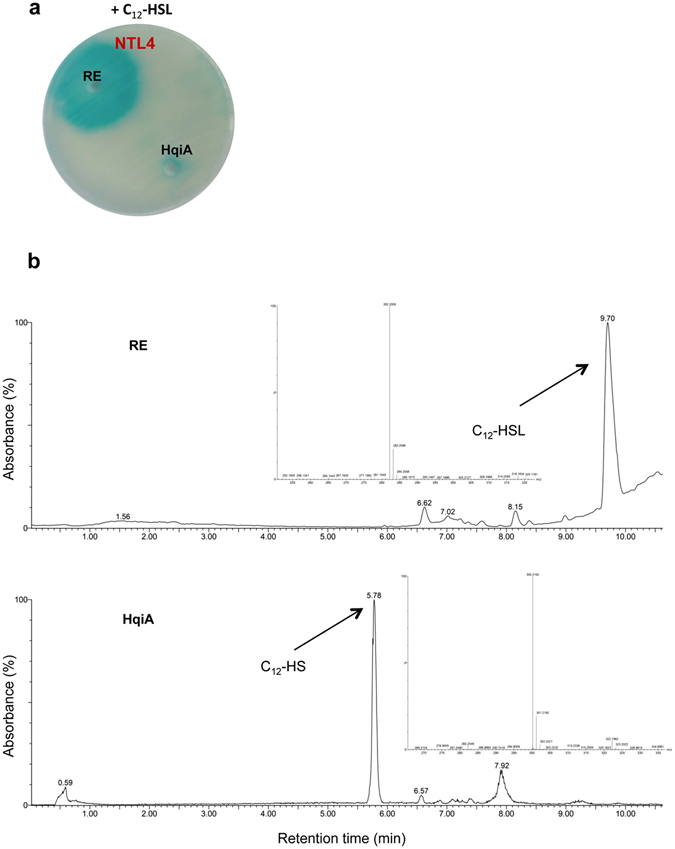
Functional characterization of purified HqiA. (a) Detection of the quorum-quenching activity against C12-HSL of the purified HqiA protein and the negative control RE (reaction buffer) using the biosensor Agrobacterium tumefaciens NTL4 (pZLR4). (b) HPLC/MS analysis of the C12-HSL degradation by HqiA. RE and HqiA sample chromatograms are shown. In the RE and HqiA samples, the main peaks masses are 282 and 300 which corresponds to the closed and open-ring form of the C12-HSL respectively, therefore indicating a lactonase activity.
Quorum-quenching activity of HqiA against virulence of Pectobacterium carotovorum CECT 225T
In the plant pathogen Pectobacterium carotovorum subsp. carotovorum CECT 225T virulence is related to maceration enzymes, such as protease or pectinase, swarming and swimming, among others, which are all regulated in a QS-dependent way and mediated by C6-HSL and 3-O-C6-HSL31–33. When the plasmid pME6010::hqiA was introduced into the pathogenic strain, the concentration of AHLs in the culture supernatant strongly decreased compared to the control performed with the empty plasmid. AHLs were no longer detected with C. violaceum CV026 and A. tumefaciens NTL4 (pZLR4) biosensors for the strain P. carotovorum carrying pME6010::hqiA, while they were clearly detected in the control conditions (Fig. 6a). The QQ ability of hqiA on P. carotovorum was then tested using the potato tuber assay. This bioassay highlighted that P. carotovorum carrying pME6010::hqiA was not able to cause soft rot symptoms (0% maceration on the surface of the potato slice), whilst the same bacterial strain carrying or not the empty plasmid caused a large maceration zone (39%) (Fig. 7). To test the impact of hqiA on the different QS regulated functions of P. carotovorum, cultures of P. carotovorum (pME6010::hqiA) and P. carotovorum carrying or not the empty plasmid (pME6010) were screened using different bioassays. This screening showed that compared to the bacterial strain carrying or not the empty plasmid, the introduction of hqiA into P. carotovorum abolished its pectinase, protease, DNase and alkaline phosphatase activities in our experimental conditions. In addition, swarming and swimming motilities were also blocked (Fig. 6b, see Supplementary Fig. S4). In all the bacterial cultures containing the plasmid with or without hqiA, cell counts were significantly similar showing that the expression of hqiA did not affect the growth of P. carotovorum.
Figure 6.
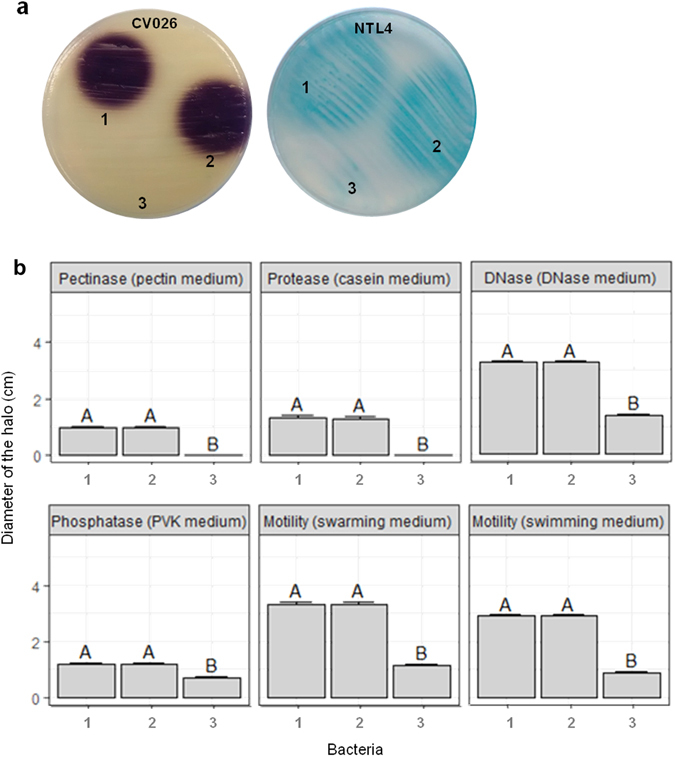
Quorum quenching of AHLs and QS-regulated functions in Pectobacterium carotovorum. (a) AHL detection using the biosensors Chromobacterium violaceum CV026 and Agrobacterium tumefaciens NTL4 (pZRL4). (b) Phenotypes tested in pectin, casein, DNase, PVK (alkaline phosphatase), swarming and swimming media. Error bars represent standard deviations. For each assay, the P. carotovorum strain without plasmid (1), with the empty plasmid (pME6010) (2) and the plasmid carrying hqiA (3) were compared. For the bioassays presented in panel b, different letters above the bars indicate that the values are significantly different according to a one-factor (treatment) ANOVA (P < 0.05) and the Tukey test.
Figure 7.
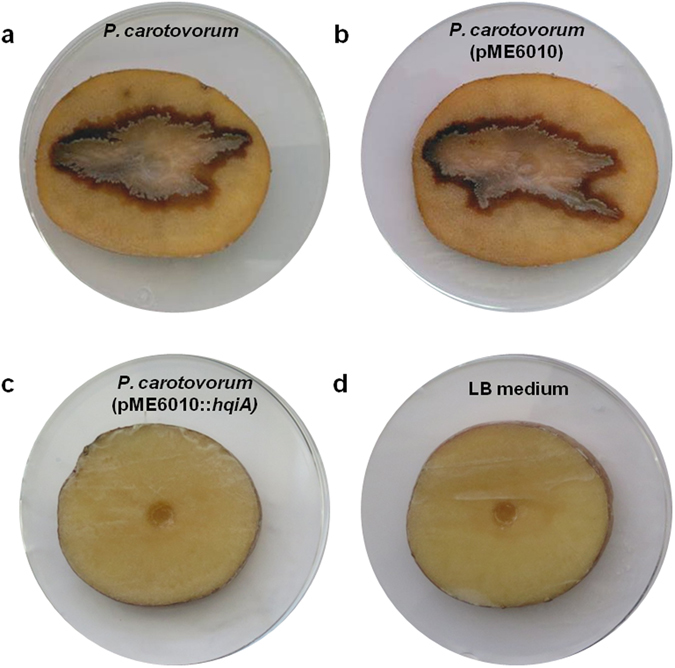
Virulence assay on potato tubers. Potato tuber slices inoculated with (a) Pectobacterium carotovorum without plasmid, (b) P. carotovorum (pME6010), (c) P. carotovorum expressing the hqiA gene and (d) cell-free LB medium.
Discussion
During the last decades, QS and QQ have been described in many bacteria inhabiting different aquatic and terrestrial environments4, 34. On the other hand, studies on QS signal communication in extremophilic bacteria are very scarce, and especially those focusing on halophilic microorganisms35, 36. The first demonstration of the existence of QS signal communication systems in bacteria inhabiting hypersaline environments was done by Llamas et al.10. They also revealed that the ability to produce AHLs was conserved in the family Halomonadaceae, one of the most frequently detected halophilic bacteria in such an environment11. These results suggest that QS signal communication systems are widespread among halophilic bacteria and may participate in their fitness in extreme environments, but they give no information about the other uncultivated bacterial taxa. Until very recently, most of our knowledge on QS and QQ relied only upon cultivation-based approaches, ignoring a large part of the microbial diversity and the functional potential of the bacterial communities occurring in complex and extreme environments37, 38. The first cultivation-dependent study focusing on the relative distribution of the bacteria from the soil capable of producing or degrading AHLs showed that 8–19% of these bacteria had the capability to produce AHLs, whilst 2–3% had the capability to degrade AHLs39. The proportion of potential AHL-producing bacteria was shown to reach 13% when considering the proteobacterial genomes available in the international sequence database NCBI, but these genes appeared far less frequently in non-proteobacterial genomes40. The metagenomic approaches based on shotgun sequencing or fosmid libraries gave very different pictures. Indeed, these approaches revealed lower frequencies but higher diversity of AHL-producing or degrading bacteria in the different environments analysed29, 41, 42. Considering only QQ homologues in various aquatic and terrestrial metagenomes, Romero et al.29 showed that the frequency of QQ homologues varied from 0.05 to 0.4 homolog per Mbp. However, QS and QQ genes are known to conserve a low sequence homology between them, consequently many homologues identified in silico in genomic and metagenomic data are non-functional29, 43. In this context, the use of fosmid libraries can be a good alternative as it permits the testing of clones for their ability to degrade or produce AHLs. New AHL-producing or degrading genes have been identified by this method9, 28, 42, 44, 45, but the frequency of obtaining positive clones remains low and varied from 4.82 × 10−4 to 3.3 × 10−3 per Mbp9, 42. In our metagenomic library generated from a hypersaline soil, a single clone was obtained after the screening of 250,000 clones, corresponding to an estimated frequency of 1 × 10−4 positive clone per Mbp. On the contrary, no genes related to the production of QS signal molecules have been detected. As AHL-producing bacteria have been evidenced in the same experimental site10, 11, we cannot exclude biased and insufficient expression of E. coli of some genes in fosmid-based metagenomic libraries46 or limited sensitivity of the screening strategy used.
The chemical characterization of the activity encoded by the hqiA gene by HPLC/MS highlighted that it encoded a lactonase activity characterized by a broad range of degradation of unsubstituted, oxo- and hydroxyl-substituted AHLs. To date, various types of prototypic AHL lactonases have been evidenced, such as the AiiA, AiiB, AidH, AttM, AhlK, QsdA or QlcA22, 23, 42, 47–51. These enzymes mostly identified by cultivation-dependent approaches can be clustered in two distinct groups: i) the Zn-metallohydrolases encompassing AiiA-like and phosphotriesterase (PTE)-like enzymes and ii) the α/β-hydrolases with only one characterized member (AidH). These enzyme types present specific functional motifs, such as HXDH-H-D for AiiA, AiiB, AttM, AhlK and QlcA, G-VL-HEH for QsdA or G-S-GG for AidH that permit their in silico identification. Notably, the only AiiA-like enzyme obtained by a fosmid screening (QlcA) was proposed as a new prototypic AHL lactonase based on phylogenetic analyses42. In our study, HqiA does not contain the structural motives typically found in the other known lactonases, such as AiiA, AttM, AhlK, QlcA, QsdA or AidH, suggesting that it represents a new family of AHL lactonases not related to the Zn-metallohydrolase (AiiA-like and PTE-like) enzymes. Moreover, a comparison of the three-dimensional structure of the different AHL lactonases and of HqiA confirmed that they did not show a similar structure. Notably, an in silico analysis revealed that HqiA presents strong homology with other types of enzymes found in the α/β-hydrolase enzyme family but not related to AidH. Indeed, HqiA appeared to present homology with enzymes grouped in the cysteine hydrolase (CHase) group. Members of this enzyme group have been shown to cleave ether, ester and amide bonds. The higher sequence homologies of HqiA were obtained with isochorismatase-like and N-carbamoylsarcosine amidase-like enzymes. Diverse biological functions have been assigned to bacterial isochorismatases, but never related to AHL degradation. As an example, HqiA presents a strong homology with PhzD, which is described as an isochorismatase involved in the phenazine biosynthesis in Pseudomonas aeruginosa PAO152. Interestingly, chemical assays identified both AHL-acylase and AHL-lactonase activities in P. aeruginosa. To date, three AHL acylase genes have been identified in P. aeruginosa PAO1, but a lactonase gene has not yet been identified34, 53–55. Even if sequence homology is not always a proof of function, our future aim is to conduct the functional characterization of PhzD, which would perhaps expand the list of QQ enzymes in P. aeruginosa PAO1 with an AHL lactonase as well as it would help to better understand the regulation of QS in this pathogenic bacterial strain.
Regarding the taxonomic origin of the environmental DNA cloned in fosmid f10/17.1H, in silico analysis (BLAST search and phylogenetic analyses) revealed that 31 out of the 46 ORFs showed high sequence homology with Deltaproteobacteria, suggesting that the environmental DNA might originate from this bacterial class. Additionally, 25 out of these 31 ORFs were related to sulphate-reducing bacteria (SRB), a group of microorganisms ubiquitous in hypersaline environments, where sulphate is a major inorganic compound56, 57. Interestingly, a large portion of the Deltaproteobacterial ORFs (n = 13) presented strong homology with chemotaxis and motility systems. These genes probably provide a competitive advantage to bacteria in the colonization of hypersaline soil. The organization of che genes in large clusters has been evidenced in other bacterial taxa, such as in Sinorhizobium, Rhizobium and Azospirillum 58, 59. Interestingly, chemotaxis was shown to be regulated by an AHL-based quorum sensing system in Sinorhizobium meliloti, but in our case we do not have evidence of such a link60. Besides the ORFs belonging to Deltaproteobacteria, our phylogenetic analysis revealed that many ORFs assigned to other phylogenetic groups (Alpha-, Beta- and Gammaproteobacteria, Clostridia and Bacteroidetes) were inserted between Deltaproteobacterial ORFs. Such gene organization without notable change of G + C% and the absence of evident mobile elements suggest that horizontal gene transfer (HGT) occurred. Horizontal gene transfer is described as a universal mechanism used by bacteria to acquire new functions that allow them to adapt to environments with different selective pressures. The presence of HGT events has already been reported in other hypersaline environments, suggesting that they also occur in the hypersaline soil of Rambla Salada61–63. The ORF29 (hqiA) encoding the AHL lactonase-like enzyme was assigned as the unique Alphaproteobacterial ORF of the cloned DNA, with a strong homology with homologous genes annotated as isochorismatases in the genomes of bacterial strains belonging to the Hyphomonas genus. Notably, homologues are also present as single copy in the genomes of Desulfobacula toluolica and D. phenolica but without evidence of HGT. In all the cases, the biological function of these other homologous genes has not yet been experimentally tested. However, the presence of inherited genes by HGT in the Deltaproteobacteria context suggests that they may confer a selective advantage. Recent observations extend the role of QQ enzymes to the detoxification of signalling molecules. Interestingly, an in silico analysis of the genome of a strongly related Deltaproteobacteria strain (Desulfatibacillum alkenivorans AK-01) revealed the presence of an annotated aiiA-like gene presenting high homology only with Firmicutes members. The identification of this other potential case of horizontal transfer of an AHL lactonase gene suggests that HGT events occur regularly between Deltaproteobacteria and other bacterial taxa and that QQ genes may be exchanged.
Although the real physiological role of the AHL-degrading enzymes remains to be determined34, such enzymes appear as a valuable alternative to chemicals and antibiotics used to treat diseases. Our QQ experiments showed that HqiA not only significantly degraded a wide range of AHLs, but that also interfered with the production of AHL molecules in the plant pathogenic bacterium P. carotovorum subsp. carotovorum. Moreover, we revealed that the expression of several QS-regulated phenotypes, such as maceration enzymes or motility were inhibited or strongly perturbed, thus confirming previous findings22, 47, 64. The QQ activity of HqiA was also verified in the AHL-producing bacteria Agrobacterium fabrum C582, Halomonas anticariensis FP35T10 and Vibrio coralliilyticus VibC-Oc-193 (unpublished data). In each case, the introduction of a plasmid carrying hqiA allowed the degradation of the AHLs (data not shown). Although the QQ results obtained with HqiA in P. carotovorum are similar to those obtained using AiiA, AttM, AidH, QlcA or QsdA, they open up new perspectives in terms of diversity of enzymes22, 47, 65. Indeed, due to variations of codon usage or the problems of expression in other genomic backgrounds, the use of very different AHL-degrading enzyme types may be a good alternative to effectively quench bacterial pathogens.
To sum up, our study represents the first fosmid-based screening of a hypersaline soil metagenome to search for genes involved in QS and QQ. In this way, we were able to a new class of AHL-degradation-enzyme encoding gene (hqiA) not previously related to the known prototypes. Further studies will be necessary to demonstrate the biological function of HqiA, its substrate specificity and its conservation among bacterial genomes.
Experimental procedures
Sampling site
The soil sample considered in this study was collected in November 2007 in Finca La Salina (Rambla Salada, Murcia, Spain) (38°07′34.44″N, 1°07′11.13″W). This soil is considered as hypersaline and its physicochemical characteristics were previously determined66.
Bacterial strains, media and growth conditions
Escherichia coli strains were grown in Luria-Bertani (LB) medium. E. coli RP437, its related chemotaxis mutants [RP9335 (RP437∆cheA) and UU2871 (RP437∆cheW)] and their transformants were grown in T-broth67, 68, they were tested for their swarming ability on T-broth 0.2% (w/v) agar. Chromobacterium violaceum CV026 and VIR07 biosensor strains and Pectobacterium carotovorum subsp. carotovorum CECT 225T were grown in LB69, 70. Agrobacterium tumefaciens NTL4 (pZLR4) was cultured in AB medium supplemented with 80 μg ml−1 of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) when needed71. All strains were grown at 28 °C.
When required, antibiotics where used at the following final concentrations: chloramphenicol (Cm) 12.5 µg ml−1, ampicillin (Amp) 50 µg ml−1, kanamycin (Km) 50 µg ml−1, streptomycin (Sm) 100 µg ml−1, gentamicin (Gm) 50 µg ml−1 and tetracycline (Tc) 10 µg ml−1.
All the AHLs used (Sigma®) [C4-HSL (N-butyryl-DL-homoserine lactone), C6-HSL (N-hexanoyl-DL-homoserine lactone), 3-O-C6-HSL (N-3-oxo-hexanoyl-DL-homoserinelactone), C8-HSL (N-octanoyl-DL-homoserine lactone), 3-O-C8-HSL (N-3-oxo-octanoyl-DL-homoserinelactone), C10-HSL (N-decanoyl-DL-homoserine lactone), 3-OH-C10-HSL (N-3-hydroxydecanoyl-DL-homoserine lactone), C12-HSL (N-dodecanoyl-DL-homoserine-lactone), 3-O-C12-HSL (N-3-oxo-dodecanoyl-DL-homoserinelactone) and C14-HSL (N-tetradecanoyl-DL-homoserine lactone)] were used at final concentration of 25 µM.
DNA extraction and preparation of the metagenomic library
Total DNA was extracted from 10 g of soil sample using the MoBio PowerMax™ Soil DNA Isolation Kit (MoBio Laboratories) following the protocol provided by the manufacturer. To generate DNA compatible with the construction of the metagenomic library, DNA was sheared manually by pipetting several times and separated using a sucrose gradient. Briefly, 100 µg of genomic DNA were loaded in 17 ml tubes and separated according to their molecular size through a 5 to 40% (w/v) sucrose gradient. This separation was done at 15,000 × g at 10 °C for 20 h with an L-80 XP Optima ultracentrifuge (Beckman Coulter) associated to a swinging bucket rotor SW28.1 Ti (Beckman Coulter). After the ultracentrifugation, fractions of 200 µl were collected and the fractions of interest (~40 kb) were pooled and used for the construction of the metagenomic library using the CopyControl Fosmid Library Production Kit (pCC1FOS vector; Epicentre).
Screening and selection of clones with quorum-quenching activity
All the experiments were conducted at pH 7 to prevent the lactonolysis of the AHLs72. The metagenomic library was diluted in a total volume of 500 ml of LB Cm to obtain a final concentration of 250 clones per ml. 200 µl of this suspension were distributed per well in 96-well microtiter plates (~50 clones per well) and incubated for 24 h at 37 °C following the procedure described by Uroz and Oger73. After this incubation time, 100 µl of each well were transferred in new 96-well microtiter plates containing fresh LB medium supplemented with C6-HSL and incubated at 28 °C for 24 h. Then the C6-HSL degradation was tested in microtiters using the biosensor C. violaceum CV02674. LB cell-free medium supplemented with C6-HSL was used as a negative control. Positive wells without violacein production were retained as wells containing clones with potential QQ activity for further analysis and fosmids were purified using the Midi Prep Purification kit (Qiagen).
To determine the AHL-degradation activity of the selected fosmids, 500 µl of an overnight culture of E. coli S17 λ pir carrying the fosmid were incubated at 28 °C for 24 h in LB medium supplemented with 25 µM of each of the synthetic AHLs. LB cell-free medium and E. coli S17 λ pir supplemented with the same concentration of AHLs were incubated at 28 °C for 24 h as negative controls. The remaining AHLs in each overnight culture were detected on AB or LB agar plates overlaid with the appropriate biosensor strains to check for the appearance of a coloured halo around each spot74.
Confirmation of AHL-degradation activity by TLC and HPLC/MS
To exclude a QQ ability linked to the production of an inhibitor, supernatants of cultures were analysed by thin-layer chromatography (TLC) and high-performance liquid chromatography/mass spectrometry (HPLC/MS). For the TLC analyses, 10 µl of supernatants of degradation assays performed in LB supplemented with C6-HSL or C12-HSL were spotted onto TLC plates (Partisil KC18 Whatman 20 × 20 cm plates) and developed with CH3OH:H20 60:40 (v/v) as mobile phase. After migration, the plates were air-dried and overlaid with top agar containing the appropriate biosensor and incubated at 28 °C for 24 h10, 75.
For the HPLC/MS analysis and quantification, 500 µl of supernatants of degradation assays of C6-HSL and C12-HSL and non-inoculated media (negative control) were extracted twice with an equal volume of dichloromethane, evaporated and resuspended in 400 µl of acetonitrile26. Analyses were carried out with an Agilent HPLC 1100 chromatograph, equipped with a C8 precolumn (2.1 × 12.5 mm, 5 μm) and a Zorbax Eclipse XDB-C18 column (2.1 × 150 mm, 5 μm) maintained at 45 °C. The mobile phase was built by HCOOH:H2O 1:1000 (v/v) and HCOOH:C2H3N 1:1000 (v/v)76. Mass spectrometry experiments were conducted on an API 4000 triple-quadrupole mass spectrometer (Applied Biosystem) equipped with a Turbolon source using the positive ion electrospray and multiple reaction monitoring (MRM) mode. The MRM signals were used to generate relative quantification information by comparison with a calibration curve constructed for molecular ion abundance, using each of the appropriate AHL synthetic standards77. The effects of the AHL chain length (C6-HSL vs C12-HSL) and of the treatment (E. coli with an empty fosmid or with the fosmid containing hqiA) on the AHL-degradation efficiency were determined by analysis of variance using a two-factor (ANOVA) test (P < 0.05) and a Tukey test using R.
Sequencing of the AHL-degrading f10/17.1H fosmid, annotation and phylogenetic analyses
Sequencing of the fosmid f1017.1H was performed on a 454 GS Junior system as recommended by the manufacturer and according to the rapid library production and sequencing method protocols (Roche). Briefly, fosmid f1017.1H was diluted in TE buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA) to reach a concentration of 5 ng µl−1 and this suspension was physically fractionated by nebulisation to generate 450 bp fragments. After ligation of the adapters, emulsion PCR was performed using the emPCR amplification kit (Lib-L, Roche). The run of pyrosequencing yielded 50,457 reads. These sequences were used to generate a de novo assembly with the Roche 454 sequencing assembly software Newbler. This assembly resulted in one contig of 50,457 bp, corresponding to 8,139 bp of the pCC1FOS fosmid and to 42,318 bp of metagenomic insert. ORFs in this insert were predicted by using the RAST and GeneMark applications78, 79. After gene prediction, nucleotide and aminoacid sequence analyses were made using BLAST80. G + C content was calculated using the web tool GC-Profile81. For each protein encoded by the fosmid f1017.1H, homologous protein sequences were retrieved from NCBI after identification by BLAST X80. Multiple sequence alignments and phylogenetic trees were constructed using Seaview82. Briefly, alignments were carried out with ClustalO83. Maximum likelihood (ML) bootstrap analyses were carried out with 1000 replicates. Phylogenetic trees were constructed with PhyML using the LG model, the NNI (nearest-neighbor interchange) and BioNJ options to improve likelihood and tree topology.
The fosmid sequence was deposited on SRA trace archive system under bioproject number PRJNA326320 and biosample number SAMN05276011.
Cloning of ORF29 and confirmation of AHL-degradation activity
To identify the gene coding for the QQ ability in the fosmid f10/17.1H, subcloning of the metagenomic insert DNA was carried out. A PCR-based strategy was applied to test the potential ORFs involved. First, 5-kb PCR fragments of the fosmid insert were cloned and tested for their ability to degrade AHLs. After this first step and on the basis of its sequence homology with a hydrolytic enzyme, ORF29 was selected. It was amplified using the specific primers ORF29 forward 5′-CATTTCAGCAAAACCATGGAGGTGA-3′ and ORF29 reverse 5′-TCAAAACGAGCTCACGGTGCAAT-3′ that include (underlined) a NcoI and SacI restriction sites respectively. Then it was cloned in pGEM-T and transferred to E. coli DH5α, where the QQ ability of the construction was tested in LB supplemented with C6-HSL as described previously. After verification of the QQ phenotype, ORF29 was digested with NcoI and SacI enzymes and transferred into the pME6010 broad-host-range cloning vector84 for construction of pME6010::hqiA (for hypersaline quorum-quenching isochorismatase). Then, it was transformed into E. coli S17 λ pir. The QQ activity of this construction was tested in LB supplemented with different AHLs as previously described in experimental procedures. E. coli S17 λ pir harbouring the empty plasmid pME6010 was used as negative control.
Cloning, expression and purification of the HqiA protein
To allow protein production, the hqiA gene was amplified using the primer pair hqiA forward 5′-GGATCCATGAGTGAAATCAGCTTGGCG-3′ and hqiA reverse 5′-AAGCTTTTACGATCCTTCGGGTAAAGAAC-3′ that include (underlined) a BamHI and HindIII restriction sites to facilitate the cloning in a modified pMAL-c2X expression vector (CmR) (New England Biolabs) where the factor Xa cleavage site has been substituted with a TEV protease cleavage site. The resulting plasmid was termed pMAL-c2TEV-ORF29 and the plasmid sequence was confirmed by automatic sequencing with universal primers.
E. coli BL21(DE3)pLysS was transformed with pMAL-c2TEV-ORF29 and cultured at 37 °C until OD600nm 0.5. Then protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 6 h at 37 °C. After centrifugation, cell pellets were resuspended in a cell-lysis buffer (CLB) (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, pH 7.5) and sonicated. The soluble fraction was clarified by centrifugation at 10,000 × g for 20 min. Supernatants were applied to MBPTrap HP columns (GE Healthcare) and washed with CLB until the protein content in the wash was negligible. Then, the recombinant protein in the column was eluted with CLB supplemented with 10 mM maltose. The eluted fractions were pooled and digested overnight at 4 °C in the same buffer with 1 mg of recombinant TEV protease for each 25 mg of protein85. The digested samples were loaded in DEAE Sephacel columns (GE Healthcare) and the protein was eluted with a NaCl linear gradient (100 to 500 mM NaCl). The eluted fractions were pooled and stored at −20 °C. Protein concentration was assayed using the Bradford method86. The potential three-dimensional structure of the purified protein was calculated using the software Phyre2, a web portal for protein modeling, prediction and analysis based on aminoacid sequences30.
Characterization of the AHL-degrading activity of purified HqiA
AHL-degradation activity of the purified HqiA was tested in reaction buffer (RE) (200 mM Na phosphate, pH 6.5) containing 16 µg ml−1 of HqiA protein and 25 µM of each of the synthetic AHLs. After incubation at 28 °C for 4 h, the remaining AHLs were detected on agar plates overlaid with A. tumefaciens NTL4 (pZLR4) as previously described above. To identify the type of QQ enzyme, the degradation products of the QQ reaction of HqiA were analyzed by HPLC/MS and by the lactone ring closure assay after acidification72, 74. HPLC/MS was conducted on a Waters Acquity UPLC equipped with an UPLCr BEH C18 reverse-phase column (100 × 2.1 mm, 1.7 µm) coupled to an ESI-MS detector (Waters Synapt G2). Samples were injected and eluted with NH4OH:H2O 1:1000 (v/v) and NH4OH:C2H3N 1:1000 (v/v). Mass spectrometry experiments were recorded with ES- Polarity. The lactone ring closure assay consisted of the acidification to pH 2 the reaction after incubation of HqiA with the different AHLs tested. This condition allows the lactone ring to restructure itself in the case that it had previously been opened by a lactonase. After neutralizing the pH, remaining AHLs were detected were detected on AB or LB agar plates overlaid with the appropriate biosensor.
Pathogenicity test and extracellular enzymatic assays
The ability of hqiA to interfere with the expression of QS-regulated functions was tested in a heterologous expression system. The construction pME6010::hqiA and the empty plasmid pME6010 (TcR), which was used as negative control, were transferred into the plant pathogen Pectobacterium carotovorum subsp. carotovorum CECT 225T by electroporation87. To confirm that P. carotovorum subsp. carotovorum contained pME6010::hqiA, PCR amplification was carried out using specific primers hqiA-T forward 5′-ATGAGTGAAATCACGTTGGC-3′ and hqiA-T reverse 5′-CTTTACCCGAAGGATCGTAA-3′. To determine the impact of the expression of hqiA on AHL-production by the pathogenic strain, an agar-diffusion assay was carried out using the biosensors C. violaceum CV026 and A. tumefaciens NTL4 (pZLR4). Briefly, 20-µl of overnight cultures (OD600nm 2.7) of the strain CECT 225T carrying or not the plasmid pME6010 or pME6010::hqiA were spotted in LB and AB agar plates previously overlayed with the biosensor strains. To check that the expression of the hqiA gene did not affect the growth of this bacterial strain, serial dilutions of overnight cultures (OD600nm 2.7) of the strain CECT 225T carrying or not the different constructions were conducted and cell counts of each culture were performed on LB agar plates supplemented or not with Tc.
To evaluate the effect of the AHL-degradation of hqiA upon some of the QS-regulated virulence factors produced by the strain P. carotovorum, 20-µl of overnight cultures (OD600nm 2.7) of the strain CECT 225T carrying or not the plasmid pME6010 or pME6010::hqiA were spotted on different enzymatic media. Proteolytic activity was determined in casein medium88. DNase activity was determined in DNase agar medium89. The swimming motility test was carried out in LB 0.3% (w/v) agar90. The swarming motility test was carried out in LB 0.5% (w/v) agar91. Pectinase activity was carried out in pectin medium92. Alkaline phosphatase activity was determined in PVK medium93. Five replicates were used for each condition. In all these enzymatic media the result was obtained by measuring haloes around the spotted area, except for the motility tests where growth due to the migration of cells away from the inoculation site was measured. The diameter of haloes in each medium was measured and statistical analyses were conducted using an ANOVA test (P < 0.05) and a Tukey test using R.
The ability of hqiA to interfere with the pathogenicity of the strain CECT 225T was also tested in a potato tuber assay12. Briefly, the strain CECT 225T carrying or not the plasmid pME6010 or pME6010::hqiA were grown at 28 °C for 24 h (OD600nm 2.7). Potato tuber slices (Solanum tuberosum) were surface sterilized using 10% (v/v) sodium hypochlorite during 15 min, rinsed with sterile distilled water, cut in slices and placed in Petri dishes with wet filter paper to keep them moist. 20 µl of each culture was inoculated on the surfaces of the potato slice and incubated at 28 °C for 48 h. LB cell-free medium was also inoculated on potatoes as a negative control. Five replicates were used for each condition. Symptoms were recorded after a 2-day incubation by visual inspection of the maceration zones. Appearance or not of maceration zones was statistically analysed using an ANOVA test (P < 0.05) and a Tukey test using R. The spatial extent of the maceration zone was estimated by image analysis using ImageJ software94 as a percentage of the potato slice surface area.
Electronic supplementary material
Acknowledgements
This research was supported by grants from the Dirección General de Investigación Científica y Técnica (CGL2011-25748, BIO2011-12879E, AGL2015-68806-R) and from INRA (MetaScreen, Projet innovant and the Labex ARBRE ANR-11-LABX-0002-01). Marta Torres is supported by a FPU fellowship from the Spanish Ministry of Science and Education (FPU13-0466) and a University of Granada-CEI BioTic International Mobility Programme. The authors thank Angela Tate for her critical reading of this manuscript, Nahid Oueriaghli for supplying the soil samples, the Functional Ecogenomics platform for their services and Pr. John S. Parkinson (University of Utah, USA) for providing the E. coli RP437 strain and its related chemotaxis mutants.
Author Contributions
S.U., E.Q. and I.L. conceived and supervised the study. M.T., S.U., R.S. and I.L. designed the experiments. M.T., S.U., R.S., L.F. and I.L. performed the experiments. M.T., S.U., R.S., E.Q. and I.L. analyzed the data. M.T., S.U., R.S. and I.L. prepared the figures and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01176-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hornby JM, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams P, Winzer K, Chan WC, Cámara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uroz S, Dessaux Y, Oger P. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. ChemBioChem. 2009;10:205–216. doi: 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- 6.González JE, Keshavan ND. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006;70:859–875. doi: 10.1128/MMBR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González JE, Marketon MM. Quorum sensing in nitrogen-fixing Rhizobia. Microbiol. Mol. Biol. Rev. 2003;67:574–592. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao Y, Winans SC, Glick BR, Charles TC. Identification and characterization of new LuxR/LuxI-type quorum sensing systems from metagenomic libraries. Environ. Microbiol. 2010;12:105–117. doi: 10.1111/j.1462-2920.2009.02049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasuno E, et al. Phylogenetically novel LuxI/LuxR-type quorum sensing systems isolated using a metagenomic approach. Appl. Environ. Microbiol. 2012;78:8067–8074. doi: 10.1128/AEM.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llamas I, et al. Quorum sensing in halophilic bacteria: detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extremophiles. 2005;9:333–341. doi: 10.1007/s00792-005-0448-1. [DOI] [PubMed] [Google Scholar]
- 11.Tahrioui A, Schwab M, Quesada E, Llamas I. Quorum sensing in some representative species of Halomonadaceae. Life. 2013;3:260–275. doi: 10.3390/life3010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uroz S, et al. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology. 2003;149:1981–1989. doi: 10.1099/mic.0.26375-0. [DOI] [PubMed] [Google Scholar]
- 13.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit. Rev. Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen TB, et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology. 2016;151:1325–1340. doi: 10.1099/mic.0.27715-0. [DOI] [PubMed] [Google Scholar]
- 15.Uroz S, Heinonsalo J. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. FEMS Microbiol. Ecol. 2008;65:271–278. doi: 10.1111/j.1574-6941.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- 16.Dong Y, Zhang L. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 2005;43:101–109. [PubMed] [Google Scholar]
- 17.Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- 18.Givskov M, et al. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bact. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manefield M, et al. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 20.Park S, et al. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 2005;71:2632–2641. doi: 10.1128/AEM.71.5.2632-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uroz S, et al. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- 22.Uroz S, et al. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 2008;74:1357–1366. doi: 10.1128/AEM.02014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y, Gusti A, Zhang Q, Xu J, Zhang L. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appli. Environ. Microbiol. 2002;68:1754–1759. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero M, Avendaño-Herrera R, Magariños B, Cámara M, Otero A. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroidetes (CFB) group. FEMS Microbiol. Lett. 2010;304:131–139. doi: 10.1111/j.1574-6968.2009.01889.x. [DOI] [PubMed] [Google Scholar]
- 25.Romero M, Diggle SP, Heeb S, Cámara M, Otero A. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 2008;280:73–80. doi: 10.1111/j.1574-6968.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 26.Romero M, Martin-Cuadrado AB, Roca-Rivada A, Cabello AM, Otero A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 2011;75:205–217. doi: 10.1111/j.1574-6941.2010.01011.x. [DOI] [PubMed] [Google Scholar]
- 27.Mayer C, Romero M, Muras A, Otero A. Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 2015;99:9523–9539. doi: 10.1007/s00253-015-6741-8. [DOI] [PubMed] [Google Scholar]
- 28.Bijtenhoorn P, et al. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. 2011;155:86–94. doi: 10.1016/j.jbiotec.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Romero M, Martin-Cuadrado A, Otero A. Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 2012;78:6345–6348. doi: 10.1128/AEM.01266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui X, Harling R. N-acyl-homoserine lactone-mediated quorum sensing blockage, a novel strategy for attenuating pathogenicity of Gram-negative bacterial plant pathogens. Eur. J. Plant Pathol. 2005;111:327–339. doi: 10.1007/s10658-004-4891-0. [DOI] [Google Scholar]
- 32.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A, Cui Y, Chakrabarty P, Chatterjee AK. Regulation of motility in Erwinia carotovora subsp. carotovora: quorum-sensing signal controls FlhDC, the global regulator of flagellar and exoprotein genes, by modulating the production of RsmA, an RNA-binding protein. Mol. Plant. Microbe. Interact. 2010;23:1316–1323. doi: 10.1094/MPMI-01-10-0017. [DOI] [PubMed] [Google Scholar]
- 34.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 2015;40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 35.Montgomery K, Charlesworth JC, LeBard R, Visscher PT, Burns BP. Quorum sensing in extreme environments. Life. 2013;3:131–148. doi: 10.3390/life3010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farah C, et al. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005;71:7033–7040. doi: 10.1128/AEM.71.11.7033-7040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson LL, et al. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 2005;71:6335–6344. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols D. Cultivation gives context to the microbial ecologist. FEMS Microbiol. Ecol. 2007;60:351–357. doi: 10.1111/j.1574-6941.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- 39.D’Angelo-Picard C, Faure D, Penot I, Dessaux Y. Diversity of N-acyl homoserine lactone-producing and -degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 2005;7:1796–1808. doi: 10.1111/j.1462-2920.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- 40.Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- 41.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riaz K, et al. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 2008;10:560–570. doi: 10.1111/j.1462-2920.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- 43.Afriat L, Roodveldt C, Manco G, Tawfik DS. The latent promiscuity of newly identified microbial lactonases is linked to recently diverged phosphotriesterase. Biochemistry. 2006;45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- 44.Schipper C, et al. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009;75:224–233. doi: 10.1128/AEM.01389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tannières M, et al. A metagenomic study highlights phylogenetic proximity of quorum-quenching and xenobiotic-degrading amidases of the AS-family. PLoS One. 2013;8:e65473. doi: 10.1371/journal.pone.0065473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uchiyama T, Miyazaki K. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 2009;20:616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Carlier A, et al. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mei G, Yan X, Turak A, Luo Z, Zhang L. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl. Environ. Microbiol. 2010;76:4933–4942. doi: 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu D, et al. Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis. Proc. Natl. Acad. Sci. 2005;102:11882–11887. doi: 10.1073/pnas.0505255102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XH, Wang L, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan K, et al. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons JF, Calabrese K, Eisenstein E, Ladner JE. Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway. Biochemistry. 2003;42:5684–5693. doi: 10.1021/bi027385d. [DOI] [PubMed] [Google Scholar]
- 53.Bokhove M, Nadal Jimenez P, Quax WJ, Dijkstra BW. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. Proc. Natl. Acad. Sci. 2010;107:686–691. doi: 10.1073/pnas.0911839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang JJ, Petersen A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2006;72:1190–1197. doi: 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wahjudi M, et al. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology. 2011;157:2042–2055. doi: 10.1099/mic.0.043935-0. [DOI] [PubMed] [Google Scholar]
- 56.Ollivier B, Caumette P, Garcia JL, Mah RA. Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 1994;58:27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foti M, et al. Diversity, activity and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 2007;73:2093–2100. doi: 10.1128/AEM.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scharf BE, Hynes MF, Alexandre GM. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 2016;90:549–559. doi: 10.1007/s11103-016-0432-4. [DOI] [PubMed] [Google Scholar]
- 59.Sourjik V, Muschler P, Scharf B, Schmitt R. VisN and VisR are global regulators of chemotaxis, flagellar and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 2000;182:782–788. doi: 10.1128/JB.182.3.782-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoang HH, Gurich N, González JE. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 2008;190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parnell J, et al. Functional biogeography as evidence of gene transfer in hypersaline microbial communities. PLoS One. 2010;5:e12919. doi: 10.1371/journal.pone.0012919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhodes ME, Spear JR, Oren A, House CH. Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evol. Biol. 2011;11:199. doi: 10.1186/1471-2148-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mongodin E, et al. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smadja B, et al. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum) Mol. Plant. Microbe. Interact. 2004;17:1269–1278. doi: 10.1094/MPMI.2004.17.11.1269. [DOI] [PubMed] [Google Scholar]
- 65.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. 2000;97:3526–3531. doi: 10.1073/pnas.97.7.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oueriaghli N, Béjar V, Quesada E, Martínez-Checa F. Molecular ecology techniques reveal both spatial and temporal variations in the diversity of archaeal communities within the athalassohaline environment of Rambla Salada, Spain. Microb. Ecol. 2013;66:297–311. doi: 10.1007/s00248-013-0176-5. [DOI] [PubMed] [Google Scholar]
- 67.Parkinson JS. CheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkinson JS. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McClean KH, et al. Quorum sensing and Chromobacteriurn violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 70.Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 2008;279:124–130. doi: 10.1111/j.1574-6968.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- 71.Chilton MD, et al. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yates EA, et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uroz, S. & Oger, P. M. In Metagenomics: Methods and Protocols (eds Streit, W. & Rolf, D.) 271–286 (Springer, 2017).
- 74.Torres M, et al. Selection of the N-acylhomoserine lactone-degrading bacterium Alteromonas stellipolaris PQQ-42 and of its potential for biocontrol in aquaculture. Front. Microbiol. 2016;7:646. doi: 10.3389/fmicb.2016.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marketon MM, Gonzalez MM. Identication of two quorum-sensing systems in Sinorhizobium meliloti. Society. 2002;184:3466–3475. doi: 10.1128/JB.184.13.3466-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ortori CA, et al. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole-linear ion trap mass spectrometry. Anal. Bioanal. Chem. 2007;387:497–511. doi: 10.1007/s00216-006-0710-0. [DOI] [PubMed] [Google Scholar]
- 77.Milton DL, et al. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl) homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 2001;183:3537–3547. doi: 10.1128/JB.183.12.3537-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:451–454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao F, Zhang C. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34:686–691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 83.Sievers F, Higgins D. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 84.Heeb S, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant. Microbe. Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 85.Parks T, Leuther K, Howard E, Johnston S, Dougherty W. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- 86.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 87.Sambrook, J. & Russell, D. Molecular Cloning: a Laboratory Manual. (Cold Spring Harbor Laboratory Press, 2001).
- 88.Barrow, G. I. & Feltham, R. K. A. Cowan and Steel’s Manual for the Identification of Medical Bacteria (eds Barrow, G. I. & Feltham, R. K. A.) (Cambridge University Press, 1993).
- 89.Jeffries CD, Holtmian DF, Guse DG. Rapid method for determining the activity of microorganisms on nucleic acids. J. Bacteriol. 1957;73:590–591. doi: 10.1128/jb.73.4.590-591.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rui H, Liu Q, Ma Y, Wang Q, Zhang Y. Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol. Lett. 2008;285:155–162. doi: 10.1111/j.1574-6968.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 91.O’May C, Tufenkji N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 2011;77:3061–3067. doi: 10.1128/AEM.02677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banakar SP, Thippeswamy B. Isolation, production and partial purification of fungal extracellular pectinolytic enzymes from the forest soils of Bhadra wildlife sanctuary. J. Biochem. Tech. 2012;3:138–143. [Google Scholar]
- 93.Pikovskaya RI. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya. 1948;17:362–370. [Google Scholar]
- 94.Rasband, W. ImageJ. US National Institutes of Health, Bethesda, Maryland, USA. http://imagej.nih.gov/ij (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


