Abstract
Resistance to the anthelmintic drug monepantel (Zolvix®) has emerged in parasitic worms infecting sheep and goats. The mechanism of resistance in these cases is unknown. The drug targets nicotinic acetylcholine receptors belonging to the nematode-specific DEG-3 subfamily. We examined the receptor gene, Hco-mptl-1, in a highly Zolvix®-resistant and a -susceptible isolate of the parasitic nematode Haemonchus contortus. cDNA coding for the full length receptor protein (Hco-MPTL-1) was present in all clones prepared from a pool of susceptible larvae (21/21 clones) and approximately 50% of those from the resistant isolate (17/33). On the other hand, the remaining clones from the resistant isolate showed various mutations that resulted in truncated predicted proteins, missing at least one transmembrane domain. The most common mutation (11/33 clones) resulted in the retention of intron 15, a premature stop codon, and a truncated protein. Sequencing of intron 15 genomic DNA showed very few SNPs in susceptible larvae and in 12/18 clones from resistant larvae, alongside the presence of at least 17 SNPs in the remaining resistant clones. The present study shows that the highly resistant isolate has a number of mutations in the drug target gene that would most-likely result in a non-functional receptor, thus rendering the larvae insensitive to the drug. The presence of many wild-type sequences in this highly-resistant population suggests that there was a significant presence of heterozygotes in the survivors of the field drench treatment from which the isolate was derived, and hence that at least some of the mutations may be dominant. Alternatively, their presence may be due to the additional influence of mutations at another locus contributing to the resistance phenotype. The presence of multiple separate mutations in the Hco-mptl-1 gene in this viable field-derived worm isolate may at least partly explain why resistance to Zolvix® has arisen rapidly in the field.
Keywords: Haemonchus contortus, Monepantel, Resistance, Mutation, Target site, Intron retention
Graphical abstract
Highlights
-
•
Hco-mptl-1 gene compared in susceptible and monepantel-resistant Haemonchus contortus.
-
•
Six separate mutations detected in the resistant isolate.
-
•
Most common mutation resulted in retention of intron and truncated predicted protein.
-
•
Many separate mutations in the drug target site in the field-derived resistant isolate.
1. Introduction
Monepantel (Zolvix®) was registered for the control of gastrointestinal nematode (GIN) parasites of small ruminants in New Zealand in 2009, and in others parts of the world in 2010. The compound is an amino-acetonitrile derivative, showing a wide spectrum of activity against GINs (Kaminsky et al., 2008). However, within several years of its release, resistance was reported in two species of worms infecting goats in New Zealand (Scott et al., 2013). More reports of resistance have followed since that time (Mederos et al., 2014, Van den Brom et al., 2015, Cintra et al., 2016, Sales and Love, 2016).
The drug targets nicotinic acetylcholine receptors of the nematode-specific DEG-3 subfamily. In Haemonchus contortus, Hco-MPTL-1 and other members of this subfamily (including Hco-DES-2H) are thought to be the targets of monepantel (Rufener et al., 2009). Resistance to the drug was examined before its release by applying in vitro drug selection pressure to the free-living larval life stages of H. contortus over eight generations, resulting in phenotypic resistance at both the adult and larval stages (Kaminsky et al., 2008). Rufener et al. (2009) examined the sequences of Hco-mptl-1 and Hco-des-2H genes in these resistant isolates and found a panel of mutations in Hco-mptl-1 that resulted in miss-spliced transcripts and premature stop codons. In addition, they identified an insertion on Hco-des-2H that resulted in the creation of out-of-frame start codons. The authors suggested that these loss-of-function mutations may be responsible for the loss of sensitivity to the drug in these laboratory-selected resistant isolates.
Raza et al. (2016) recently described the use of in vitro larval development assays to examine the sensitivity of an isolate of H. contortus (MPL-R) showing resistance to monepantel in vivo. The MPL-R isolate was initiated as the survivors of a drench with a recommended dose of Zolvix® that had shown a 99.2% efficacy. The progeny of these survivors were used to infect a housed animal, which was then drenched with Zolvix®. The progeny of the survivors were collected and used to establish infections in two further housed animals for use in the study of Raza et al. (2016). The isolate therefore represents a field-derived laboratory-propagated isolate in which only the survivors of a drench treatment have been propagated further. A recommended dose of Zolvix® shows zero efficacy against this isolate in vivo (unpublished data). The present study aimed to determine whether mutations of the type detected by Rufener et al. (2009) in laboratory-selected isolates could also be responsible for the monepantel resistance shown by the field-derived MPL-R isolate. We report on a number of loss-of-function mutations in the Hco-mptl-1 gene which were likely responsible for the presence of survivors following the original field drench treatment.
2. Materials and methods
2.1. Parasites
Two isolates of H. contortus were used in this study;
-
i)
Kirby: isolated from the field at the University of New England Kirby Research Farm in 1982; susceptible to all commercial anthelmintics (Albers and Burgess, 1988).
-
ii)
MPL-R: isolated from a property in southwest Queensland, Australia, in 2014. H. contortus larvae were cultured from faeces collected from sheep that had shown clinical signs of scouring after a Zolvix® drench treatment (drench efficacy measured as 99.2%). These larvae were subsequently used to establish infections in a housed animal. This animal was treated with a recommended dose of Zolvix®, and larvae were collected and used to infect two further animals. Larvae collected from these animals were used for the present study. The recommended dose of Zolvix® shows zero efficacy against the isolate.
Sheep infected with the Kirby isolate were housed at the CSIRO McMaster laboratory, Armidale, NSW. Sheep infected with the MPL-R isolate were housed at the Invetus animal house facility in Armidale, NSW. Faeces were collected and sent by overnight courier to the CSIRO laboratory in Brisbane, QLD, and used to establish faecal cultures. Third-stage larvae (L3) were collected from faecal cultures which had been incubated for 7 days. The L3 were allowed to pass through a 20 μm filter, and stored at 15 °C until used for molecular analyses within 2 weeks.
2.2. Preparation of RNA/cDNA/genomic DNA, and sequence analysis
RNA was isolated from pools of 10,000 Kirby or MPL-R L3 stage larvae using the RNeasy® mini kit (Qiagen, Hilden, Germany), with an on-column DNase (Qiagen) digest step during the extraction process. cDNA was prepared using the Superscript® III First-Strand Synthesis Kit (Invitrogen, Carlsbad, USA), and diluted 1/5 with water before using in PCR. PCR was carried out with primers designed from H. contortus acetylcholine receptor monepantel-1 mRNA sequence (Acc No. FJ807280). Primers were designed, using Primer 3, to cover the whole coding sequence (Hc.mon-1.F1/R1), as well as various internal primers for sequencing (Hc.mon-1.F2-F5, R3) (Table 1).
Table 1.
Primers used for PCR of full length Hco-mptl-1 cDNA, sequencing of cDNA, and PCR of genomic DNA intron 15.
| Primer use | Notation | 5′-3′ Sequence |
|---|---|---|
| PCR of full length cDNA | Hc.mon-1.F1 | GAGGGCAACAATTTCCTTCA |
| Hc.mon-1.R1 | GAACTCCAATTAATTTATC | |
| cDNA sequencing | Hc.mon-1.F2 | CATGGATGACCACGACACTC |
| Hc.mon-1.R3 | AAATGTCCAACTGCCGAAAG | |
| Hc.mon-1.F3 | GGGTTCTTCAGCTCATCGTC | |
| Hc.mon-1.F4 | GCCAAAGAACAGGCACAAAT | |
| Hc.mon-1.F5 | TCGTCCTTTCCAGACATTGA | |
| PCR of intron 15 | Hc.intron15.F1 | CAAGTGCCGAAGAAAAGTTGCA |
| Hc.intron15.R1 | ATGCGCACGTACTCATTTTG |
PCR reactions contained 50 ng of cDNA, 200 μM each dNTP and 1.0 unit of Taq DNA polymerase (Advantage 2 Polymerase mix, Takara Bio, USA) in reaction buffer, with primers at a concentration of 0.25 μM. Cycling conditions were an initial denaturation of 95 °C × 1 min, followed by 40 cycles of 95 °C × 15 s, 51–64 °C (temperature dependent on specific primer set) × 15 s, 68 °C × 1 min and a final extension at 70 °C × 10 min. PCR products were cloned directly into pGEM-T Easy vector (Promega, Madison, USA) according to the manufacturers' instructions. Clones were prepared with the QIAprep® spin miniprep kit (Qiagen) and sequenced using Big Dye Terminator v3.1 on an ABI PRISM 3700 genetic analyser with M13F/R primers and the internal primers listed in Table 1. BLAST was used to confirm the cloned sequence was acetylcholine receptor monepantel-1 (Hco-mptl-1). Sequences were manually observed for overlapping sequence, trimmed, pasted together, and aligned in MEGA6 (Tamura et al., 2013).
Genomic DNA was isolated from pools of 10,000 Kirby or MPL-R L3 stage larvae using the QIAamp® DNA Mini kit (Qiagen). PCR reactions contained 50 ng of genomic DNA, 200 μM each dNTP and 1.0 unit of taq (Advantage 2 Polymerase mix) in reaction buffer, with primers at a concentration of 0.25 μM. Primers are shown in Table 1. Cycling conditions were an initial denaturation of 95 °C × 1 min, followed by 40 cycles of 95 °C × 15 s, 68 °C × 15 s, 68 °C × 1 min and a final extension at 70 °C × 10 min. PCR products were cloned, then sequenced using M13F/R primers, and aligned in MEGA6, as described above.
3. Results and discussion
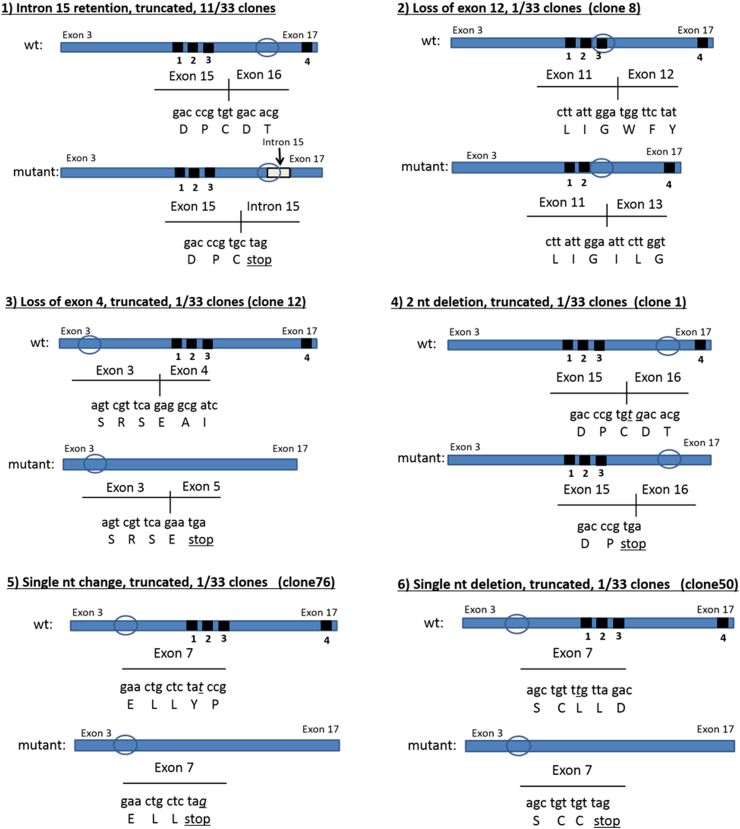
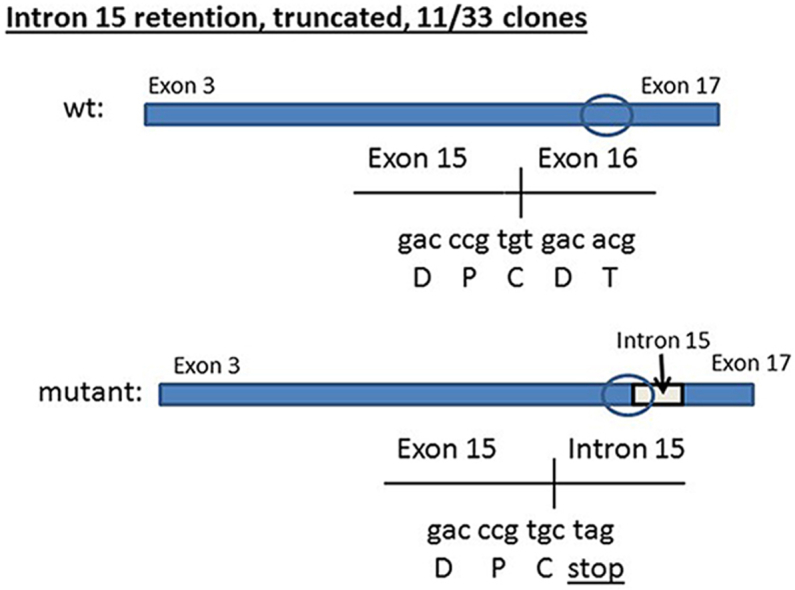
Sequencing of the Hco-mptl-1 gene in 21 cDNA clones prepared from a pooled sample of Kirby isolate larvae resulted in sequences showing the presence of some SNPs, resulting in altered predicted amino acid residues in a number of instances, however all sequences were predicted to code for full length MPTL-1 proteins (564 amino acids (AA) (GenBank Acc. No. KY983245–KY983253). cDNA sequence coding for predicted full length proteins was also present in 17 out of 33 clones derived from a pooled sample of MPL-R larvae. On the other hand, the remaining 16 MPL-R clones showed the presence of various mutations resulting in truncated predicted proteins (Fig. 1):
-
1.
intron 15 retained in 11/33 clones; resulting in a premature stop codon near the start of the intron sequence; protein length = 448 AA; missing the predicted transmembrane 4 domain (KY983239).
-
2.
loss of exon 12 in 1/33 clones; protein length = 537 AA; missing the predicted transmembrane 3 domain (KY983240).
-
3.
loss of exon 4 in 1/33 clones; resulting in a premature stop codon at the start of exon 5; protein length = 18 AA; missing all four predicted transmembrane domains (KY983241); identical to the m2 mutant described by Rufener et al. (2009).
-
4.
2 nucleotide (nt) deletion in 1/33 clones; 2 nt deletion at the exon 15/16 junction, resulting in a premature stop codon, protein length = 447 AA; missing the predicted transmembrane 4 domain (KY983242).
-
5.
single nt change; ‘t’ to ‘g’ transversion in exon 7, resulting in a premature stop codon; protein length = 148 AA; missing all four predicted transmembrane domains (KY983243).
-
6.
single nt deletion in exon 7, resulting in a premature stop codon; protein length = 158 AA; missing all four predicted transmembrane domains (KY983244).
Fig. 1.
Schematic of mutations identified in Hco-mptl-1 cDNA from the sequencing of clones derived from a pooled sample of MPL-R isolate larvae (wt = wild type). The approximate positions of the four predicted transmembrane domains (numbered 1–4) are shown on each of the wt cDNA sequences. For the mutant cDNA sequences, only the transmembrane domains that would be present on the predicted proteins are shown.
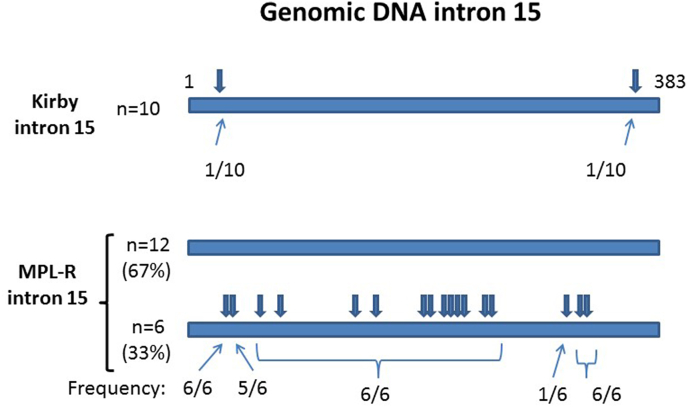
We looked more closely at the retention of intron 15 (number 1. above) by sequencing the genomic DNA across this intron from Kirby and MPL-R larvae (KY983254, KY983255). The intron sequence was almost identical in 10 clones from Kirby larvae, with presence of just 1 SNP in two separate clones (Fig. 2). This intron sequence was also present in 12/18 clones from MPL-R larvae. However, SNPs at 17 different positions were present in the remaining 6 clones from MPL-R larvae. The SNPs at 14 of the positions occurred in all 6 sequences, while 1 SNP was present in 5/6 sequences, and another was present in only 1/6. The SNPs occurred in positions at least 30 nucleotides from the 5′ and 3’ ends of the intron, and, hence do not interfere directly with the splice acceptor or splice donor sites. The possibility that these mutations could create or remove motifs important for alternative splicing exists, but the gene has not yet been characterised sufficiently for us to predict whether this occurs.
Fig. 2.
Schematic of Hco-mptl-1 intron 15 genomic DNA from the sequencing of clones derived from separate pooled samples of Kirby and MPL-R isolate larvae. Arrows and fractions indicate approximate position and frequency of SNPs.
It is clear that there are a number of mutations present in the Hco-mptl-1 gene in the MPL-R isolate of H. contortus that would most-likely result in the generation of non-functional drug receptors. Most of the observed mutations resulted in truncation of the protein, with the loss of predicted transmembrane 4 domain or domains 1–4, while a single mutation resulted in the skipping of exon 12, with the loss of transmembrane domain 3. Half of the clones derived from MPL-R larvae showed the presence of one of these mutations, while the most frequent mutation, retention of intron 15, occurred in one-third of the clones. The present study therefore indicates that changes to the monepantel receptor, Hco-MPL-1, are likely to be at least partly responsible for the resistance shown by the MPL-R isolate towards this drug. Only one of the mutations observed here (number 3: loss of exon 4, truncated) was also reported by Rufener et al. (2009) in their two laboratory-selected resistant isolates.
Raza et al. (2016) showed that LDA assays measuring the response of MPL-R larvae to monepantel resulted in a 2-step dose response curve that subdivided the population into two separate sub-populations showing low- and high-level resistance, with resistance factors of approximately 7- and 1000-fold, respectively, compared to Kirby larvae. The high level resistant component represented approximately 60% of the total worm population. It is unclear how the presence of mutations in the Hco-mptl-1 gene observed in the present study relate to the reduced drug sensitivity measured in the two worm sub-populations. However, the presence of two sub-populations suggests that at least two loci, including one containing the Hco-mptl-1 gene, are involved in the observed resistance. Mutations at one or more of the loci would appear to confer a much higher degree of resistance compared to others. The loss of a drug receptor would be expected to have a more profound effect on drug action than a mutation that reduces binding affinity (for example, beta-tubulin SNPs and benzimidazole drugs; Kwa et al., 1994, Ghisi et al., 2007) or mutations resulting in increased detoxification activity that reduce the amount of drug reaching its target site. It is therefore likely that the high level resistance shown by sub-population 2 in larval development assays (resistance factor > 1000-fold) is associated with the mutations in the Hco-mptl-1 gene reported here, however, this requires confirmation. Resistance shown by aphids towards pyrethroid and neonicotinoid insecticides have shown a pattern of low level detoxification-based resistance, followed by much higher levels of resistance appearing in field populations as a result of target site mutations (Bass et al., 2014).
We found that approximately 50% of the Hco-mptl-1 alleles were wild type in the MPL-R population. This may be interpreted in several ways: firstly, the presence of many wild type alleles may be a result of a significant component of the surviving adult worm population after the initial field drench treatment being heterozygous at the Hco-mptl-1 locus, with the resistance-causing mutations being dominant. A further complexity may exist where a dominant negative effect might be induced by incompletely penetrant alternative splicing, so that a resistance allele may at times produce an alternate protein which interferes with channel formation (for example), whilst not at other times; in this way, the summation of the effect across cells in two individuals of the same genotype might differ. Alternatively, if 2 loci are involved in the resistance phenotype, as suggested by the larval development assay data of Raza et al. (2016), then the presence of many wild type Hco-mptl-1 alleles in the drench survivors may be a result of a mutation at the second locus conferring resistance in some individuals, allowing them to survive the drench despite possessing a functional MPTL-1 receptor. These individuals who survived the drench due to the mutation at the second locus could have been homozygous wild type or heterozygous at the Hco-mptl-1 locus, with the Hco-mptl-1 mutations being recessive. Another factor that may be important when considering the relative numbers of wild type and mutant alleles is the fact that the presence of just one or two truncated subunits in the pentamer forming the homomeric channel may be enough to disrupt the functioning of the channel. In addition, we did not measure the expression levels of the Hco-mptl-1 gene in the present study, and hence have no information as to whether downregulation of the full length gene may be a component of the resistance. Further study will be required to examine these, and other, possibilities.
The most frequent mutation was the retention of intron 15, resulting in a premature stop codon immediately after exon 15. Our analysis of genomic sequences of intron 15 showed the presence of many polymorphisms in one-third of the sequences from the MPL-R larvae. Hence, at present it is not possible to identify specific nucleotide changes that may be responsible for interfering with the splicing mechanism leading to retention of the intron. The presence of many wild type intron 15 sequences in MPL-R larvae is in agreement with our observations of the coding region.
The present study has important implications for drug resistance diagnostics and anthelmintic discovery:
1) Many separate mutations in the Hco-mptl-1 gene in H. contortus may confer resistance due to the loss of the drug target site. We sequenced only 33 clones from a pooled larval sample of the MPL-R isolate and hence our study is certainly not exhaustive. It is likely that more mutations would have been revealed by sequencing of further clones from this pooled larval sample, or by the sequencing of individual larvae. Rufener et al. (2009) detected additional mutations in laboratory-selected isolates. A molecular diagnostic for drug resistance that focuses on mutations in the Hco-mptl-1 gene must therefore cover the entire cDNA sequence. An alternative diagnostic approach could use western-blotting with an antibody directed against the amino terminus of the Hco-MPTL-1 protein to detect truncated versions.
2) Rufener et al. (2009) suggested that the presence of many mutations in the Hco-mptl-1 gene in their laboratory-selected isolates indicated that the gene may not be essential in H. contortus. Similarly, the MPL-R isolate remains viable in the presence of many mutations to the monepantel receptor gene, indicating that the Hco-MPTL-1 protein is most likely not essential for the worm. Given that H. contortus is known to be highly polymorphic (reviewed in Gilleard and Redman, 2016), it is perhaps not unexpected that many mutations will exist in a gene that codes for a non-essential protein as the mutations will have very little or no negative impact on the fitness of the worm. Hence, these mutations which confer drug resistance through the loss of a functional receptor will be rapidly selected for by drug exposure. On the other hand, where a receptor is essential for the worms’ fitness, mutations that interfere with drug binding while retaining receptor function will likely be scarce compared to mutations for which there is no functional constraint. In the former case, selection for resistance would be expected to be slower, starting from a lower resistance allele frequency, and possibly involving selection at other loci to compensate for losses in fitness. This suggests that it may be important in the future to focus on drugs which interact with essential target proteins in anthelmintic discovery programmes.
Footnotes
Nucleotide sequence data reported in this paper are available in the GenBank, EMBL and DDBJ databases under accession numbers: KY983239–KY983255.
References
- Albers G.A., Burgess S.K. Serial passage of Haemonchus contortus in resistant and susceptible sheep. Vet. Parasitol. 1988;28:303–306. doi: 10.1016/0304-4017(88)90077-5. [DOI] [PubMed] [Google Scholar]
- Bass C., Puinean A.M., Zimmer C.T., Denholm I., Field L.M., Foster S.P., Gutbrod O., Nauen R., Slater R., Williamson M.S. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 2014;51:41–51. doi: 10.1016/j.ibmb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Cintra M.C., Teixeira V.N., Nascimento L.V., Sotomaior C.S. Lack of efficacy of monepantel against Trichostrongylus colubriformis in sheep in Brazil. Vet. Parasitol. 2016;216:4–6. doi: 10.1016/j.vetpar.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Mäser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Gilleard J.S., Redman E. Genetic diversity and population structure of Haemonchus contortus. Adv. Parasitol. 2016;93:31–68. doi: 10.1016/bs.apar.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Ducray P., Jung M., Clover R., Rufener L., Bouvier J., Weber S.S., Wenger A., Wieland-Berghausen S., Goebel T., Gauvry N., Pautrat F., Skripsky T., Froelich O., Komoin-Oka C., Westlund B., Sluder A., Mäser P. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Mederos A.E., Ramos Z., Banchero G.E. First report of monepantel Haemonchus contortus resistance on sheep farms in Uruguay. Parasit. Vectors. 2014;7:598. doi: 10.1186/s13071-014-0598-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Lamb J., Chambers M., Hunt P.W., Kotze A.C. Larval development assays reveal the presence of sub-populations showing high- and low-level resistance in a monepantel (Zolvix®)-resistant isolate of Haemonchus contortus. Vet. Parasitol. 2016;220:77–82. doi: 10.1016/j.vetpar.2016.02.031. [DOI] [PubMed] [Google Scholar]
- Rufener L., Mäser P., Roditi I., Kaminsky R. Haemonchus contortus acetylcholine receptors of the DEG-3 subfamily and their role in sensitivity to monepantel. PLoS Pathog. 2009;5:e1000380. doi: 10.1371/journal.ppat.1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales N., Love S. Resistance of Haemonchus sp. to monepantel and reduced efficacy of a derquantel/abamectin combination confirmed in sheep in NSW, Australia. Vet. Parasitol. 2016;228:193–196. doi: 10.1016/j.vetpar.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Scott I., Pomroy W.E., Kenyon P.R., Smith G., Adlington B., Moss A. Lack of efficacy of monepantel against Teladorsagia circumcincta and Trichostrongylus colubriformis. Vet. Parasitol. 2013;198:166–171. doi: 10.1016/j.vetpar.2013.07.037. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Brom R., Moll L., Kappert C., Vellema P. Haemonchus contortus resistance to monepantel in sheep. Vet. Parasitol. 2015;209:278–280. doi: 10.1016/j.vetpar.2015.02.026. [DOI] [PubMed] [Google Scholar]