Abstract
The chloroplast genomes of many algae and almost all land plants carry two identical copies of a large inverted repeat (IR) sequence that can pair for flip-flop recombination and undergo expansion/contraction. Although the IR has been lost multiple times during the evolution of the green algae, the underlying mechanisms are still largely unknown. A recent comparison of IR-lacking and IR-containing chloroplast genomes of chlorophytes from the Ulvophyceae (Ulotrichales) suggested that differential elimination of genes from the IR copies might lead to IR loss. To gain deeper insights into the evolutionary history of the chloroplast genome in the Ulvophyceae, we analyzed the genomes of Ignatius tetrasporus and Pseudocharacium americanum (Ignatiales, an order not previously sampled), Dangemannia microcystis (Oltmannsiellopsidales), Pseudoneochloris marina (Ulvales) and also Chamaetrichon capsulatum and Trichosarcina mucosa (Ulotrichales). Our comparison of these six chloroplast genomes with those previously reported for nine ulvophyceans revealed unsuspected variability. All newly examined genomes feature an IR, but remarkably, the copies of the IR present in the Ignatiales, Pseudoneochloris, and Chamaetrichon diverge in sequence, with the tRNA genes from the rRNA operon missing in one IR copy. The implications of this unprecedented finding for the mechanism of IR loss and flip-flop recombination are discussed.
Introduction
Descendants from cyanobacteria, the chloroplasts (plastids) of photosynthetic eukaryotes possess their own genome that encodes components necessary for replication and expression as well as for diverse functions of the chloroplasts1, 2. Chloroplast genes exhibit a predominantly uniparental mode of inheritance and their substitution rates are moderate3, thus providing useful markers for phylogenetic investigations. The low complexity and high copy number of organelle genomes greatly facilitate their characterization. Owing to these properties and the advent of next-generation sequencing, the pace at which chloroplast genome sequences are being produced has dramatically increased during the last five years4. As of December 2016, 1382 chloroplast genome sequences of green plants were publicly available in the Organelle Genome Resources of the NCBI reference sequence project, the majority of which (1276/1382) are derived from land plants. In addition to clarifying phylogenetic relationships, comparative chloroplast genome studies in green plants (green algae + land plants) have shown that this genome experienced a variety of changes through time, with the green algae exhibiting a much wider range of architectural diversity compared to their land plant homologs5–13.
Although much has been learned about chloroplast genome evolution in green plants, several questions still need to be addressed. One of them concerns the loss of the rRNA operon-encoding inverted repeat (IR), a structural feature conserved in many algae2 and almost all land plants3, 14. The two copies of the IR sequence, which varies in size from 6.0 kb (in Pseudendoclonium akinetum 15) to 76 kb (in Pelargonium X hortorum 16) and contains five to 40 genes, divide the genome into two distinct single copy (SC) regions and thus create a quadripartite structure. Considering its presence across all major green plant lineages, the IR has undoubtedly functional importance, with suggested roles in replication initiation17, genome stabilization18 and gene conservation18, 19. The two IR copies undergo frequent intramolecular recombination to produce isomeric forms that differ in the relative orientations of the SC regions20. This flip-flop recombination may help to maintain the gene complement of each SC region by reducing illegitimate recombination between opposite SC regions, especially if short dispersed repeats are not abundant21, 22. When mutations occur within the IR, sequence homogeneity of the two IR copies is maintained by gene conversion. In a variety of land plants, with the notable exception of Pelargonium 23, genes in the IR evolve three-fold more slowly than those in the SC regions, supporting the hypothesis that the copy-correction mechanism is biased against new mutations19, 24–26.
The IR has also the ability to undergo slight expansion or contraction, a phenomenon called the ebb and flow27. Each event of expansion/contraction usually involves no more than a few hundred nucleotides. Despite frequent variations in IR/SC boundaries, the overall quadripartite structure shows a high degree of conservation in land plants, as shown by the retention of similar gene contents in the large and small SC regions (LSC and SSC regions) of different taxa and the absence of the IR in only a few lineages (e.g. gymnosperms, legumes, Erodium)3, 14. But the situation differs sharply in green algae, especially in the Chlorophyta — the lineage sister to the Streptophyta (charophytes + land plants) — where the quadripartite structure has been remodeled extensively5, 8, 10, 13, 15, 28, 29 and the IR has been lost in multiple lineages6, 8, 11, 13, 30, 31. IR losses took place at least six times in charophytes9, four times in prasinophytes8, seven times in the Trebouxiophyceae13, twice in the Ulvophyceae31 and once in the Chlorophyceae6. Currently, there exists no convincing evidence for the creation of an IR de novo from an IR-less chloroplast genome.
Although the mechanisms that led to IR loss are still largely unknown, three possible models have been envisioned. First, IR loss might be the ultimate consequence of repeated events of IR contraction; however, no compelling evidence supports this hypothesis. Indeed, all green plant IRs, with a single known exception (the IR of the angiosperm Monsonia 32), contain the complete set of genes making up the rRNA operon, implying that erosion of the IR appears to be impeded when the IR/SC boundaries reach the 5′ or 3′ ends of this operon. Second, complete excision of one of the IR sequence might occur in a single step through intramolecular recombination between short direct repeats at the endpoints of this IR sequences. Comparative analyses of some IR-lacking chloroplast genomes with their IR-containing relatives are consistent with this mechanism of IR loss (e.g. in Coleochaete 9). Third, a more recent study in which gene orders of the IR-lacking genome of the ulvophycean green alga Gloeotilopsis planctonica (Ulotrichales) and the IR-containing genome of Pseudendoclonium akinetum (Ulotrichales) were compared led to the hypothesis that IR loss might occur through differential elimination of the gene sequences making up the rRNA operon in the two IR copies31. Obviously, identifying ulvophycean genomes carrying IR copies with differing gene content would provide strong support for this hypothesis.
In the present study, we analyzed the chloroplast genomes of six additional representatives of the Ulvophyceae, with the goals of gaining deeper insights into the mechanism of IR loss, the variability of the quadripartite structure, the dynamics of intron contents, and the phylogeny of chlorophytes. The freshwater unicellular Ignatius tetrasporus and Pseudocharacium americanum are members of the Ignatiales, an order that has not previously been sampled for genome analysis, while the other taxa are the marine unicellular flagellate Dangemannia microcystis (Oltmannsiellopsidales), the coccoid unicellular Pseudoneochloris marina (Ulvales) and the filamentous Chamaetrichon capsulatum and Trichosarcina mucosa (Ulotrichales). Here, we describe these ulvophycean genomes and compare them with nine previously reported genomes of the Ulvophyceae. We also present the phylogenomic trees we inferred from the chloroplast genome sequences of 100 chlorophytes. Our results have broad implications for the evolution of the IR and the chloroplast genome in general. All six newly examined genomes feature an IR, but remarkably the IR copies of the Ignatiales, Pseudoneochloris, and Chamaetrichon carry divergent sequences. The consequences of this unprecedented finding for the mechanism of IR loss and the recombinational processes to which the IR is subjected are discussed in this report.
Results
General features
The chloroplast genome sequences of the six newly sampled taxa were assembled as circular-mapping molecules, with sizes ranging from 135 kb (for Pseudoneochloris, the representative of the Ulvales) to 239 kb (for the members of the Ignatiales) (Table 1 and Supplementary Figs S1–S6). The summary statistics of these sequence assemblies are reported in Supplementary Table S1 and the general features of the genomes are compared to those of previously examined ulvophyceans in Table 1. All six genomes contain more than one copy of a sequence that encodes the genes making up the rRNA operon; this sequence is designated hereafter as the IR even though the copies differ in both size and sequence in four of the taxa: the two representatives of the Ignatiales (Ignatius and Pseudocharacium), the ulvalean Pseudoneochloris and the ulotrichalean Chamaetrichon (Table 1 and Fig. 1a). Remarkably, the latter taxon boasts three copies of the IR, instead of two. Most of the genome size variation observed among ulvophycean taxa can be attributed to variations in intron content and lengths of intergenic and coding regions (Fig. 1b) as well as to differences in IR size (Fig. 1a). The genomes of Ignatius, Pseudocharacium and the ulotrichalean Trichosarcina, which are the largest among the six newly examined taxa, have a moderate number of introns but the highest amount of intergenic sequences (Table 1 and Fig. 1b). Moreover, they exhibit the highest G + C content and the greatest proportion of dispersed repeats ≥30 bp (Table 1 and Fig. 1b).
Table 1.
General features of ulvophycean chloroplast genomes.
| Taxona | Accession | Size (bp) | A + T (%) | Genesb (no.) | Introns (no.)c | Repeatsd (%) | ||
|---|---|---|---|---|---|---|---|---|
| Genome | IRA/IRB/IRC | GI | GII | |||||
| Bryopsidales | ||||||||
| Tydemania expeditionis FL1151 | NC_026796 | 105,200 | — | 67.2 | 109 | 8 | 3 | 0.4 |
| Bryopsis hypnoides | NC_013359 | 153,426e | — | 66.9 | 108f | 6 | 6 | 9.9 |
| Bryopsis plumosa West4718 | NC_026795 | 106,859 | — | 69.2 | 108 | 7 | 6 | 2.4 |
| Ignatiales | ||||||||
| Ignatius tetrasporus UTEX 2012* | KY407659 | 239,387 | 7,848/7,431 | 63.0 | 107 | 7 | 2 | 13.3 |
| Pseudocharacium americanum UTEX 2112* | KY407658 | 239,448 | 7,848/7,427 | 63.0 | 107 | 7 | 2 | 13.3 |
| Oltmannsiellopsidales | ||||||||
| Oltmannsiellopsis viridis NIES 360 | NC_008099 | 151,933 | 18,510/18,510 | 59.5 | 104 | 5 | 0 | 11.1 |
| Dangemannia microcystis SAG 2022* | KY407660 | 166,355 | 12,407/12,407 | 66.3 | 106 | 7 | 1 | 6.0 |
| Ulvales | ||||||||
| Pseudoneochloris marina UTEX 1445* | KY407657 | 134,753 | 7,524/5,784 | 70.7 | 102 | 7 | 8 | 1.4 |
| Ulva sp. UNA00071828 | KP720616 | 99,983 | — | 74.7 | 100 | 4 | 1 | 0.5 |
| Ulva fasciata | NC_029040 | 96,005 | — | 75.1 | 100 | 4 | 1 | 0.5 |
| Ulotrichales | ||||||||
| Chamaetrichon capsulatum UTEX 1918* | KY407661 | 189,599 | 5,189/4,765/5191 | 69.2 | 104 | 15 | 1 | 2.7 |
| Pseudendoclonium akinetum UTEX 1912 | NC_008114 | 195,867 | 6,039/6,039 | 68.5 | 105 | 27 | 0 | 5.3 |
| Trichosarcina mucosa SAG 4.90* | KY407656 | 227,181 | 8,979/8,979 | 62.8 | 103 | 7 | 7 | 14.4 |
| Gloeotilopsis planctonica SAG 29.93 | KX306824 | 221,431 | — | 68.5 | 104 | 14 | 17 | 3.7 |
| Gloeotilopsis sarcinoidea UTEX 1710 | KX306821 | 262,888 | — | 68.5 | 104 | 15 | 12 | 11.6 |
aThe taxa newly examined in this study are denoted by asterisks. bIntron-encoded genes and freestanding ORFs not usually found in green plant chloroplast genomes were not considered. Duplicated genes were counted only once. cNumbers of group I (GI) and group II (GII) introns are given. dNonoverlapping repeat elements were mapped on each genome with RepeatMasker using as input sequences the repeats of at least 30 bp identified with REPuter. eThis value is based on the re-annotated version of Leliaert & Lopez-Bautista35. fThis value includes four genes with frameshift mutations (rpoB, rpoC1, ycf20 and ycf47).
Figure 1.
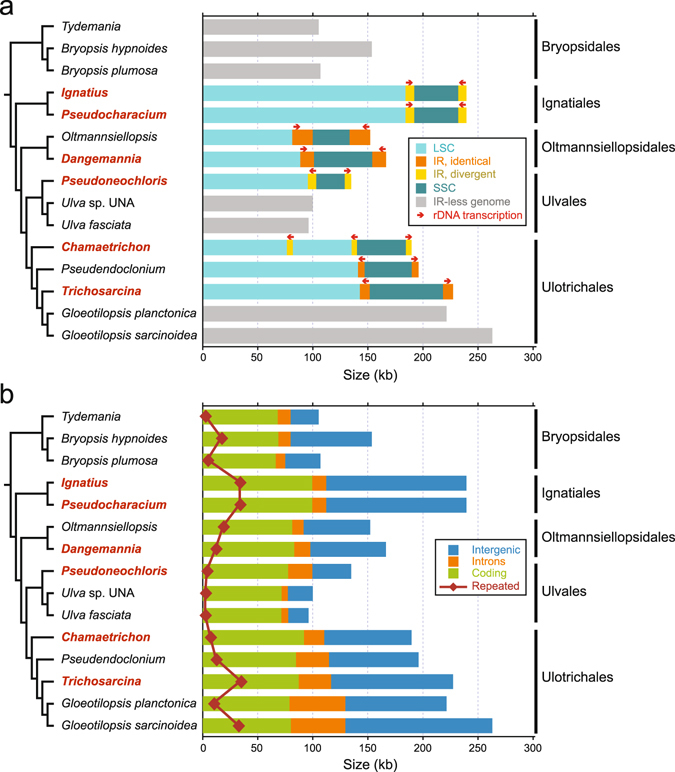
Sequence coverages in the 15 ulvophycean chloroplast genomes compared in this study. (a) Sizes of the SSC, IR and LSC regions. Red arrows indicate the direction of transcription of the rRNA operon in IR-containing genomes. Genomes lacking the IR are represented in grey. The names of the newly examined taxa are indicated in red. (b) Amounts of coding, intronic, intergenic and small repeated sequences (≥30 bp). Note that intron-encoded genes were not considered as coding sequences but rather as intron sequences. The phylogenetic relationships among the taxa examined are derived from Fig. 2b.
The genomes of the Ignatiales were found to be the most similar in our study group: they can be aligned over their entire length and differ only at a few sites. This observation raises some doubt about the classification of Ignatius and Pseudocharacium into separate genera. These two taxa were initially distinguished from each other by the fact that cells of Pseudocharacium grow on green algal substrates like Characium spp.; however, Watanable and Nakayama33 detected no ultrastructural difference between these algae and observed high sequence identity between their 18S rDNAs.
Phylogenomic analyses
Before comparing the gene content and gene organization of the examined genomes, we present here the phylogenetic context required to interpret these results. Our chloroplast phylogenomic analyses were carried out using amino acid and nucleotide data sets that included 102 green algal taxa (100 chlorophytes and the streptophytes Mesostigma and Chlorokybus). The amino acid data set (PCG-AA, 14,144 sites) was generated using 79 protein-coding genes, whereas the nucleotide data set (PCG12RNA, 31,893 sites) was assembled from the same set of protein-coding genes (first two codon positions) plus 29 RNA-coding genes (three rRNA and 26 tRNA genes) (see Methods for the gene list). To reduce among-site compositional heterogeneity and thus minimize systematic errors of phylogenetic reconstructions, the most rapidly evolving sites were eliminated from the two data sets. This strategy has recently been reported to produce more robust chloroplast phylogenomic inferences of deep divergences among green algae34. The gene locations of the sites that were removed are indicated in Supplementary Fig. S7; they account for 8.4% and 12.8% of the original data from which the PCG-AA and PCG12RNA data sets were derived. Each data set comprises only a small proportion of missing data (8.1% for the PCG-AA and 6.9% for the PCG12RNA data sets).
The inferred topologies were dependent upon the data set and the method of analysis, differing mainly with respect to the relative positions of the major lineages in the core Chlorophyta, i.e. the clade sister to prasinophyte lineage VIIA (Fig. 2a and Supplementary Figs S8–S11). Analyses of the PCG12RNA data set using RAxML and PhyloBayes revealed that the class Ulvophyceae is sister to the Chlorophyceae, albeit with no statistical support. In contrast, both the RAxML and PhyloBayes analyses of the PCG-AA data set identified the Ulvophyceae as non-monophyletic, again with no statistical support, with either the Bryopsidales or the clade formed by the Ignatiales, Oltmansiellopsidales, Ulvales, Ulotrichales as sister to the Chlorophyceae. Identical relationships were recovered for the 12 taxa forming the latter clade in the RAxML trees inferred from the two data sets as well as the Bayesian tree inferred from the nucleotide data set (Fig. 2b). In these trees, the Ignatiales was the earliest-diverging lineage, immediately followed by the Oltmannsiellopsidales, and then by the Ulvales and Ulotrichales. The Oltmannsiellopsidales was instead recovered as the earliest divergence in the Bayesian tree inferred from the amino acid data set.
Figure 2.
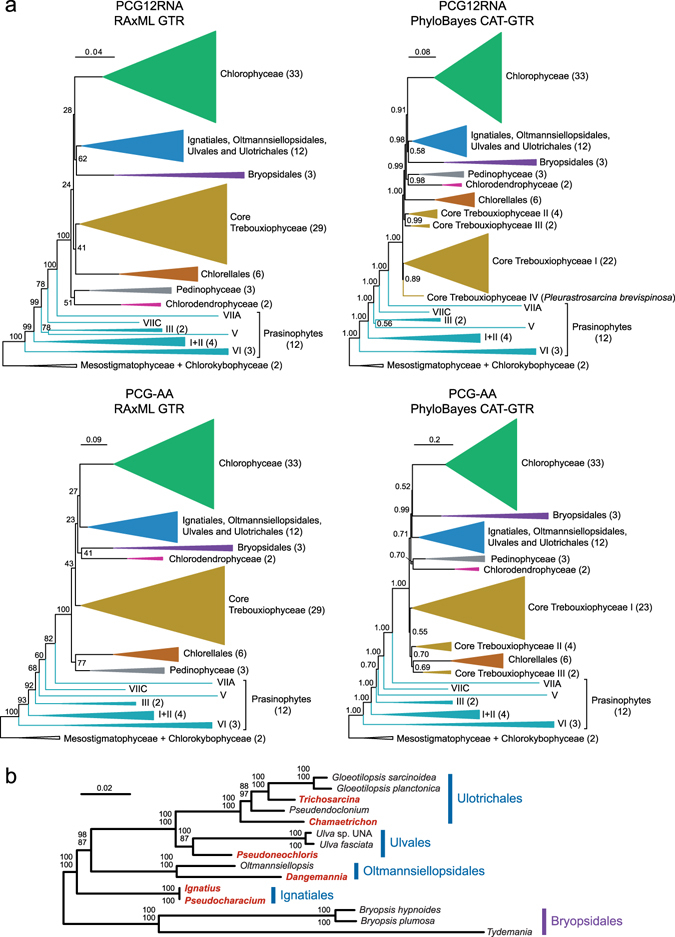
Chloroplast phylogenomic trees of chlorophytes inferred from the PCG-AA and PCG12RNA data sets using RAxML and PhyloBayes. (a) Relationships among the major lineages of the Chlorophyta. Strongly supported clades are represented as triangles with sizes proportional to the number of taxa (indicated in parentheses). Bootstrap support and posterior probability values are reported on the nodes. (b) Relationships among ulvophycean taxa. The best-scoring RAxML tree inferred from the PCG12RNA data set is presented. Bootstrap support values are reported on the nodes: from top to bottom are shown the values for the RAxML analyses of the PCG12RNA and PCG-AA data sets. The names of the newly examined taxa are indicated in red.
Gene content
The compared ulvophycean genomes share 95 genes coding for 67 proteins, three rRNAs (rrs, rrl and rrf), and 25 tRNAs (see legend of Supplementary Fig. S12 for the list of common genes). Although rpoB, rpoC1 and ycf20 sequences were detected in every taxon, these genes were not included in the set of shared genes because frameshift mutations were present in the Bryopsis hypnoides genome (Supplementary Fig. S12). Instead of being the consequence of pseudogeneization, these nucleotide changes could be the result of sequencing errors, as such errors were previously detected in other genes of the same genome sequence35. From the gene complements of the 15 compared genomes, we predicted that 113 canonical genes were present in the common ancestor of ulvophycean algae and that 15 of them (nine protein-coding genes and six tRNA genes) experienced total loss from the chloroplast in the course of evolution (Supplementary Fig. S12). Seven protein-coding genes (chlB, chlL, chlN, cysA, cysT, rpl12 and ycf47) were lost only once, with these loss events mapping at two deep nodes and a terminal branch of the ulvophycean phylogeny. The two remaining protein-coding genes (minD and tilS) were lost on two or three occasions.
Besides canonical genes, we identified freestanding open reading frames (ORFs) showing similarities (E-value threshold of 1e-06) with recognized protein domains or previously reported ORFs of unknown function in four of the newly sequenced genomes (Supplementary Table S2). These ORFs encode proteins with domains characteristic of HNH endonucleases, group II intron maturases, reverse transcriptases, and DNA breaking-rejoining enzymes (recombinase/integrase).
Quadripartite architecture and genome rearrangements
By comparing the gene contents of the LSC and SSC regions in the investigated ulvophycean genomes, one can observe that the Ignatiales, Oltmannsiellopsidales, and the Ulvales + Ulotrichales exhibit distinct quadripartite architectures (Fig. 3). As previously shown for Oltmannsiellopsis 29, the longest SC regions in the genomes of Dangemannia and the two representatives of the Ignatiales correspond to the SSC region of the ancestral core chlorophyte genome. Although the Oltmannsiellopsis and Dangemannia LSC regions have exactly the same gene complement, the SSC region of Dangemannia differs from its Oltmannsiellopsis counterpart by the presence of three extra genes (psbA, petA and petB) which are located in the IR of the latter taxon, thus indicating that the IR underwent contraction/expansion towards the SSC in the Oltmannsiellopsidales. The IRs of the Oltmannsiellopsidales are currently the only known ulvophycean IRs housing protein-coding genes. The IR-containing genomes of the Ulvales and Ulotrichales most closely resemble the ancestral core chlorophyte genome in term of gene partitioning pattern: for instance, only seven of the 23 genes (psaA, the atpA-atpF-atpH-atpI-rps2 cluster, and psaB) in the SSC region of the ulvalean Pseudoneochloris are missing from the SSC of the core chlorophyte ancestor. In the ulotrichalean Chamaetrichon, the IRB and IRC copies reside at the same locations as the IR sequences in Pseudoneochloris and other ulotrichaleans, while IRA is inserted between psbB and trnR(ucu), two genes forming a conserved pair in the Ulvales and Ulotrichales. With regards to the IR-lacking genomes from the latter lineages, we note that the gene partitioning pattern characteristic of the IR-containing genomes has been more highly preserved in the Ulotrichales than in the Ulvales.
Figure 3.
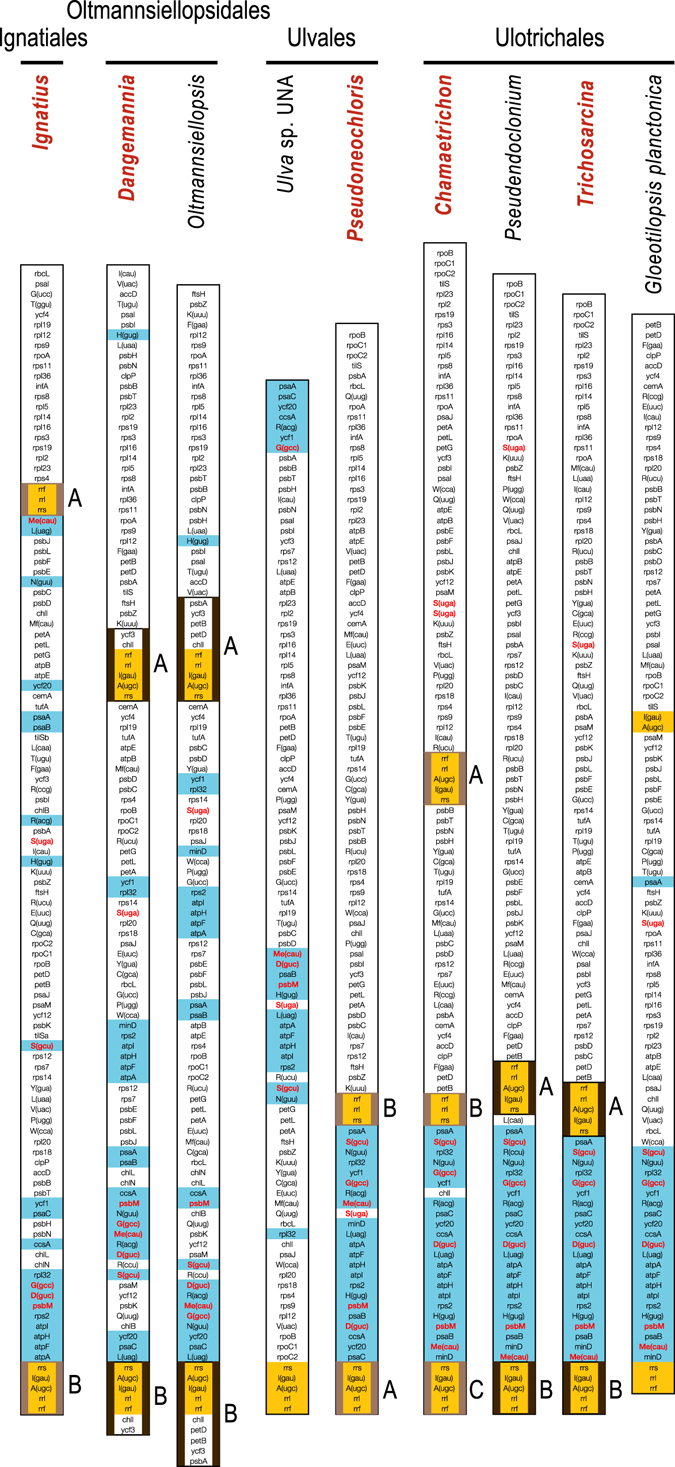
Gene partitioning patterns of ulvophycean chloroplast genomes. The suite of genes in each IR-containing genome is displayed so that the SC region with the gene content the most similar to that predicted for the ancestral SSC region of core chlorophytes is presented at the bottom of the figure. Thick vertical lines delimit the genes encoded in the IR (thick black lines, identical IR copies; thick brown lines, divergent IR copies). The genes making up the rDNA operon are highlighted in yellow whereas those present in the SSC region of Trichosarcina are highlighted in blue. Red letterings designate the genes of ancestral LSC origin that have been acquired by the IRs of core chlorophytes.
To compare the overall gene organization among ulvophyceans chloroplast genomes, we estimated, using MGR v2.0336 and a data set of 98 genes, the numbers of reversals that would be required to interconvert gene order in all possible pairs of genomes. The resulting reversal distances were used to construct the gene rearrangement tree shown in Fig. 4, which is based on the ulvophycean phylogeny reported in Fig. 2b. The results revealed that the chloroplast genomes of the Ignatiales are the least rearranged relative to those of the Bryopsidales. Consistent with the results reported above, the IR-containing genomes sharing the same quadripartite architecture are the most similar in overall gene order. Moreover, gene order is more conserved between IR-lacking and IR-containing genomes in the Ulotrichales than in the Ulvales.
Figure 4.
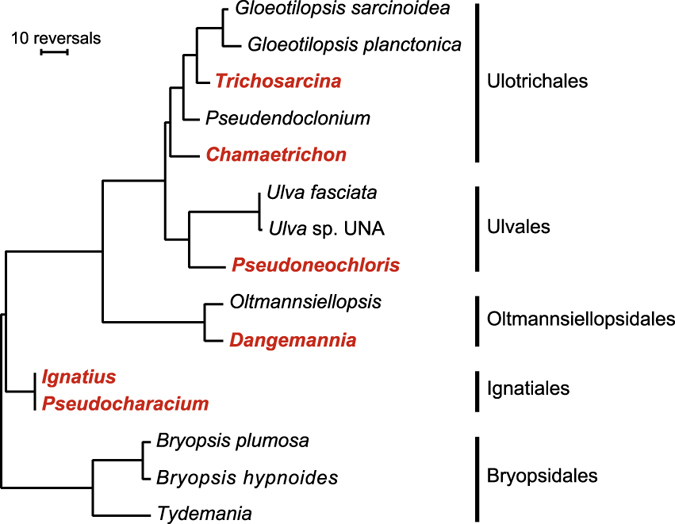
Rearrangements of gene order among ulvophycean chloroplast genomes. The numbers of gene reversals were estimated using MGR v2.03 and the tree topology shown in Fig. 2b. The gene order data set contained 98 genes and included three conserved genes carrying frameshift mutations in Bryopsis hypnoides (rpoB, rpoC1 and ycf20). The names of the newly examined taxa are indicated in red.
Divergent IR copies and their Influence on flip-flop recombination
Among the newly sequenced ulvophycean genomes, identical IR copies are found only in the Oltmannsiellopsidales and the ulvalean Trichosarcina. The ulvophycean genomes carrying non-identical IR copies, i.e. those of the Ignatiales, Pseudoneochloris and Chamaetricon, are all missing trnI(gau) and trnA(ugc) in one of their IR copies (Figs 3 and 5). IR sequence divergence, however, is not limited to deletion of these tRNA genes. In the Ignatiales, the entire region between rrs and rrl in the IR copy devoid of tRNA genes (IRA) lacks sequence similarity with the corresponding region in the other IR copy (IRB). Moreover, these regions are substantially larger than their homologs in most other algal chloroplast rRNA operons. In the ulvalean Pseudoneochloris, the IR copy with the deleted tRNA genes (IRB) lacks the three intron ORFs present in rrs and rrl; in addition, the rrs/rrl spacer of 7 bp in the same copy reflects the deletion of a non-coding sequence relative to the corresponding region in IRA. In the ulotrichalean Chamaetrichon, the IR copy missing the tRNA genes (IRB) has clearly lost sequences in the intergenic regions surrounding these genes and the rrl/rrf spacer has diverged considerably from the corresponding sequences in the two other IR copies. Finally, the IR copies exhibit nucleotide differences in the rRNA genes: we detected a single nucleotide polymorphism in the Ignatiales (5′ end of rrs), 16 in Pseudoneochloris (10 at the 5′end of rrs and the others at the edges of the rrl exons), and 8 to 21 nucleotide differences in Chamaetrichon (IRA and IRB are the most similar, with six polymorphisms located in rrl, while the IRA/IRC and IRB/IRC comparisons revealed 17 and 21 polymorphisms, respectively, most occurring in rrs).
Figure 5.
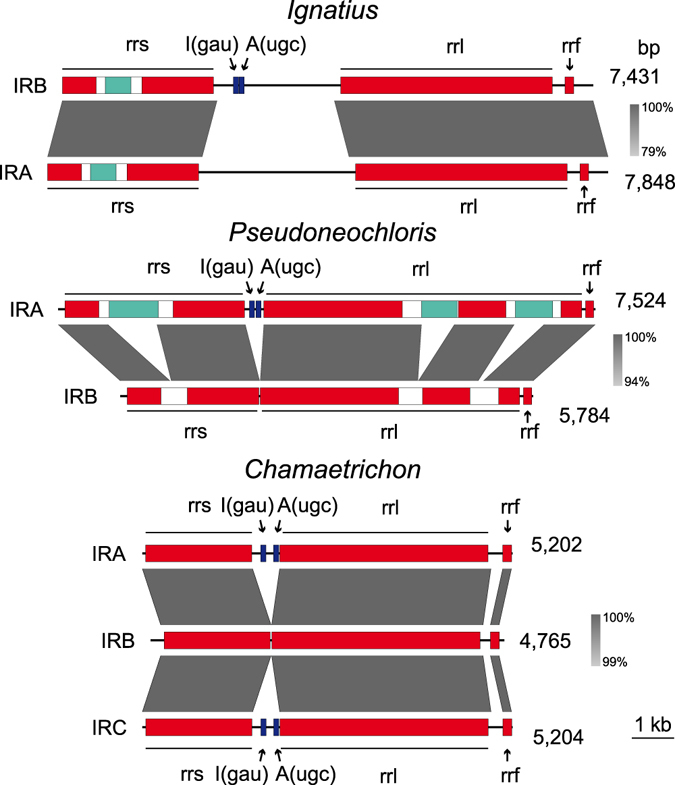
Comparison of IR sequences in the ulvophycean chloroplast genomes carrying non-identical IR copies. Regions displaying similar sequences are connected by shaded areas, with sequence identity denoted by the grey scale. Red and dark blue boxes represent coding regions of rRNA and tRNA genes, respectively; turquoise and white boxes represent ORFs and noncoding regions within introns, respectively.
We undertook a PCR approach to determine whether the divergent IR copies of Ignatius and Pseudoneochloris participate in flip-flop recombination (Fig. 6). For the PCR assays with Ignatius, two primers specific to opposite ends of the SSC region (primers 4 and 8) were used in combination with primers specific to internal sites within the IRA and IRB (primers 3 and 7), while two primers specific to opposite ends of the LSC region (primers 1 and 5) were used in combination with primers specific to IRA and IRB sites (primers 2 or 6). All eight assays yielded products of the expected sizes, indicating that flip-flop recombination occurs in the chloroplast of Ignatius. For Pseudoneochloris, four PCR assays were designed to generate products that span both the IR/SSC and IR/LSC junctions using combinations of primers specific to genes within the SSC (primers 1 and 3) and LSC (primers 2 and 4) regions. Only two of these essays yielded products, indicating that each IR copy is surrounded by SC regions with a fixed orientation and hence that flip-flop recombination does not take place. Four additional PCR assays, each carried out using a SC-specific primer and a primer complementary to a sequence shared by the IRA and IRB (primers 5 or 6), yielded essentially the same conclusions and confirmed the identities of the genes on either side of the two divergent IR copies.
Figure 6.
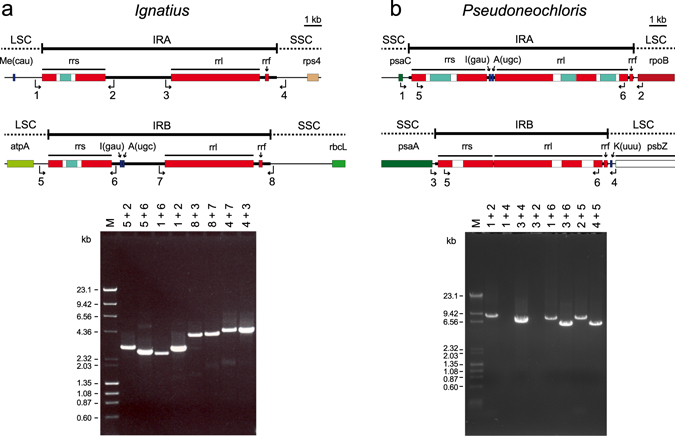
Analysis of flip-flop recombination in the Ignatius (a) and Pseudoneochloris (b) chloroplast genomes. PCR assays using eight different pairs of primers were carried out to test whether the non-identical IRs in each algal genome undergo homologous recombination. Primer locations and polarities are indicated by numbered arrows on the diagrams showing the organizations of the IR copies (see Supplementary Table S3 for the primer sequences). PCR products were analyzed by electrophoresis on agarose gels; the numbers above the gel lanes indicate the combinations of primers used.
Intron distribution
All six newly examined ulvophycean taxa have seven group I introns in their chloroplast genome, except Chamaetrichon which holds 15 (Table 1). Most of these introns occupy insertion sites that have been previously reported (Fig. 7a). Although two introns in the Ignatiales (chlL_210 and rbcL_462) and two in Chaemaetrichon (psaB_1769 and rpl16_324) represent novel insertion sites for the Ulvophyceae, only the position of the Chaemaetrichon rpl16_324 intron has not been described in other groups of chlorophytes. The intron distribution is irregular, with numerous sites shared between distant lineages of the Ulvophyceae. Homing endonucleases of the LAGLIDADG, GIY-YIG, and HNH families are encoded by ulvophycean group I introns, with the LAGLIDADG genes being the most represented. It is noteworthy that introns sharing a given site always carry the same type of homing endonuclease gene.
Figure 7.
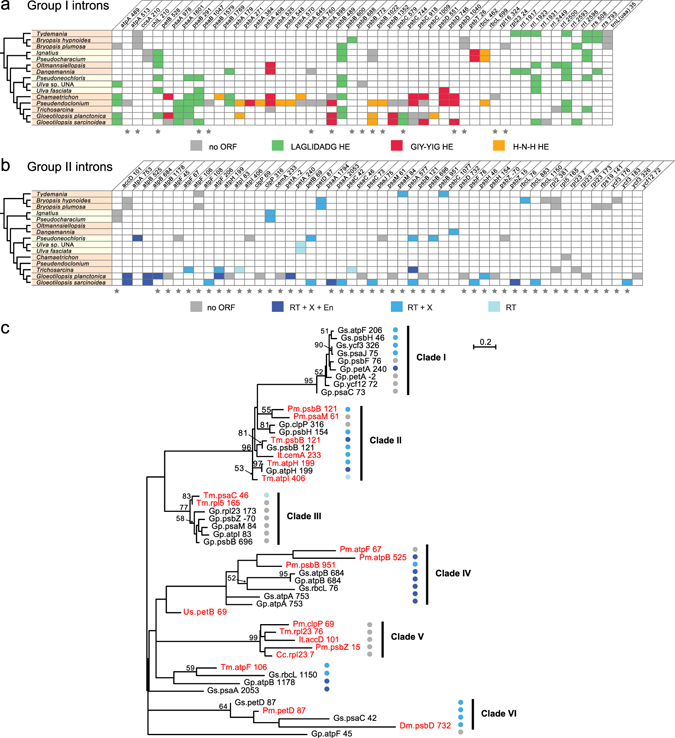
Introns in ulvophycean chloroplast genomes. (a) Distribution of group I introns. (b) Distribution of group II introns. A grey box denotes an intron lacking an ORF, whereas a colored box represents an intron containing an ORF (see the color code for the type of intron-encoded protein). Stars denote the intron insertion sites that have not been observed in other groups of chlorophytes. Intron insertion sites in protein-coding and tRNA genes are given relative to the corresponding genes in the Mesostigma chloroplast genome67; insertion sites in rrs and rrl are given relative to E. coli 16S and 23S rRNAs, respectively. For each insertion site, the position corresponding to the nucleotide immediately preceding the intron is reported. Abbreviations: EN, H-N-H endonuclease; RT, reverse transcriptase; X, intron maturase. (c) Phylogenetic relationships among group II introns of the Ignatiales, Oltmannsiellopsidales, Ulvales and Ulotrichales. The tree shown here was inferred by RAxML analysis of an alignment of 124 nucleotides corresponding to domains IA, IVB, V and VI of the core secondary structure. Bootstrap support values higher than 50% are reported on the nodes. Colored circles denote the types of intron ORF (see the color code on panel b). Note that clades I through IV were identified in a previous phylogenetic study of Gloeotilopsis group II introns31. Names of the taxa newly examined in the present investigation are indicated in red. Abbreviations: Cc, Chamaetrichon capsulatum; Dm, Dangemannia microcystis; Gp, Gloeotilopsis planctonica; Gs, Gloeotilopsis sarcinoidea; It, Ignatius tetrasporus; Pm, Pseudoneochloris marina; Tm, Trichosarcina mucosa; Us, Ulva sp. UNA00071828.
Of the six newly examined ulvophyceans, Pseudoneochloris and Trichosarcina are the only taxa carrying more than two group II introns (Table 1). The 21 group II introns we annotated represent 19 insertion sites (only the ignatialean taxa share introns at the same sites): 15 and 13 of these intron positions have not been previously observed in the Ulvophyceae and chlorophytes, respectively (Fig. 7b). All six ulvophycean taxa contain introns with ORFs; in total, 12 introns encode proteins with reverse transcriptase, intron maturase and/or H-N-H endonuclease domains (Fig. 7b).
To delineate the relationships among the 49 introns currently known in the Ignatiales, Oltmannsiellopsidales, Ulvales and Ulotrichales, a global alignment of 124 nucleotides corresponding to their core secondary structures (domains IA, IVB, V and VI) was submitted to phylogenetic analysis using RAxML under the GTR + G4 model (Fig. 7c). All 21 introns uncovered in the present study, with the exception of the five clustering in clade V (all are ORF-less introns), fall within clades identified in our recent analysis of Gloeotilopsis introns31. Aside from clades I and III which contain exclusively ulotrichalean introns, all others include members from two or three distinct lineages.
Discussion
The six newly sequenced IR-containing chloroplast genomes we analyzed in this study highlight the diversity of genome architectures found in the Ulvophyceae. Prior to our investigation, only the IR-containing chloroplast genomes of Oltmannsiellopsis 29 and of the ulotrichalean Pseudendoclodium 15 were available for the Ulvophyceae. Early on, it was recognized that their quadripartite structures were distinct from one another and from those of all other chlorophyte genomes that were completely sequenced at the time29. In both ulvophycean chloroplast genomes, the SC region encoding the genes usually found in the SSC region of ancestral-type prasinophyte genomes exhibits extra genes, with this SC region being the shortest of the unique regions in Pseudendoclonium but the longest in Oltmannsiellopsis. Here we report that the ignatialean chloroplast genomes differ substantially in gene partitioning compared to their ulvophycean relatives (Fig. 3). Sampling of a second member of the Oltmannsiellopsidales (Dangemannia) revealed that the IR underwent expansion/contraction in this lineage, an event that involved three protein-coding genes but no further modification to the gene contents of the SC regions. In addition, characterization of the first ulvalean IR-containing chloroplast genome (Pseudoneochloris) and of two IR-containing chloroplast genomes from the Ulotrichales (Chamaetrichon and Trichosarcina) disclosed a quadripartite structure identical or highly similar to that of Pseudendoclonium. The gene partitioning pattern observed for the Ulvales/Ulotrichales is the closest to that predicted for the common ancestor of all core chlorophytes10, 13. As observed for some lineages of the Trebouxiophyceae (Prasiola and Parietochloris clades)13, psbM and a set of five tRNA genes (trnD(guc), trnG(gcc), trnMe(cau) trnS(gcu), trnS(uga)) predicted to have been present in the IR of this ancestor10 were transferred to the adjacent SSC region early during the evolution of the Ulvophyceae as a result of IR contraction.
Our finding of divergent IR sequences in the chloroplast genomes of the Ignatiales, Pseudoneochloris (Ulvales) and Chamaetrichon (Ulotrichales) is an unprecedented observation for the Viridiplantae. In these ulvophycean genomes, one of the IR copies contains all five genes making up the standard rRNA operon, whereas a second copy is missing both trnI(gau) and trnA(ugc) in the ribosomal intergenic spacer (Fig. 5). The latter copy of the Pseudoneochloris IR is also missing three LAGLIDADG endonuclease genes in the group I introns of the rrs and rrl genes. Non-identical IR copies featuring indels have been previously observed in the chloroplast genomes of haptophytes belonging to the Prymnesiales and Phaeocystales and as reported here for the Ulvophyceae, the genes they encode are restricted to the rRNA operon37, 38. Each IR copy of Chrysochromulina tobin (Primnesiales) lacks a single tRNA gene (trnI(gau) or trnA(ugc)) in the ribosomal intergenic spacer37, whereas the situation for the IRs of Phaeocystis antartica and Phaeocystis globosa is identical to what we uncovered in our study, i.e. a standard rRNA operon in one IR copy and only the rRNA genes in the other38.
Despite the divergence of the IR sequences, we detected intramolecular recombination between the IR copies of Ignatius; however, no isomers were identified for the Pseudoneochloris genome (Fig. 6). The absence of flip-flop recombination in the latter genome is correlated with the accumulation of nucleotide polymorphisms in the IR copies. Similar observations (i.e. presence of polymorphisms and absence of recombination) were reported for the haptophyte Chrysochromulina 37, supporting the view that pairing of the two IR copies for recombination provides a copy-correction mechanism. What triggered the independent losses of the tRNA genes from the IR in the three distinct lineages of the Ulvophyceae? Why were the functional copies of these genes not used as templates for copy-correction of the non-canonical sequences? Was a mutation in a nuclear-encoded gene participating in DNA recombination or DNA repair involved? Even though further investigations are required to answer these questions, it appears that the events linked to the degeneration of the rRNA operon were complex and involved multiple steps.
Aside from the divergent IR copies located at the same positions as in other ulotrichalean chloroplast genomes (copies B and C), the Chamaetrichon genome contains a third IR copy (copy A) that is inserted between two genes forming a syntenic pair in the Ulotrichales and Ulvales (Fig. 3). To our knowledge, this is the first time that three copies of the rRNA genes are reported in a chloroplast genome of the Viridiplantae. Although compelling evidence for de novo creation of an IR from an IR-less chloroplast genome has not been documented, our finding of a third copy of the rDNA operon in Chamaetrichon makes this evolutionary scenario plausible.
The chloroplast IR has been entirely lost multiple times during the evolution of green algae6, 8, 9, 13, 31. In the cases of ulvophycean and haptophyte chloroplast genomes carrying short IRs, the process of IR sequence divergence and degeneration of the rRNA operon likely represents an intermediate step towards the complete loss of the IR. But to provide unambiguous evidence for or against this hypothesis, it will be necessary to investigate IR-less genomes from close relatives of taxa carrying divergent IR copies. Note, however, that the data reported here for the Ulotrichales are consistent with our recent comparison of gene order between the Pseudendoclonium and Gloeotilopsis genomes, which suggested that differential elimination of sequences within the rRNA operon from the two IR copies led to IR loss31.
Our study also highlights the diversity of both group I and group II introns in ulvophycean chloroplast genomes. Novel insertion sites were found to be more abundant for the group II introns, especially in the Ulvales and Ulotrichales (Fig. 7). Our phylogenetic analysis of ulvophycean group II introns uncovered a number of clades containing introns originating from different species and insertion sites. This observation suggests that several introns arose by intragenomic proliferation of existing introns, thus echoing our recent conclusions regarding the mobility of group II introns in Gloeotilopsis 31.
The Ignatiales affiliated with the Oltmansiellopsiales, Ulvales and Ulotrichales to form a strongly supported clade in all phylogenomic trees inferred in this study, but whether the Ignatiales or the Oltmansiellopsidales is the most basal lineage could not be identified unambiguously (Fig. 2). These results are not congruent with the ten-gene phylogenetic analyses of Cocquyt et al.39, which recovered Ignatius with the TBCD (Trentepohliales-Bryopsidales-Cladophorales-Dasycladales) clade, a large assemblage that contains the bulk of the green seaweeds. Previously reported phylogenies based on the nuclear small-subunit rRNA gene33 had revealed that Ignatius is either embedded in the Ulvales–Ulotrichales clade or clustered with the TBCD clade depending on the inference method used.
Although the major clades of core chlorophytes received high support in all trees we inferred, their precise placements were dependent upon the phylogenetic methods and data sets employed (Fig. 2a). As we pointed out earlier10, inference of more robust and reliable trees will probably require a broader sampling of chlorophytes and improved models of sequence evolution. Although the relationships among core chlorophyte lineages remain ambiguous, the newly sequenced ulvophycean genomes reported here strengthen the database of available genomes for future studies aimed at deciphering the phylogenetic relationships among members of specific lineages.
Materials and Methods
Strains and culture conditions
Ignatius tetrasporus UTEX 2012, Pseudocharacium americanum UTEX 2112, Pseudoneochloris marina UTEX 1445 and Chamaetrichon capsulatum UTEX 1918 were obtained from the culture collection of algae at the University of Texas in Austin, while Dangemannia microcystis SAG 2022 and Trichosarcina mucosa SAG 4.90 originated from the culture collection at the University of Goettingen. Ignatius and Pseudocharacium were grown in medium C40, while the remaining ulvophyceans were grown in medium K41. All cultures were incubated at 18 °C under alternating 12 h-light/12-h dark periods.
DNA isolation, sequencing and de novo assemblies
The chloroplast genomes of Ignatius, Pseudocharacium, Dangemannia, and Pseudoneochloris were sequenced using the Roche 454 method, whereas those of Chamaetricon and Trichosarcina were sequenced using the Illumina method.
For 454 sequencing, A + T-rich organellar DNA was separated from nuclear DNA by CsCl-bisbenzimide isopycnic centrifugation8. Shotgun libraries (700-bp fragments) of A + T-rich DNA were constructed using the GS-FLX Titanium Rapid Library Preparation Kit of Roche 454 Life Sciences (Branford, CT, USA). Library construction and 454 GS-FLX DNA Titanium pyrosequencing were carried out by the “Plateforme d’Analyses Génomiques de l’Université Laval” (http://pag.ibis.ulaval.ca/seq/en/). Following trimming of adapter and low-quality sequences with CUTADAPT42 and PRINTSEQ43, respectively, reads were assembled using Newbler v2.544 with default parameters, and contigs were visualized, linked and edited using CONSED v2245. Contigs of chloroplast origin were identified by BlastN and BlastX searches46 against a local database of green plant chloroplast genomes. Regions spanning gaps in the assemblies were amplified by polymerase chain reaction (PCR) with primers specific to the flanking sequences. Purified PCR products were sequenced using Sanger chemistry with the PRISM BigDye Terminator Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on ABI model 373 or 377 DNA sequencers (Applied Biosystems).
For Illumina sequencing, total cellular DNA was isolated using the EZNA HP Plant Mini Kit of Omega Bio-Tek (Norcross, GA, USA). Libraries of 500-bp fragments were constructed using the TrueSeq DNA Sample Prep Kit (Illumina, San Diego, CA, USA) and paired-end reads were generated on the Illumina HiSeq 2000 (100-bp reads) or the MiSeq (300-bp reads) sequencing platforms by the Innovation Centre of McGill University and Génome Québec (http://gqinnovationcenter.com/index.aspx) and the “Plateforme d’Analyses Génomiques de l’Université Laval”, respectively. Reads were trimmed to remove adapter and low-quality sequences with CUTADAPT42 and PRINTSEQ43, respectively, and the paired-end sequences were merged using FLASH47. The reads were then assembled using Ray v2.3.148 and contigs were visualized, linked and edited using CONSED v2245. Identification of chloroplast contigs and gap filling were performed as described above for the 454 sequence assemblies.
Genome annotations
We used a custom-built suite of bioinformatics tools allowing the automated execution of the following three steps: (1) ORFs were found using GETORF in EMBOSS49, (2) their translated products were identified by BlastP searches46 against a local database of chloroplast-encoded proteins or the nr database at the National Center for Biotechnology Information, and (3) consecutive 100-bp segments of the genome sequence were analyzed with BlastN and BlastX to determine the approximate positions of RNA-coding genes, introns and exons. Only the ORFs that revealed identities with genes of known functions or previously reported ORFs were annotated. The precise positions of rRNA and tRNA genes were identified using RNAmmer50 and tRNAscan-SE51, respectively. Intron boundaries were determined by manual modelling of intron secondary structures52, 53 and by comparing the sequences of intron-containing genes with those of intronless homologs. Circular genome maps were drawn with OGDraw54.
Genome-scale sequence comparisons were carried out with LAST v7.1.455. Comparisons of IR sequences were performed using EasyFig v2.2.256. To estimate the proportion of small repeated sequences, repeats with a minimal size of 30 bp were retrieved using REPFIND of REPuter v2.7457 and were masked on the genome sequence using RepeatMasker (http://www.repeatmasker.org/) running under the Crossmatch search engine (http://www.phrap.org/).
Phylogenomic analyses
The GenBank accession codes used to generate the amino acid and nucleotide data sets (PCG-AA and PCG12RNA, respectively) are provided in Supplementary Table S4. The PCG-AA data set was assembled from 79 protein-coding genes: accD, atpA, B, E, F, H, I, ccsA, cemA, chlB, I, L, N, clpP, cysA, T, ftsH, infA, minD, petA, B, D, G, L, psaA, B, C, I, J, M, psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, rbcL, rpl2, 5, 12, 14, 16, 19, 20, 23, 32, 36, rpoA, B, C1, C2, rps2, 3, 4, 7, 8, 9, 11, 12, 14, 18, 19, tufA, ycf1, 3, 4, 12, 20, 47, 62. It was prepared as follows: the deduced amino acid sequences from the individual genes were aligned using MUSCLE v3.758, the ambiguously aligned regions in each alignment were removed using TrimAl v1.359 with the options block = 6, gt = 0.7, st = 0.005 and sw = 3, and the protein alignments were concatenated using Phyutility v2.2.660. The 15,447 characters in the concatenated matrix were then sorted into 30 bins according to their rate of variation using TIGER v1.0261, and the 1,303 fastest evolving characters identified in bins 29 and 30 were removed from the matrix in an attempt to reduce among-site compositional heterogeneity in the data set.
Phylogenies were inferred from the PCG-AA data set using the maximum likelihood (ML) and Bayesian methods. ML analyses were carried out using RAxML v8.2.662 and the GTR + Γ4 model of sequence evolution; in these analyses, the data set was partitioned by individual gene, with the model applied to each partition. Confidence of branch points was estimated by bootstrap analysis with 100 replicates. Bayesian analyses were performed with PhyloBayes v4.163 using the site-heterogeneous CATGTR + Γ4 model64. Five independent chains were run for 2,000 cycles and consensus topologies were calculated from the saved trees using the BPCOMP program of PhyloBayes after a burn-in of 500 cycles. Note that the chains failed to converge under these conditions (maxdiff = 0.86), indicating that at least one of the chains was stuck in a local maximum.
The PCG12RNA data set was prepared from the first and second codon positions of the 79 protein-coding genes abovementioned and from three rRNA and 26 tRNA genes. The multiple sequence alignment of each protein was first converted into a codon alignment, poorly aligned and divergent regions in each codon alignment were excluded using Gblocks v0.91b65 with the −t = c, −b3 = 5, −b4 = 5 and −b5 = half options, and the individual gene alignments were concatenated using Phyutility v2.2.660. The third codon positions of the resulting alignment were then excluded using Mesquite v3.0466 to produce the PCG12 data set. To obtain the PCG12RNA, the PCG12 matrix was merged with the concatenated alignment of the following RNA genes: rrf, rrl, rrs, trnA(ugc), C(gca), D(guc), E(uuc), F(gaa), G(gcc), G(ucc), H(gug), I(cau), I(gau), K(uuu), L(uaa), L(uag), Me(cau), Mf(cau), N(guu), P(ugg), Q(uug), R(acg), R(ucu), S(gcu), S(uga), T(ugu), V(uac), W(cca), Y(gua). The latter genes were aligned using MUSCLE 3.758, the ambiguously aligned regions in each alignment were removed using TrimAl v1.359 with the options block = 6, gt = 0.9, st = 0.4 and sw = 3, and the individual alignments were concatenated using Phyutility v2.2.660. The fastest evolving sites in the resulting concatenated alignment of 36,385 nucleotide characters were then identified and removed essentially as described above for the PCG-AA data set. A total of 4,492 characters were eliminated during this step.
ML analysis of the PCG12RNA data set was performed using RAxML v8.2.6 and the GTR + Γ4 model of sequence evolution. The data set was partitioned into gene groups, with the model applied to each partition. The partitions included two RNA gene groups (rRNA and tRNA genes) in addition to the protein-coding gene partitions. Confidence of branch points was estimated by bootstrap analysis with 100 replicates. The Bayesian analyses were performed under the same conditions as those described above for the PCG-AA data set. Here again, the five independent chains failed to converge (maxdiff = 0.78), indicating that at least one of the chains was stuck in a local maximum.
Analysis of gene rearrangements
A gene reversal tree was inferred using a gene order matrix of 98 genes from 15 ulvophycean chloroplast genomes. The branch lengths of this tree were computed on the tree topology inferred from the RAxML analyses of the sequence data using the -t option of MGR v2.0336. Because MGR cannot handle duplicated genes, only one copy of the IR and of each duplicated gene was included in the matrix.
Phylogenetic analyses of group II introns
Group II intron sequences were aligned manually on the basis of their secondary structure models, and poorly aligned and divergent regions were removed. The data set of 124 sites corresponding to domains IA, IVB, V and VI of the core secondary structure52 was analyzed using RAxML v8.2.662 and the GTR + Γ4 model. Confidence of branch points was estimated by bootstrap analysis with 1000 replicates.
Analyses of chloroplast genome isomers
PCR analyses were performed to test whether the Ignatius and Pseudoneochloris IRs undergo flip-flop recombination. For each algal IR, multiple pairs of oligonucleotide primers were designed to yield products that overlap all possible boundaries of the IR sequences with the flanking SC regions (see Supplementary Table S3 for the sequences of these primers). PCR assays were carried out using the GeneAmp XL PCR kit (ABI Applied Biosystems, Foster City, CA, USA) and the conditions recommended by the manufacturer.
Electronic supplementary material
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (http://www.nserc-crsng.gc.ca/index_eng.asp) (Grant No. 2830-2007 to MT and CL). We thank Professor Shin Watanabe (University of Toyama) for providing us with the Ignatius tetrasporus and Pseudocharacium americanum strains.
Author Contributions
C.L. and M.T. conceived the study, C.O. generated DNA sequence data, C.L. and C.O. carried out the genome assemblies and annotations, C.L., and M.T. performed the genomic and phylogenetic analyses, C.L. and M.T. wrote the manuscript and generated the figures. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01144-1
Accession codes: The complete chloroplast genome sequences generated in this study have been deposited under the GenBank accession numbers KY407656-KY407661.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- 2.Lang, B. F. & Nedelcu, A. M. In Genomics of Chloroplasts and Mitochondria Vol. 35 Advances in Photosynthesis and Respiration (eds Ralph Bock & Volker Knoop) Ch. 3, 59–87 (Springer Netherlands, 2012).
- 3.Jansen, R. K. & Ruhlman, T. A. In Genomics of Chloroplasts and Mitochondria Vol. 35 Advances in Photosynthesis and Respiration (eds Ralph Bock & Volker Knoop) Ch. 5, 103–126 (Springer Netherlands, 2012).
- 4.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouard JS, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouard JS, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leliaert F, et al. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci Rep. 2016;6:25367. doi: 10.1038/srep25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemieux C, Otis C, Turmel M. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics. 2014;15:857. doi: 10.1186/1471-2164-15-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemieux C, Otis C, Turmel M. Comparative chloroplast genome analyses of streptophyte green algae uncover major structural alterations in the Klebsormidiophyceae, Coleochaetophyceae and Zygnematophyceae. Front Plant Sci. 2016;7:697. doi: 10.3389/fpls.2016.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turmel M, de Cambiaire JC, Otis C, Lemieux C. Distinctive architecture of the chloroplast genome in the chlorodendrophycean green algae Scherffelia dubia and Tetraselmis sp. CCMP 881. PLoS One. 2016;11:e0148934. doi: 10.1371/journal.pone.0148934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- 12.Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitaria reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol. 2009;26:2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- 13.Turmel M, Otis C, Lemieux C. Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol Evol. 2015;7:2062–2082. doi: 10.1093/gbe/evv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pombert JF, Otis C, Lemieux C, Turmel M. The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol Biol Evol. 2005;22:1903–1918. doi: 10.1093/molbev/msi182. [DOI] [PubMed] [Google Scholar]
- 16.Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 17.Heinhorst S, Cannon GC. DNA replication in chloroplasts. J Cell Sci. 1993;104:1. [Google Scholar]
- 18.Palmer JD, Thompson WF. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982;29:537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer JD. Chloroplast DNA exists in two orientations. Nature. 1983;301:92–93. doi: 10.1038/301092a0. [DOI] [Google Scholar]
- 21.Blazier JC, et al. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 2016;117:1209–1220. doi: 10.1093/aob/mcw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer JD, Osorio B, Aldrich J, Thompson WF. Chloroplast DNA evolution among legumes: Loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr Genet. 1987;11:275–286. doi: 10.1007/BF00355401. [DOI] [Google Scholar]
- 23.Weng, M. L., Ruhlman, T. A. & Jansen, R. K. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol, doi:10.1111/nph.14375 (2016). [DOI] [PubMed]
- 24.Perry AS, Wolfe KH. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. Journal of Molecular Evolution. 2002;55:501–508. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- 26.Wu CS, Chaw SM. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol Evol. 2015;7:2000–2009. doi: 10.1093/gbe/evv125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 28.de Cambiaire JC, Otis C, Lemieux C, Turmel M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol Biol. 2006;6:37. doi: 10.1186/1471-2148-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pombert JF, Lemieux C, Turmel M. The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol. 2006;4:3. doi: 10.1186/1741-7007-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bélanger AS, et al. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Genet Genomics. 2006;276:464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- 31.Turmel M, Otis C, Lemieux C. Mitochondrion-to-chloroplast DNA transfers and intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae) Genome Biol Evol. 2016;8:2789–2805. doi: 10.1093/gbe/evw190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe S, Nakayama T. Ultrastructure and phylogenetic relationships of the unicellular green algae Ignatius tetrasporus and Pseudocharacium americanum (Chlorophyta) Phycol Res. 2007;55:1–16. doi: 10.1111/j.1440-1835.2006.00439.x. [DOI] [Google Scholar]
- 34.Sun L, et al. Chloroplast phylogenomic inference of green algae relationships. Sci Rep. 2016;6:20528. doi: 10.1038/srep20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leliaert F, Lopez-Bautista JM. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics. 2015;16:204. doi: 10.1186/s12864-015-1418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourque G, Pevzner PA. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 2002;12:26–36. [PMC free article] [PubMed] [Google Scholar]
- 37.Hovde BT, et al. The mitochondrial and chloroplast genomes of the haptophyte Chrysochromulina tobin contain unique repeat structures and gene profiles. BMC Genomics. 2014;15:604. doi: 10.1186/1471-2164-15-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith DR, Arrigo KR, Alderkamp AC, Allen AE. Massive difference in synonymous substitution rates among mitochondrial, plastid, and nuclear genes of Phaeocystis algae. Mol Phylogenet Evol. 2014;71:36–40. doi: 10.1016/j.ympev.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. Evolution and cytological diversification of the green seaweeds (Ulvophyceae) Mol Biol Evol. 2010;27:2052–2061. doi: 10.1093/molbev/msq091. [DOI] [PubMed] [Google Scholar]
- 40.Andersen, R. A. Algal culturing techniques. (Elsevier/Academic Press, 2005).
- 41.Keller MD, Seluin RC, Claus W, Guillard RRL. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23:633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x. [DOI] [Google Scholar]
- 42.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 43.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 50.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns - a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 53.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 54.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 55.Frith MC, Hamada M, Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:1–14. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24:715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 61.Cummins CA, McInerney JO. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. Syst Biol. 2011;60:833–844. doi: 10.1093/sysbio/syr064. [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 64.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 65.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 66.Mesquite: a modular system for evolutionary analysis. Version 3.02. http://mesquiteproject.org/ (2015).
- 67.Lemieux C, Otis C, Turmel M. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature. 2000;403:649–652. doi: 10.1038/35001059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


