Abstract
Background
Fusarium head blight (FHB) of wheat caused by Fusarium graminearum species complex (FGSC) is a devastating disease worldwide. The pathogens not only reduce the yield of wheat, but also impact the quality of wheat by contamination with trichothecene mycotoxins. A systematic investigation on the pathogens of FHB in Taiwan is lacking. Here, molecular and morphological approaches were used to identify species of the Taiwanese FGSC isolates and determine their trichothecene genotypes.
Results
In this study, a total of 195 isolates of FGSC from diseased wheat were collected from 8 areas of northern and central Taiwan. All isolates were subjected to seedling inoculation for verification of pathogenicity. The pathogenic isolates were genetically characterized by sequence characterized amplified region (SCAR), PCR- restriction fragment length polymorphism (RFLP), phylogenetic analysis and fixed nucleotides to clarify their phylogenetic species, and by PCR assays of TRI genes to determine trichothecene genotypes. They were identified as F. asiaticum, F. graminearum sensu stricto, F. meridionale and an unknown species. Isolates of F. asiaticum were the major causal agents (98%) in this investigated population and were comprised of SCAR type 5 (75%), SCAR type 4 (21%) and SCAR type 3 (2%). Their trichothecene genotypes were either 15-acetyl-deoxynivalenol (15-ADON) (83%) or nivalenol (NIV) genotype (17%). These genetic characterizations indicated that F. asiaticum (15-ADON SCAR type 5) accounts for 60% of this Taiwanese population. Virulence assay on wheat heads indicated virulence of F. asiaticum isolates in subpopulations divided by SCAR types or trichothecene genotypes were comparable, suggesting other factors influence the unequal subpopulation sizes.
Conclusions
This is the first study that FGSC isolates in Taiwan were systematically collected and characterized. In addition to F. graminearum sensu stricto and F. meridionale, F. asiaticum with 15-ADON genotype was identified as the predominate species in Taiwan. In contrast to Chinese and Japanese populations that F. asiaticum isolates were typically of 3-ADON or NIV genotype, the predominate 15-ADON genotype in Taiwanese population was unique among F. asiaticum populations and represented the southernmost 15-ADON genotype population in East Asia.
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-016-0156-4) contains supplementary material, which is available to authorized users.
Keywords: Fusarium head blight, Fusarium asiaticum, Fusarium graminearum, Fusarium meridionale, Deoxynivalenol, Nivalenol
Background
Wheat was an important crop that grew in many counties of Taiwan in 1960s. Due to the impact of world trade, wheat production and plantation areas have shrunk dramatically over the years. There were only limited areas in central Taiwan growing wheat for malt production in the past 40 years. In recent years, the acreages of wheat were gradually increased from central Taiwan to southern and northern Taiwan because of the advocacy of eating locally grown crops and a governmental policy of raising food self-sufficiency rates. In an informal survey, Fusarium head blight (FHB) was found to be prevalent in many counties after the anthesis stage of wheat (Kuo et al. 2014).
FHB could be caused by a number of Fusarium species. Some species played a more important role than others, depending on geographic regions. Among them, F. graminearum sensu lato was reported as an economically devastating pathogen (Windels 2000). The morphologically characterized F. graminearum sensu lato was revealed as a species complex since 2000 (O’Donnell et al. 2000). To date, at least 15 species were contained within F. graminearum species complex (FGSC), isolated from various hosts and substrates that have been established based on the genealogical concordance phylogenetic species recognition (O’Donnell et al. 2004, 2008; Sarver et al. 2011; Starkey et al. 2007; Yli-Mattila et al. 2009). Except for F. graminearum sensu stricto (s. str.) that distributed worldwide, most species of FGSC were mainly restricted to few geographic regions (O’Donnell et al. 2004). Fusarium austroamericanum, for example, was distributed in South America. Fusarium asiaticum was reported in Brazil, USA, and mainly in Asian countries. In spite of numerous causal agents of FHB to wheat, particular species usually were more prevalent than others. For instance, among 623 isolates of FHB pathogens from 7 states of United States, 523 isolates belonged to F. graminearum s. str. (Zeller et al. 2004). Fusarium graminearum s. str. represented 76% of a genotyped Louisiana population (154 isolates) (Gale et al. 2011). These studies illustrated that F. graminearum s. str. was the predominate species in the United States. Fusarium asiaticum (77%) and F. graminearum s. str. (23%) were identified from 299 isolates collected from 12 provinces in China that were mainly located in the Yangtze River valleys (Zhang et al. 2007). Among 530 isolates collected from 6 provinces (Jiangsu, Anhui, Henan, Hebei, Shandong and Hubei) in China, 65% were F. asiaticum and 34% were F. graminearum s. str. (Shen et al. 2012). In a broader sampling from 15 provinces in China, F. graminearum s. str. (36%) and F. asiaticum (58%) were the major FHB pathogens of 469 isolates though the other 6 Fusarium species were also found (Zhang et al. 2012). From 35 prefectures in Japan, 246 F. asiaticum and 50 F. graminearum s. str. were identified in 298 collected isolates (Suga et al. 2008). In South Korea, of 356 isolates from rice grown in 5 provinces, 333 were F. asiaticum and 23 were F. graminearum s. str. (Lee et al. 2009). Of 568 isolates from maize at eight locations in South Korea, F. graminearum s. str. (75%) was most common species followed by F. asiaticum (12%) (Lee et al. 2012). Above information also indicated that F. asiaticum and F. graminearum were the major FHB pathogens on wheat in China and Japan, and the major FGSC species in South Korea.
In addition to yield loss, diseased grains of FHB usually accumulated trichothecenes that were associated with toxicoses in humans and farm animals (Desjardins 2006). Species of FGSC were trichothecene B-producing fungi that could be classified into three strain-specific chemotypes based on their trichothecene production: the 3-ADON chemotype isolates produced deoxynivalenol (DON) and 3-acetyl-deoxynivalenol (3-ADON); the 15-ADON chemotype isolates produced DON and 15-acetyl-deoxynivalenol (15-ADON); and the NIV chemotype isolates produced nivalenol (NIV) and 4-acetylnivalenol (Ward et al. 2002; Miller et al. 1991). Individual trichothecene may differ in toxicological effects on plants and animals. DON inhibited root growth and coleoptiles elongation of wheat, but NIV had no effect at similar concentration (Eudes et al. 2000; Shimada and Otani 1990). DON was shown to be a virulent factor of FHB to wheat whereas NIV was a virulent factor of ear rot to maize (Maier et al. 2006; Jansen et al. 2005; Proctor et al. 1995). Compared to DON, some studies observed higher cytotoxicity for NIV (Minervini et al. 2004). These differences in toxic effects made chemotypes of isolates important in geographic distribution and population composition of fungal pathogens.
The conventional method for chemotyping the trichothecenes of Fusarium species was the HPLC or GC/MS analysis of culture extracts from individual isolates. PCR methods for genotyping the trichothecene chemotypes of Fusarium species were developed according to allelic variants of trichothecene biosynthesis genes in isolates with strain-specific chemotypes. Sequences of TRI7, TRI12 and TRI13 genes, for instances, were distinct in DON- and NIV-producing isolates (Lee et al. 2001; Chandler et al. 2003; Ward et al. 2002; Quarta et al. 2006). Primers designed on TRI3 and TRI12 were used to assess 3-ADON- and 15-ADON-producing isolates (Jennings et al. 2004; Quarta et al. 2006; Ward et al. 2002). These studies also demonstrated that results of genotyping were highly correlated to these of chemical chemotyping. The trichothecene genotyping by PCR assays allowed rapid screening of a large number of isolates, and identification of the increasing occurrence of high toxigenic population such as 3-ADON- and NIV-producing isolates (Pasquali et al. 2010; Puri and Zhong 2010; Zhang et al. 2012).
In Taiwan, FHB of wheat caused by F. graminearum sensu lato was first reported in 1919 (Sawada 1919). Although other species of potential FHB pathogens such as F. poae, F. avenaceum and F. culmorum were also reported, none of them were isolated from wheat (Huang and Sun 1997). These earlier studies were based on the morphological characterization which was not able to resolve species in a species complex. Along with the significant occurrence of FHB, there was an increasing need to clarify the composition of the causal agents, contributing to the formulation of control strategies and for evaluation of mycotoxin threat. Trichothecenes, especially the DON (also known as vomitoxin), produced in the infected grains will become a concern for consumers. Unfortunately, there was no systematic survey carried out regarding these concerns in Taiwan. This study was aimed to identify the species of FHB pathogens including species of FGSC, and to determine their trichothecene genotypes. Results will also improve our understanding of the distribution and trichothecene genotypes of FGSC species in wheat production areas in East Asia.
Methods
Strain isolation and morphological identification
Wheat samples were collected from 10 areas of 6 cities and counties from northern Taiwan to southern Taiwan. Potential Fusarium head blight pathogens of wheat were isolated from seeds and wheat heads showing head blight symptoms. In areas of southern Taiwan where Fusarium head blight was not observed, around 100 wheat heads were randomly collected from each field for fungal isolation. For isolating fungi from diseased or symptomless wheat heads, 10 every other kernels were picked from bottom to top of a head, and incubated on 2% (w/v) water agar (WA) at room temperature for 3–5 days. After fungi grew out, one isolate was picked from each head for further study. Therefore, all isolates were obtained from different plants. A modified blotter method (De Tempe 1953) was used for fungal isolation from seeds. Briefly, seeds were treated with 1% sodium chloride for 1 min, rinsed with sterilized water for 3 times, and then incubated on a piece of wet autoclaved filter paper in 9-cm Petri dishes at 24 °C for 7 days. Ten seeds were plated in each plate, and 400 seeds were randomly sampled from each bag of seeds. Fungal colonies secreting carmine red pigment in WA or filter paper and producing characterized spindle-sickle macroconidia were selected for further purification. Single macroconidium of Fusarium was isolated for analyses. For morphological analysis, Fusarium isolates were grown on potato dextrose agar (PDA; Difco, NJ, USA) to observe culture characteristics and carnation leaf agar (CLA) to examine the morphology of macroconidia (Leslie and Summerell 2006). Thirty macroconidia with 4–5 septa from each isolate were measured to determine the conidial size. Perithecia were induced on carrot medium (5 g of Bacto agar, 400 g freshly sliced carrots, per liter of distilled water) (Cavinder et al. 2012). Ascospores released from asci were measured and characterized.
Molecular identification on FGSC
For genomic DNA isolation, fungal isolates were grown in yeast extract peptone dextrose broth (0.3 g of yeast extract, 1.0 g of peptone and 1.0 g of dextrose in 100 ml of distilled water) at 25 °C for 3 days. Fungal mycelia were harvested with miracloth (pore size: 22–25 µm) (Calbiochem, CA, USA) and DNAs were extracted following the standard procedures (Sambrook and Russell 2001). Primers Fg16F (5′-CTCCG-GATATGTTGCGTCAA-3′) and Fg16R (5′-GGTAGGTATCCGACATGGCAA-3′) were used as a pair of specific PCR primers to determine if a Fusarium isolate belongs to a FGSC isolate, and sizes of this PCR amplicons were used to assign the sequence characterized amplified region (SCAR) types to each isolate (Carter et al. 2000; Nicholson et al. 1998; Carter et al. 2002). The PCR program was set according to Nicolson et al. (1998). Except where indicated otherwise all PCR reactions in this study were carried out in a total volume of 25 µl containing 19.7 µl of MilliQ water, 2.5 µl of 10× buffer, 0.5 µl of dNTP (200 µM each of dATP, dTTP, dGTP and dCTP), 0.5 µl of each primer (10 µM), 1 µl of DNA template (100 ng/µl), and 0.3 µl of DNA polymerase (Pro Taq plus; Protech Technology Enterprise, Taipei, Taiwan). PCR amplification was conducted in a thermal cycler (Applied Biosystems 2720; Life Technologies, CA, USA). Electrophoresis was performed in 1.5% (w/v) TAE agarose gel, and ethidium bromide was used for gel staining. PCR-restriction fragment length polymorphism (PCR–RFLP) of partial histone gene was applied to identify F. graminearum s. str. and F. asiaticum. The partial histone H3 gene fragment with size of 223 bp was amplified using primers H3dStyI (5′-AGCATCACCYGAACATCGCATCATCCCATG-3′) and H3R1 (5′-TTGGACTGG-ATRGTAACACGC-3′), and purified with Plus DNA Clean/Extraction Kit (GeneMark, Taichung, Taiwan). The purified PCR products were digested with EcoRV and StyI, respectively (Suga et al. 2008). The digestion results were visualized by electrophoresis in 2.5% (w/v) TAE (Tris-acetate-EDTA) agarose gel.
Trichothecene genotyping
Primers locating at TRI13 gene were used to differentiate the genotypes of DON or NIV. Primers Tri13F (5′-CATCATGAGACTTGTKCRAGTTTGGG-3′) and Tri13DONR (5′-GCTAGATCGATTGTTGCATTGAG-3′) would amplify a PCR band from DON genotype isolates, but not from NIV genotype isolates. In contrast, primers Tri13NIVF (5′-CCAAATCCGAAAACCGCAG-3′) and Tri13R (5′-TTGAAAGCTCCAATGTCGTG-3′) would amplify a band from NIV genotype isolates, but not from DON genotype isolates (Chandler et al. 2003). Primers Tri303F (5′-GATGGCCGCAAGTGGA-3′) and Tri303R (5′-GCCGGACTGCCCTATTG-3′) were used to detect 3-ADON genotype whereas primers Tri315F (5′-CTCGCTGAAGTTGGACGTAA-3′) and Tri315R (5′-GTCTATGCTCTCAACGGACAAC-3′) were used to determine 15-ADON genotype (Jennings et al. 2004).
Pathogenicity assays
To induce the production of conidia, agar blocks containing mycelia of each isolate were cultured in mung bean broth (40 g of mung beans was steeped in 1 l of just boiled distilled water for 10 min. After beans were removed by cheesecloth, the filtrate was autoclaved for use.) at 25 °C and 100 rpm for 3 days (Wu et al. 2005). Conidia were filtrated through miracloth and counted by hemocytometer. The suspension was centrifuged and adjusted to 105 conidia/ml with sterilized water and used for pathogenicity tests. The local wheat cultivar, Taichung Sel. 2, was used as host plants. A seedling coleoptile inoculation method (Wu et al. 2005) was modified to quickly determine the pathogenicity of all isolates. Briefly, seedling coleoptiles were wounded by a needle, and then immersing in a conidia suspension (105 conidia/ml) for 30 min. Five seedlings were inoculated with each isolate. Inoculated seedlings were placed on wet paper towel in Petri dishes and put in moister chambers at 24 °C with 12 h light cycle. To evaluate the virulence of each isolate, 5 µl conidia suspension (105 conidia/ml) was inoculated at the interior of the two central spikelets of each head at the anthesis stage (Malbrán et al. 2012). The inoculated wheat heads were covered with plastic bags for 48 h to maintain moisture. Three replicates were conducted for each isolate. Virulence was evaluated by the number of symptomatic spikelets at the 12 days post inoculation (dpi).
Phylogenetic analysis
The partial sequences of RED gene and TEF-1α gene of 19 selected representative isolates were used for resolving their phylogenetic relationships with reference strains of all known species of FGSC. A PCR product of RED sequence in a range from 900 to 1000 bp was amplified with primers RED1d (5′- TCTCAGAAAGACGCATATATG-3′) and RED2 (5′- CGTAACTGCGTCATTCGGC-3′). TEF-1α sequence in a range from 600 to 700 bp was amplified with primers TEF1 (5′- ATGGGTAAGGAGGACAAGAC-3′) and TEF2 (5′- GGAAGTACCAGTGATCATGTT-3′) (O’Donnell et al. 2000). PCR products were purified for sequencing with the forward and reverse primers. Phylogenetic analysis was performed by MEGA 6 (Tamura et al. 2013). ClustalW software was applied to align the two gene sequences from the 19 isolates in this study and other reference strains belonging to species of FGSC. The concatenate sequences of RED and TEF-1α genes were analyzed by the maximum likelihood method with 1000 bootstrap replications.
Results
Species diagnosis and SCAR types of FGSC isolates from wheat
Based on secretion of carmine red pigment and sickle-shaped macroconidia of Fusarium, a total of 203 potential causal agents of Fusarium head blight were isolated. Almost 100% of fungi that produce the feature pigment and macroconidia could be isolated from wheat heads with typical symptom of FHB from most areas such as Daya, Pingzhen, Guanying and Xinwu where the occurrence of FHB was more frequent than other areas. In areas of southern Taiwan (Chiayi county and Tainan city), the FHB was not observed. Wheat heads with suspicious symptoms were collected for isolation. In this case, the isolation rate of carmine red pigment colony was rare. A seedling coleoptile inoculation method was adapted to quickly assay the pathogenicity of each isolate. Pathogenic isolates would cause necrotic lesions on leaf sheath or leaf blade while the water control or non-pathogenic Fusarium isolates to wheat such as the Fusarium oxysporum f. sp. lycopersici did not produce any lesion on wheat seedlings (Fig. 1a). Among them, 195 isolates (186 isolates from diseased plants and 9 isolates from seeds) collected from 8 areas of northern and central Taiwan were determined as wheat pathogens whereas 8 isolates obtained from 600 wheat heads with suspicious symptoms produced in southern Taiwan (Chiayi county and Tainan city) were not FHB pathogens. Of these 195 isolates, 193 isolates but 2 isolates (Dacheng 1-2 and Dacheng 2-1) showed the positive band in the PCR results using primers Fg16F and Fg16R, indicating that most of them belonged to FGSC isolates. Different sizes of the PCR bands were isolated and cloned for sequencing to determine the exact number of nucleotide base pairs (Fig. 1b). The 193 isolates were assigned into different SCAR types based on their band sizes. The SCAR type 5 (497 bp) isolates (75%) were most prevalent in population. The SCAR type 4 (557 bp) isolates represented 21% of the population. The SCAR type 3 (527 bp) isolates was only 1.5% of this studied population. Both SCAR type 1 and SCAR type 2 were found only in one isolate. Isolates from Daya area (Taichung city, central Taiwan) were most diverse in SCAR types. Isolates of SCAR type 5 and SCAR type 4 consistently presented in 3- 3.5–1 ratio in most geographic areas (Table 1). A PCR–RFLP method that was based on fixed nucleotide character states specific to F. graminearum s. str. and F. asiaticum has been reliable to diagnose 296 isolates of the two species in Japan (Suga et al. 2008), 530 isolates of the two species in China (Shen et al. 2012), and 41 isolates of F. asiaticum in US (Gale et al. 2011). Since F. graminearum s. str. and F. asiaticum were the most common species of FGSC in Asian countries (e.g. China, South Korea and Japan) (review see van der Lee et al. 2015; Wang et al. 2011), we used the method to diagnose if any of these Taiwanese FGSC isolates belong to the two species. In an earlier report, the partial histone H3 gene fragment of F. asiaticum would be cut into 191 and 32 bp by StyI whereas the partial histone H3 gene fragment of F. graminearum s. str. would be cut into 195 and 28 bp by EcoRV (Suga et al. 2008). The band shifted to a reduced size, after the restriction enzyme treatments, indicating a positive digestion (Fig. 1c). DNAs of a total of 192 out of the 195 isolates were digested into smaller fragments by StyI, suggesting that F. asiaticum (99%) was the predominate species of FGSC in Taiwan. Only one isolate (Daya211-13) was diagnosed as F. graminearum s. str. Two isolates (Fanyuan1-11 and Daya272-3) that failed to be digested by neither StyI nor EcoRV were selected for further identification by phylogenetic analysis.
Fig. 1.
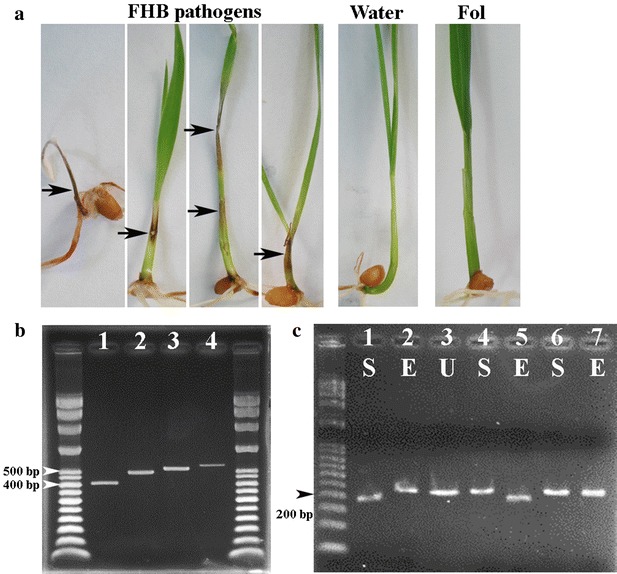
Representative results of seedling pathogenicity assays, SCAR types, and PCR–RFLP diagnosis. a The pathogenicity assay with seedling coleoptile inoculation method was used to test if all isolates are pathogenic to wheat. Necrotic leaf sheath lesions (arrows) indicated that the tested isolates were FHB pathogens. No symptom displayed after inoculation with sterilized water or Fusarium oxysporum f.sp. lycopersici (Fol, not a FHB pathogen), respectively. b The variation of SCAR types in studied population. The first lane is the DNA ladder; Lane 1 is SCAR type 1 (410 bp); Lane 2 is SCAR type 2 or type 5 (497 bp); Lane 3 is SCAR type 3 (527 bp); Lane 4 is SCAR type 4 (557 bp). The last lane is another DNA ladder. c A PCR–RFLP method for diagnosis of F. asiaticum and F. graminearum s. str. through a partial histone H3 fragment. A F. asiaticum isolate will be digested by StyI but not EcoRV whereas a F. graminearum s. str. isolate will be digested by EcoRV but not StyI. Other species will not be digested by the two enzymes. The first lane is the DNA ladder; Lanes 1 to 3 are a F. asiaticum isolate (Pingzhen1-49); Lanes 4 and 5 are a F. graminearum s. str. isolate (Daya211-13); Lanes 6 and 7 are an unknown species isolate (Daya272-3). Different restriction enzyme treatments are indicated: for “S” this is StyI, for “E” this is EcoRV, and for “U” this is the uncut control
Table 1.
The geographic distribution of identified species and SCAR types of all isolates
| Collection area | No. of isolates | F. asiaticum | F. graminearum s. str. | F. meridionale | Unknown species | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Typea 3 | Type 4 | Type 5 | Type 1 | Type 5 | Type 2 | |||||
| Northern Taiwan | ||||||||||
| Taoyuan city | ||||||||||
| Guanying | 30 | 7 | 23 | |||||||
| Xinwu | 30 | 8 | 22 | |||||||
| Pinzhen | 30 | 3 | 27 | |||||||
| Central Taiwan | ||||||||||
| Miaoli county | ||||||||||
| Yuanli | 3 | 1 | 2 | |||||||
| Taichung city | ||||||||||
| Daya | 70 | 3 | 15 | 50 | 1 | 1 | ||||
| Waipu | 4 | 4 | ||||||||
| Changhua county | ||||||||||
| Fanyuan | 23 | 5 | 17 | 1 | ||||||
| Dacheng | 5b | 2 | 1 | |||||||
| Total | 195b | 3 | 41 | 146 | 1 | 1 | 1 | |||
aThe SCAR types were defined by Carter et al. (2002). Type 1 is 410 bp; Type 2 is 479 bp; Type 3 is 527 bp; Type 4 is 557 bp; Type 5 is 479 bp. Type 2 and Type 5 are different in nucleotide sequences
bThe SCAR patterns were not amplified from two F. asiaticum isolates (Dacheng 1-2 and Dacheng 2-1)
Phylogenetic analysis
To further confirm the diagnostic results of PCR–RFLP and determine the 2 unidentified isolates, a total of 19 representative isolates including 16 isolates of F. asiaticum, 1 isolate of F. graminearum s. str., and the 2 unidentified isolates were further investigated for their phylogenetic relationships with reference strains and type strains of all known FGSC species. The 16 isolates of F. asiaticum were randomly selected from each SCAR type and trichothecene genotype. For phylogenetic analysis, RED gene and TEF-1α gene of the 19 representative isolates were sequenced and deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank/) with accession numbers (Table 2). A total of 1296 informative nucleotides were used for analysis including 775 nucleotides of RED gene and 521 nucleotides of TEF-1α gene. The 16 isolates of F. asiaticum were well clustered with 3 reference F. asiaticum strains with a high bootstrap value (99%). The F. graminearum s. str. isolate was also placed in the F. graminearum s. str. clade with a bootstrap value of 91%. These results not only confirmed the PCR–RFLP method was reliable, but also illustrated this phylogenetic tree was able to resolve the phylogenetic relationship at species level to some extent. One of the unidentified isolates (Fanyuan1-11) was clearly clustered with 2 reference strains and the type strain of F. meridionale with a bootstrap value of 92%. The other unidentified isolate (Daya272-3) was closest to the F. cortaderiae clade, but had a low bootstrap value of 52% and this did not support the relationship (Fig. 2).
Table 2.
The list of representative isolates used for phylogenetic analysis
| Species of FGSC and isolate | SCAR typea | Trichothecene genotype | Accession number | |
|---|---|---|---|---|
| Reductase | TEF-1α | |||
| F. asiaticum | ||||
| Daya350-5 | Type 5 | NIV | KT372157 | KT380119 |
| Daya211-2 | Type 5 | NIV | KT372158 | KT380120 |
| Fanyuan1-13 | Type 5 | NIV | KT372159 | KT380121 |
| R-p-1 | Type 5 | 15-ADON | KT372149 | KT380111 |
| Daya350-3 | Type 5 | 15-ADON | KT372162 | KT380124 |
| Daya211-6 | Type 5 | 15-ADON | KT372164 | KT380126 |
| Xinwu1-1 | Type 5 | 15-ADON | KT372163 | KT380125 |
| Daya350-11 | Type 3 | 15-ADON | KT372147 | KT380109 |
| Daya350-12 | Type 3 | 15-ADON | KT372152 | KT380114 |
| Daya272-14 | Type 3 | 15-ADON | KT372153 | KT380115 |
| Daya350-2 | Type 4 | NIV | KT372154 | KT380116 |
| Daya272-6 | Type 4 | NIV | KT372155 | KT380117 |
| Pingzhen1-13 | Type 4 | NIV | KT372156 | KT380118 |
| Daya350-13 | Type 4 | 15-ADON | KT372160 | KT380122 |
| k-r-1 | Type 4 | 15-ADON | KT372150 | KT380112 |
| Guanying 1-5 | Type 4 | 15-ADON | KT372161 | KT380123 |
| F. graminearum s. str. | ||||
| Daya211-13 | Type 1 | 15-ADON | KT334553 | KT380127 |
| F. meridionale | ||||
| Fanyuan1-11 | Type 5 | NIV | KT372148 | KT380110 |
| Unknown species | ||||
| Daya272-3 | Type 2 | NIV | KT372151 | KT380113 |
aThe SCAR types were defined by Carter et al. (2002). Type 1 is 410 bp; Type 2 is 479 bp; Type 3 is 527 bp; Type 4 is 557 bp; Type 5 is 479 bp. Type 2 and Type 5 are different in nucleotide sequences
Fig. 2.
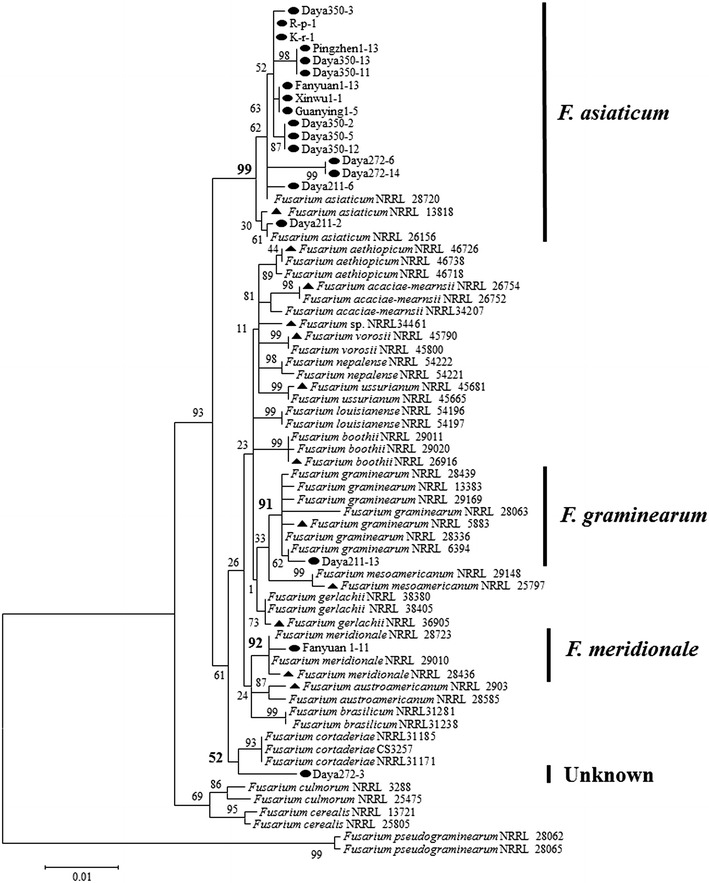
The phylogenetic analysis. The phylogenetic tree of representative strains of FHB pathogens in Taiwan based on partial sequences of RED and TEF-1α genes. The maximum likelihood method was used for the analysis with 1000 bootstrap repeats (black circle isolates in this study, filled triangle type strains of species of FGSC)
All sequenced isolates of identified F. asiaticum were further compared with the species type strain NRRL 13818 on the defined fixed nucleotides of RED sequence, and the results showed that they were identical at all fixed nucleotides. The RED and TEF-1α sequences of identified F. graminearum s. str. and F. meridionale were aligned with reference strains F. graminearum s. str. NRRL 5883 and F. meridionale NRRL 28436, respectively. All fixed nucleotides were also conserved in the investigated strains in the 2 gene alignments. In addition, morphology of all sequenced isolates was characterized and was indistinguishable from each other. All isolates produced carmine red pigment on PDA and CLA, and formed macroconidia and no microconidia. Macroconidia from CLA were gradually curved and 52–63 × 5–6 μm with 5–6 septa. All isolates were homothallic and produced various numbers of dark blue perithecia containing a number of asci on carrot medium. Each ascus bore 8 ascospores. Ascospores were 21–24 × 3–5 μm with 1–3 septa, hyaline in color and fusiform with rounded ends (Additional file 1: Figure S1). These morphological characteristics conformed to the defined characteristics of FGSC.
Trichothecene genotypes
The allelic nucleotide polymorphism of TRI13 gene was used to distinguish DON genotype or NIV genotype. A PCR band of about 227 bp indicating DON genotype was amplified from 162 isolates (83%) by primers Tri13F and Tri13DONR. On the other hand, a PCR band of about 312 bp indicating NIV genotype was amplified from 33 isolates (17%) by primers Tri13NIVF and Tri13R (Fig. 3a). The DON genotype isolates were further differentiated as 3-ADON or 15-ADON isolates by primers designed from the different alleles of TRI3 gene. All DON genotype isolates generated a 864 bp band in the PCR reactions with primers Tri315F and Tri315R, but not a 586 bp band with primers Tri303F and Tri303R, indicating that all DON genotype isolates were of 15-ADON genotype (Fig. 3b). In northern Taiwan, all isolates but one belonged to 15-ADON (99%). In central Taiwan, NIV isolates accounted for 32 and 19% of populations in Taichung city and Changhua county, respectively (Fig. 3c). As with SCAR types, trichothecene genotypes were more diverse in central Taiwan than northern Taiwan.
Fig. 3.
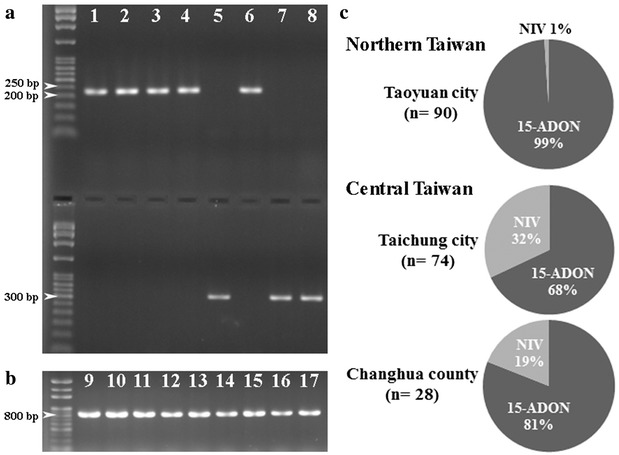
The identification and distribution of trichothecene genotypes of FGSC isolates. a Identification of DON or NIV genotype by the amplification of TRI13 gene with primers Tri13F and Tri13DONR (upper panel), and primers Tri13NIVF and Tri13R (lower panel), respectively. The first lane is the DNA ladder; Lane 1 is Daya 350-12; Lane 2 is Daya 350-13; Lane 3 is Daya 350-14; Lane 4 is Daya 350-15; Lane 5 is Daya 350-16; Lane 6 is Daya 211-1; Lane 7 is Daya 211-2; Lane 8 is Daya 211-3. b Identification of 15-ADON genotype by the amplification of TRI3 gene with primers Tri315F and Tri315R. The first lane is the DNA ladder; Lane 9 is Daya 350-11; Lane 10 is Daya 350-12; Lane 11 is Daya 350-13; Lane 12 is Daya 211-6; Lane 13 is Xinwu 1-1; Lane 14 is Pingzhen1-13; Lane 15 is R-P-1; Lane 16 is Guanying 1-5; Lane 17 is Daya 211-13. c Distributions of FGSC isolates regarding trichothecene genotypes in northern (Taoyuan city) and central (Taichung city and Changhua county) Taiwan
Combined with the above results of isolate identity and SCAR types, one would conclude that the most predominate species and genotype is F. asiaticum 15-ADON SCAR type 5 (60%). Fusarium asiaticum 15-ADON SCAR type 4 isolates and F. asiaticum NIV SCAR type 5 isolates contributed 19 and 15% of the studied population, respectively. The single F. graminearum s. str. isolate was of 15-ADON genotype whereas the single F. meridionale isolate and the unknown species isolate were of NIV genotype (Table 3).
Table 3.
The trichothecene genotypes of all species with reference to SCAR types
| Genotype | No. of isolates | F. asiaticum | F. graminearum s.s. | F. meridionale | Unknown | ||
|---|---|---|---|---|---|---|---|
| Type 5a | Type 3 | Type 4 | Type 1 | Type 5 | Type 2 | ||
| DON | 162b | ||||||
| 15-ADON | 162b | 118 (60.5%) | 3 (1.5%) | 38 (19.4%) | 1 (0.5%) | ||
| NIV | 33 | 30 (15.3%) | 3 (1.5%) | 1 (0.5%) | 1 (0.5%) | ||
| Total | 195b | 146 | 3 | 41 | 1 | 1 | 1 |
aThe SCAR types were defined by Carter et al. (2002). Type 1 is 410 bp; Type 2 is 479 bp; Type 3 is 527 bp; Type 4 is 557 bp; Type 5 is 479 bp. Type 2 and Type 5 are different in nucleotide sequences
bThe SCAR types of two F. asiaticum isolates (Dacheng 1-2 and Dacheng 2-1) were not available
Virulence comparison among isolates with different SCAR types or trichothecene genotypes
To determine if virulence variation could account for the unequal subpopulation sizes of F. asiaticum 15-ADON SCAR type 5 isolates (60%), F. asiaticum 15-ADON SCAR type 4 isolates (19%) and F. asiaticum NIV SCAR type 5 isolates (15%), 10 isolates from each of the 3 subpopulations were randomly selected to compare their virulence on wheat heads. Conidial suspension of each isolate was inoculated on the 2 central spikelets of heads. Number of the discolored spikelet was counted to evaluate the virulence of each isolate at the 12 dpi. Although isolates of the F. asiaticum 15-ADON SCAR type 5 subpopulation caused slightly more symptomatic spikelets in average than isolates in the other two subpopulations, the differences did not statistically distinguish them in virulence to wheat (Fig. 4). This result suggests that the difference of trichothecene genotypes (15-ADON and NIV) or SCAR types (SCAR type 4 and SCAR type 5) did not explain the unequal subpopulation sizes.
Fig. 4.
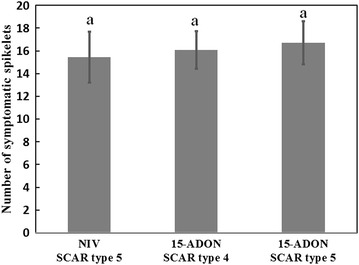
The virulence comparison of F. asiaticum isolates with different genotypes or SCAR types. The pathogenicity assays were performed by point inoculation on central spikelets of wheat heads. Data were analyzed by LSD method with the Software SPSS
Discussion
Fungal isolates collected in this study were from areas of northern and central Taiwan located from North 23°50′ to North 25°02′ which represented one of the wheat production areas at the lowest latitude in the world. FHB pathogens were not detected from samples collected from southern Taiwan near or lower than the Tropic of Cancer. It would be interesting to dissect the reasons of no detection in southern Taiwan, including the presence or absence of pathogens, pathogen survival ability in high temperature, alternative hosts and environmental factors for disease development. In this study, all pathogenic isolates were considered to be members of FGSC by PCR amplification with primers Fg16F and Fg16R and morphological characteristics of asexual and sexual stages. Because species of FGSC were established based on the molecular characteristics and cannot be identified through the morphological characteristics (O’Donnell et al. 2004; Sarver et al. 2011), we used molecular methods to determine the species of each isolate. As mentioned earlier, F. asiaticum and F. graminearum s. str. have been reported as predominate species of FGSC in China and Japan (Shen et al. 2012; Zhang et al. 2007; Suga et al. 2008; Zhang et al. 2012). A PCR–RFLP method has been reliable to diagnose 867 isolates of F. graminearum s. str. and F. asiaticum in Japan, China and US (Shen et al. 2012; Gale et al. 2011; Suga et al. 2008). We, therefore, applied the PCR–RFLP diagnosis method to detect F. asiaticum and F. graminearum s. str. from collected isolates. A phylogenetic analysis based on RED and TEF-1α sequences complied with the diagnosed results, and further identified a F. meridionale isolate in our collection. As shown in studies, RED and TEF-1α sequences were able to separate most species of FGSC and used at the initial stage to discover a novel species (O’Donnell et al. 2004; Starkey et al. 2007). These identifications of F. asiaticum, F. graminearum s. str. and F. meridionale were also supported by the fixed nucleotides presenting in the 2 gene sequences and by the morphology of the sexual and conidial stages. Nevertheless, there was still one isolate in query. More informative genes such as genes in the mating-type locus are required to resolve the relationship between the unknown isolate and other species of FGSC (O’Donnell et al. 2004).
Fusarium asiaticum was identified as a predominate species with 98% of the population in this study. Fusarium asiaticum and F. graminearum s. str. were the major species in many Asian countries (van der Lee et al. 2015; Wang et al. 2011). Factors like temperatures and hosts have been ascribed to species distribution. In China, F. asiaticum was mainly distributed in the southern provinces, whereas F. graminearum s. str. was mostly in the northern provinces (Qu et al. 2008b; Shen et al. 2012). The same trend was observed in an analysis of Japanese isolates (Suga et al. 2008). From a study of FHB of wheat, Qu et al. (2008b) concluded that F. asiaticum was mainly obtained from warmer regions where the annual average temperature were above 15 °C, and the majority of F. graminearum s. str. was isolated from cooler regions with the annual average temperature at 15 °C or lower. A BIOCLIM analysis suggested that areas where the warmest quarter with the mean temperature above 22 °C and the precipitation over 320 nm favored the occurrence of F. asiaticum (Backhouse 2014). In addition, the host preference was implicated by a study of rice population in South Korea in which F. asiaticum (FGSC lineage 6) dominated over F. graminearum s. str. (FGSC lineage 7) and F. boothii (FGSC lineage 3) (Lee et al. 2009). The study showed that F. asiaticum produced more perithecia on rice straw than the other two species in an experiment of mixed-species inoculum. Zhang et al. (2012) found a strong association between the predominate crops and the occurrence of F. asiaticum and F. graminearum s. str. They showed that F. asiaticum was more frequently obtained from the middle and low valleys of Yangtze River (southern China) where rice acreages were higher than maize acreages. Fusarium graminearum s. str. was prevalent in northern China with higher maize acreages (Zhang et al. 2012). In our study, the average annual temperatures of sampling areas in Taiwan were 22 °C and higher, and the precipitation of the warmest quarter were from 322 to 608 nm. All areas where samples were collected were operated under the rice–wheat rotation system. The result of predominate F. asiaticum in Taiwan was consistent with the notion of those previous studies.
Fg16F and Fg16R were designed as specific primers for FGSC isolates (Nicholson et al. 1998). The various amplicons from Fg16F/R PCR were used for the SCAR analysis of FGSC isolates to reveal the genetic variation (Carter et al. 2000, 2002; Qu et al. 2008a, b; Desjardins et al. 2004). Later studies showed that SCAR type 1 and type 5 were fully congruent with F. graminearum s. str. and F. asiaticum, respectively, and were used to screen these 2 phylogenetic species (Chandler et al. 2003; Qiu et al. 2014; Zhang et al. 2007). SCAR type 2 was congruent with F. meridionale, but contained the same PCR band size as SCAR type 5 (Chandler et al. 2003). In this study, isolates of SCAR type 1 and most SCAR type 5 were confirmed as F. graminearum s. str. and F. asiaticum, respectively, by PCR–RFLP and phylogenetic analysis. Some F. asiaticum isolates were of SCAR type 3 and SCAR type 4, which has been reported elsewhere (Chandler et al. 2003). Notably, the one F. meridionale isolate (Fanyuan1-11) was determined to be SCAR type 5, rather than SCAR type 2. The unknown species isolate (Daya272-3) was of SCAR type 2, but was phylogenetically closest to F. cortaderiae rather than F. meridionale. These results suggested that additional caution was needed to be taken to infer FGSC species by SACR types.
It was surprising to find that F. asiaticum isolates were either 15-ADON or NIV genotype in this Taiwanese population. The 3-ADON genotype isolate was not detected in current collection. The 15-ADON genotype isolates were more prevalent than NIV genotype isolates in all geographic areas, especially in northern Taiwan where 15-ADON genotype isolates accounted for 99% of the population. In Japanese and Chinese populations of F. asiaticum, all 3 trichothecene genotypes (3-ADON, 15-ADON and NIV genotypes) were detected. Both 3-ADON and NIV genotype isolates usually dominated over 15-ADON genotype isolates (Qiu et al. 2014; Suga et al. 2008). Outside of Asia, NIV genotype isolates of F. asiaticum were prevalent in southern Louisiana, United States and southern Brazil (Gale et al. 2011; Gomes et al. 2015). The 15-ADON genotype isolates of F. asiaticum were a typical minority and may be absent in culture collections (Karugia et al. 2009; Shen et al. 2012). In China, the 3-ADON and NIV genotype isolates were prevalent in the warmer southern regions whereas the 15-ADON genotype was mainly presented in the cooler northern regions. The geographic difference of trichothecene genotypes were explained by the unevenly distribution of phylogenetic species. F. asiaticum, the warmer-region inhabitant, mainly belonged to 3-ADON and NIV genotypes based on 1903 isolates while F. graminearum s. str., the cooler-region inhabitant, contained more 15-ADON genotype isolates based on 383 isolates, according to several studies in China and Japan (Karugia et al. 2009; Qiu et al. 2014; Shen et al. 2012; Suga et al. 2008; Zhang et al. 2007). Regarding the geographic locations of 15-ADON genotype isolates which were mostly in the north of East Asia, this Taiwanese F. asiaticum 15-ADON isolates represented a unique population locating in the south of East Asia. This result suggested that 3-ADON genotype isolates were possibly not existed in Taiwanese populations or several factors may favor the 15-ADON genotype isolates to establish in Taiwan. It is notable that the wheat cultivar Taichung Sel. 2 derived from the wheat line Au-Maya74“S” was the only wheat cultivar used for wheat production in Taiwan currently. All FGSC isolates were isolated from diseased plants of the cultivar. The pathogenicity assay, however, indicated that the host cultivar was equally susceptible to 15-ADON and NIV genotype isolates. It needed further evaluations if the Taichung Sel. 2 cultivar favors F. asiaticum 15-ADON genotype isolates for growth, survival, sporulation, or other biological traits that would facilitate these 15-ADON genotype isolates to outcompete other genotype isolates.
Conclusions
This study was the first report on phylogenetic species and trichothecene genotypes for a systematically collected FGSC population in Taiwan. Fusarium graminearum s. str., F. meridionale, F. asiaticum, and an unknown species were identified, and both F. meridionale and F. asiaticum were first reported in Taiwan. Of 195 isolates, F. asiaticum (n = 192) was the predominate species of FGSC wheat isolates and prevalent in northern and central Taiwan. Most Taiwanese F. asiaticum isolates contained 15-ADON trichothecene genotype (n = 162), and the rest of them contained NIV trichothecene genotype (n = 33). The high 15-ADON genotype of F. asiaticum population in Taiwan was a sharp contrast to the high 3-ADON genotype of F. asiaticum reported in the neighbor countries. The above information also provided valuable information for the global distribution of FGSC in East Asia (van der Lee et al. 2015).
Authors’ contributions
CLW carried out writing and revising the manuscript, and provide the concepts of experimental designs. YHC performed experiments. CLW and YHC participated the discussion of the experimental results. Both authors read and approved the final manuscript.
Acknowledgements
The authors thank Dr. Chung-Jan Chang (University of Georgia, USA) and Dr. Wen-Hsiung Ko (National Chung Hsing University, Taiwan) for his critical reading with English editing, Mr. Chien-Chih Kuo (Taichung District Agricultural Research and Extension Station, Taiwan) for sharing 10 isolates, and Mr. Hong-Yue Chen (National Chung Hsing University, Taiwan) for technical assistance. Research grants to CLW were supported by the National Science Council (NSC 101-2313-B-005-049-MY2) and the Ministry of Science and Technology (MOST 103-2313-B-005-028-MY3), Taiwan.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- FHB
Fusarium head blight
- FGSC
Fusarium graminearum species complex
- SCAR
sequence characterized amplified region
- RFLP
restriction fragment length polymorphism
- DON
deoxynivalenol
- 15-ADON
15-acetyl-deoxynivalenol
- 3-ADON
3-acetyl-deoxynivalenol
- NIV
nivalenol
- PDA
potato dextrose agar
- CLA
carnation leaf agar
Additional file
Additional file 1: Figure S1. The morphology of FGSC species with different SCAR types and genotypes. a An unknown species (Daya 272-3). b F. meridionale (Fanyuan 1-11). c F. graninearum s.str. (Daya 211-13). d F. asiaticum SCAR type 3 DON (Daya 350-11). e F. asiaticum SCAR type 4 NIV (Daya 350-2). f F. asiaticum SCAR type 5 NIV (Daya 350-5). g F. asiaticum SCAR type 4 DON (K-r-1). h F. asiaticum SCAR type 5 DON (R-p-1). The column-1 and column-2 were the 10-day PDA cultures. The column-3 was the 7-day CLA culture. The column-4 and column-5 showed the perithecia and ascospores, respectively. The column-6 showed macroconidia produced on CLA cultures (scale bar = 10 μm).
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-016-0156-4) contains supplementary material, which is available to authorized users.
Contributor Information
Chih-Li Wang, Phone: +886-4-22840780, Email: clwang@nchu.edu.tw.
Yi-Hong Cheng, Email: chengyihong0317@gmail.com.
References
- Backhouse D. Global distribution of Fusarium graminearum, F. asiaticum and F. boothii from wheat in relation to climate. Eur J Plant Pathol. 2014;139:161–173. doi: 10.1007/s10658-013-0374-5. [DOI] [Google Scholar]
- Carter J, Rezanoor H, Desjardins A, Nicholson P. Variation in Fusarium graminearum isolates from Nepal associated with their host of origin. Plant Pathol. 2000;49:452–460. doi: 10.1046/j.1365-3059.2000.00482.x. [DOI] [Google Scholar]
- Carter J, Rezanoor H, Holden D, Desjardins A, Plattner R, Nicholson P. Variation in pathogenicity associated with the genetic diversity of Fusarium graminearum. Eur J Plant Pathol. 2002;108:573–583. doi: 10.1023/A:1019921203161. [DOI] [Google Scholar]
- Cavinder B, Sikhakolli U, Fellows KM, Trail F. Sexual development and ascospore discharge in Fusarium graminearum. J Vis Exp. 2012;61:e3895. doi: 10.3791/3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler EA, Simpson DR, Thomsett MA, Nicholson P. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterisation of chemotypes of Fusarium graminearum, Fusarium culmorum and Fusarium cerealis. Physiol Mol Plant Pathol. 2003;62:355–367. doi: 10.1016/S0885-5765(03)00092-4. [DOI] [Google Scholar]
- De Tempe J. The blotter method of seed health testing. Proc Int Seed Test Assoc. 1953;21:133–151. [Google Scholar]
- Desjardins AE (2006) Fusarium mycotoxins: chemistry, genetics, and biology. American Phytopathological Society. APS Press
- Desjardins AE, Jarosz AM, Plattner RD, Alexander NJ, Brown DW, Jurgenson JE. Patterns of trichothecene production, genetic variability, and virulence to wheat of Fusarium graminearum from smallholder farms in Nepal. J Agric Food Chem. 2004;52:6341–6346. doi: 10.1021/jf040181e. [DOI] [PubMed] [Google Scholar]
- Eudes F, Comeau A, Rioux S, Collin J. Phytotoxicity of eight mycotoxins associated with Fusarium in wheat head blight. Can J Plant Pathol. 2000;22:286–292. doi: 10.1080/07060660009500477. [DOI] [Google Scholar]
- Gale LR, Harrison SA, Ward TJ, O’Donnell K, Milus EA, Gale SW, Kistler HC. Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology. 2011;101:124–134. doi: 10.1094/PHYTO-03-10-0067. [DOI] [PubMed] [Google Scholar]
- Gomes LB, Ward TJ, Badiale-Furlong E, Del Ponte EM. Species composition, toxigenic potential and pathogenicity of Fusarium graminearum species complex isolates from southern Brazilian rice. Plant Pathol. 2015;64:980–987. doi: 10.1111/ppa.12332. [DOI] [Google Scholar]
- Huang JW, Sun SK. The Genus Fusarium from Taiwan. Taichung: Shih Way Publishers; 1997. [Google Scholar]
- Jansen C, von Wettstein D, Schafer W, Kogel KH, Felk A, Maier FJ. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc Natl Acad Sci USA. 2005;102:16892–16897. doi: 10.1073/pnas.0508467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P, Coates M, Turner J, Chandler E, Nicholson P. Determination of deoxynivalenol and nivalenol chemotypes of Fusarium culmorum isolates from England and Wales by PCR assay. Plant Pathol. 2004;53:182–190. doi: 10.1111/j.0032-0862.2004.00985.x. [DOI] [Google Scholar]
- Karugia GW, Suga H, Gale LR, Nakajima T, Ueda A, Hyakumachi M. Population structure of Fusarium asiaticum from two Japanese regions and eastern China. J Gen Plant Pathol. 2009;75:110–118. doi: 10.1007/s10327-009-0153-5. [DOI] [Google Scholar]
- Kuo CC, Lin HS, Liao CT, Huang DC, Cheng YH, Wang CL. The investigation of wheat Fusarium head blight caused by Fusarium graminearum:pathogen identification and fungicide screening assay. Plant Pathol Bull. 2014;23:192–193. [Google Scholar]
- Lee T, Oh DW, Kim HS, Lee J, Kim YH, Yun SH, Lee YW. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae by using PCR. Appl Environ Microbiol. 2001;67:2966–2972. doi: 10.1128/AEM.67.7.2966-2972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Chang IY, Kim H, Yun SH, Leslie JF, Lee YW. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl Environ Microbiol. 2009;75:3289–3295. doi: 10.1128/AEM.02287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim H, Jeon JJ, Kim HS, Zeller KA, Carter LL, Leslie JF, Lee YW. Population structure of and mycotoxin production by Fusarium graminearum from maize in South Korea. Appl Environ Microbiol. 2012;78:2161–2167. doi: 10.1128/AEM.07043-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium laboratory manual. Oxford: Blackwell publishing; 2006. [Google Scholar]
- Maier FJ, Miedaner T, Hadeler B, Felk A, Salomon S, Lemmens M, Kassner H, Schaefer W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol Plant Pathol. 2006;7:449–461. doi: 10.1111/j.1364-3703.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- Malbrán I, Mourelos C, Girotti J, Aulicino M, Balatti P, Lori G. Aggressiveness variation of Fusarium graminearum isolates from Argentina following point inoculation of field grown wheat spikes. Crop Prot. 2012;42:234–243. doi: 10.1016/j.cropro.2012.05.025. [DOI] [Google Scholar]
- Miller JD, Greenhalgh R, Wang Y, Lu M. Trichothecene chemotypes of three Fusarium species. Mycologia. 1991;83:121–130. doi: 10.2307/3759927. [DOI] [Google Scholar]
- Minervini F, Fornelli F, Flynn KM. Toxicity and apoptosis induced by the mycotoxins nivalenol, deoxynivalenol and fumonisin B1 in a human erythroleukemia cell line. Toxicol In Vitro. 2004;18:21–28. doi: 10.1016/S0887-2333(03)00130-9. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Simpson D, Weston G, Rezanoor H, Lees A, Parry D, Joyce D. Detection and quantification of Fusarium culmorum and Fusarium graminearum in cereals using PCR assays. Physiol Mol Plant Pathol. 1998;53:17–37. doi: 10.1006/pmpp.1998.0170. [DOI] [Google Scholar]
- O’Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Geiser DM, Corby Kistler H, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Aberra D, Kistler HC, Aoki T, Orwig N, Kimura M, Bjornstad S, Klemsdal SS. Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet Biol. 2008;45:1514–1522. doi: 10.1016/j.fgb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Pasquali M, Giraud F, Brochot C, Cocco E, Hoffmann L, Bohn T. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. Int J Food Microbiol. 2010;137:246–253. doi: 10.1016/j.ijfoodmicro.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM, McCormick SP. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact. 1995;8:593–601. doi: 10.1094/MPMI-8-0593. [DOI] [PubMed] [Google Scholar]
- Puri KD, Zhong S. The 3ADON population of Fusarium graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology. 2010;100:1007–1014. doi: 10.1094/PHYTO-12-09-0332. [DOI] [PubMed] [Google Scholar]
- Qiu J, Xu J, Shi J. Molecular characterization of the Fusarium graminearum species complex in Eastern China. Eur J Plant Pathol. 2014;139:811–823. doi: 10.1007/s10658-014-0435-4. [DOI] [Google Scholar]
- Qu B, Li H, Zhang J, Huang T, Carter J, Liao Y, Nicholson P. Comparison of genetic diversity and pathogenicity of Fusarium head blight pathogens from China and Europe by SSCP and seedling assays on wheat. Plant Pathol. 2008;57:642–651. doi: 10.1111/j.1365-3059.2008.01824.x. [DOI] [Google Scholar]
- Qu B, Li H, Zhang J, Xu Y, Huang T, Wu A, Zhao C, Carter J, Nicholson P, Liao Y. Geographic distribution and genetic diversity of Fusarium graminearum and F. asiaticum on wheat spikes throughout China. Plant Pathol. 2008;57:15–24. doi: 10.1111/j.1365-3059.2008.01824.x. [DOI] [Google Scholar]
- Quarta A, Mita G, Haidukowski M, Logrieco A, Mule G, Visconti A. Multiplex PCR assay for the identification of nivalenol, 3- and 15-acetyl-deoxynivalenol chemotypes in Fusarium. FEMS Microbiol Lett. 2006;259:7–13. doi: 10.1111/j.1574-6968.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: a laboratory manual. In. Cold Spring Harbor, New York, Cold Spring Harbor Laboratory
- Sarver BA, Ward TJ, Gale LR, Broz K, Kistler HC, Aoki T, Nicholson P, Carter J, O’Donnell K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet Biol. 2011;48:1096–1107. doi: 10.1016/j.fgb.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Sawada K (1919) Descriptive Catalogue of the Formosan Fungi Part 1. Formosa agricultural experiment station special bulletin 19
- Shen CM, Hu YC, Sun HY, Li W, Guo JH, Chen HG. Geographic distribution of trichothecene chemotypes of the Fusarium graminearum species complex in major winter wheat production areas of China. Plant Dis. 2012;96:1172–1178. doi: 10.1094/PDIS-11-11-0974-RE. [DOI] [PubMed] [Google Scholar]
- Shimada T, Otani M. Effects of Fusarium mycotoxins on the growth of shoots and roots at germination in some japanese wheat cultivars. Cereal Res Commun. 1990;18:229–232. [Google Scholar]
- Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, Geiser DM, Suga H, Toth B, Varga J, O’Donnell K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol. 2007;44:1191–1204. doi: 10.1016/j.fgb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Suga H, Karugia GW, Ward T, Gale LR, Tomimura K, Nakajima T, Miyasaka A, Koizumi S, Kageyama K, Hyakumachi M. Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology. 2008;98:159–166. doi: 10.1094/PHYTO-98-2-0159. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee T, Zhang H, van Diepeningen A, Waalwijk C. Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Addit Contam Part A. 2015;32:453–460. doi: 10.1080/19440049.2014.984244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Ndoye M, Zhang JB, Li HP, Liao YC. Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins (Basel) 2011;3:1020–1037. doi: 10.3390/toxins3081020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci USA. 2002;99:9278–9283. doi: 10.1073/pnas.142307199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels CE. Economic and social impacts of Fusarium head blight: changing farms and rural communities in the northern great plains. Phytopathology. 2000;90:17–21. doi: 10.1094/PHYTO.2000.90.1.17. [DOI] [PubMed] [Google Scholar]
- Wu AB, Li HP, Zhao CS, Liao YC. Comparative pathogenicity of Fusarium graminearum isolates from China revealed by wheat coleoptile and floret inoculations. Mycopathologia. 2005;160:75–83. doi: 10.1007/s11046-005-1153-4. [DOI] [PubMed] [Google Scholar]
- Yli-Mattila T, Gagkaeva T, Ward TJ, Aoki T, Kistler HC, O’Donnell K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia. 2009;101:841–852. doi: 10.3852/08-217. [DOI] [PubMed] [Google Scholar]
- Zeller KA, Bowden RL, Leslie JF. Population differentiation and recombination in wheat scab populations of Gibberella zeae from the United States. Mol Ecol. 2004;13:563–571. doi: 10.1046/j.1365-294X.2004.02098.x. [DOI] [PubMed] [Google Scholar]
- Zhang JB, Li HP, Dang FJ, Qu B, Xu YB, Zhao CS, Liao YC. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol Res. 2007;111:967–975. doi: 10.1016/j.mycres.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Zhang H, Van der Lee T, Waalwijk C, Chen W, Xu J, Xu J, Zhang Y, Feng J. Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. Plos ONE. 2012;7:e31722. doi: 10.1371/journal.pone.0031722. [DOI] [PMC free article] [PubMed] [Google Scholar]


